Research Article |
|
Corresponding author: Varpu Vahtera ( varpu.vahtera@gmail.com ) Academic editor: Marzio Zapparoli
© 2020 Varpu Vahtera, Pavel Stoev, Nesrine Akkari.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Vahtera V, Stoev P, Akkari N (2020) Five million years in the darkness: A new troglomorphic species of Cryptops Leach, 1814 (Chilopoda, Scolopendromorpha) from Movile Cave, Romania. ZooKeys 1004: 1-26. https://doi.org/10.3897/zookeys.1004.58537
|
Abstract
A new species of Cryptops Leach, 1814, C. speleorex sp. nov., is described from Movile Cave, Dobrogea, Romania. The cave is remarkable for its unique ecosystem entirely dependent on methane- and sulfur-oxidising bacteria. Until now, the cave was thought to be inhabited by the epigean species C. anomalans, which is widespread in Europe. Despite its resemblance to C. anomalans, the new species is well-defined morphologically and molecularly based on two mitochondrial (cytochrome c oxidase subunit I COI and 16S rDNA) and one nuclear (28S rDNA) markers. Cryptops speleorex sp. nov. shows a number of troglomorphic traits such as a generally large body and elongated appendages and spiracles, higher number of coxal pores and saw teeth on the tibia of the ultimate leg. With this record, the number of endemic species known from the Movile Cave reaches 35, which ranks it as one of the most species-rich caves in the world.
Keywords
Biospeleology, Cryptops speleorex sp. nov., Dobrogea, molecular phylogenetics, new species, troglomorphism
Introduction
Located in the southeastern part of Romania not far from the Black Sea Coast, Movile Cave is the first known subterranean chemosynthesis-based ecosystem (
Despite its harsh living conditions, Movile Cave ecosystem is known to harbor a diverse and unique fauna. The cave hosts 51 invertebrate species, of which 34 species are endemic (
Five species of myriapods are hitherto discovered from the innermost parts of Movile viz. Archiboreoiulus serbansarbui Giurginca, Vănoaica, Šustr, & Tajovský, 2020 (Diplopoda), Symphylella Silvestri, 1902 sp. (Symphyla), Geophilus alpinus Meinert, 1870 and Clinopodes carinthiacus (Latzel, 1880) (Geophilomorpha) and a troglobitic population of Cryptops anomalans Newport, 1844 (
Recently, we had the occasion to study freshly collected specimens of an undetermined species of the genus Cryptops Leach, 1814 from Movile Cave. Using both, morphological and molecular evidence, the cave specimens were compared with those of C. anomalans living on the surface, outside the cave. A phylogenetic analysis of 29 Cryptops specimens from different parts of Europe, including two from inside Movile Cave, based on two mitochondrial (cytochrome c oxidase subunit I COI and 16S rDNA) and one nuclear (28S rDNA) markers was performed. Morphological and molecular analyses confirmed that the cave specimens from Movile correspond to a new species, Cryptops speleorex sp. nov., that we describe herein. Additionally, we provide an annotated list and a key to the troglobitic Cryptops species in the world.
Material and methods
All Cryptops specimens from Movile Cave were hand-collected by the biospeleologists Serban Sarbu and A. Hillebrand and preserved in 70% or 96% ethanol. Microphotographs were obtained with a Nikon DS-Ri-2 camera mounted on a Nikon SMZ25 stereomicroscope using NIS-Elements Microscope Imaging Software with an Extended Depth of Focus (EDF) patch. Images were edited in Photoshop CS6 and assembled in InDesign CS6. Material is shared between the
Morphological terminology follows
Abbreviations: T – tergite, S – sternite.
Molecular methods
Altogether 29 specimens from both inside and outside the Movile Cave were included in the phylogenetic analysis. Of these, 14 were sequenced in this study. Total DNA was extracted from the legs using NucleoSpinTissue kit (Macherey-Nagel) according to the standard protocol for human or animal and cultured cells. Samples were incubated overnight. One nuclear (28S rRNA) and two mitochondrial (cytochrome c oxidase subunit I, COI, and 16S rRNA) fragments were chosen for amplification since they have proven informative between closely related taxa (
Polymerase chain reaction (PCR) amplifications were performed with MyTaqTM HS Red Mix. PCR was performed in a total volume of 23 μL containing 7.5 μL of MQ, 12.5 μL of MyTaq HS Red Mix, 2×, 0.5 μL of each primer (10 μM) and 2 μL of DNA template. PCR started with initial denaturation at 95 °C for 1 min and was followed by denaturation at 95 °C for 15 s. Annealing temperature for 28S rRNA and COI was 49 °C and 43 °C for 16S rRNA. Annealing lasted for 15 s and was followed by extension at 72 °C for 10 s. The last three steps were repeated 35 times. A negative control was included. PCR products were run in electrophoresis on 1% Agarose gel using Midori Green Advanced DNA Stain (Nippon Genetics). Samples were purified with an A’SAP PCR clean-up kit (ArcticZymes). Sequencing was performed by Macrogen Europe. The resulting chromatograms were visualized and assembled using the software Sequencher 5 (Gene codes corporation, USA). All new sequences are deposited in GenBank (See Table
Specimens used in the molecular phylogeny and their GenBank accession numbers (specimens sequenced in this study in bold). Institutional abbreviations:
| Species | Lab code | Voucher ID number | Voucher | Country | COI | 16S | 28S |
|---|---|---|---|---|---|---|---|
| Cryptops speleorex sp. nov. | K3 | http://mus.utu.fi/ZMUT.MYR-TYPE001 | ZMUT | Romania | MW240507 | MW243978 | MW243648 |
| C. speleorex sp. nov. | K4 |
|
Romania | MW240508 | MW243977 | MW243649 | |
| C. anomalans | 1a |
|
Serbia | MW240504 | MW243967 | MW243651 | |
| C. anomalans | 1b |
|
Serbia | MW240505 | MW243968 | MW243652 | |
| C. anomalans | 2 |
|
Serbia | MW240511 | – | MW243642 | |
| C. anomalans | 3 |
|
Serbia | MW240515 | MW243970 | MW243643 | |
| C. anomalans | 4 |
|
Serbia | MW240503 | MW243979 | MW243654 | |
| C. anomalans | 7 |
|
Serbia | MW240506 | MW243969 | MW243653 | |
| C. anomalans | 8 |
|
Serbia | MW240512 | MW243971 | MW243644 | |
| C. anomalans | 9 |
|
Serbia | MW240514 | MW243973 | MW243645 | |
| C. anomalans | 12 |
|
Serbia | MW240516 | MW243974 | MW243646 | |
| C. anomalans | 13 |
|
Serbia | MW240513 | MW243972 | MW243647 | |
| C. anomalans | 54a |
|
Romania | MW240510 | MW243975 | MW243650 | |
| C. anomalans | 57a |
|
Romania | MW240509 | MW243976 | MW243641 | |
| C. anomalans |
|
|
Germany | KM491639 | – | – | |
| C. anomalans |
|
|
Germany | KM491699 | – | – | |
| C. anomalans |
|
|
Germany | KM491703 | – | – | |
| C. anomalans |
|
|
Germany | KM491706 | – | – | |
| C. anomalans |
|
|
Germany | KU497151 | – | – | |
| C. anomalans |
|
|
Germany | KU497158 | – | – | |
| C. anomalans |
|
|
Germany | KU497159 | – | – | |
| C. anomalans | IZ-131458 |
|
UK | KF676499 | KF676457 | KF676353 | |
| Cryptops sp. |
|
|
Austria | KM491620 | – | – | |
| Cryptops sp. |
|
|
Germany | KU342042 | – | – | |
| Cryptops sp. |
|
|
Slovenia | KU497143 | – | – | |
| Cryptops sp. |
|
|
Croatia | KU497153 | – | – | |
| C. croaticus |
|
|
Austria | KU342049 | – | – | |
| C. hortensis | IZ-130582 |
|
UK | JX422662 | JX422684 | JX422582 | |
| C. parisi | IZ-130592 |
|
UK | KF676502 | KF676460 | KF676356 | |
| Scolopendra cingulata | IZ-131446 |
|
Spain | HM453310 | HM453220 | AF000782 |
Phylogenetic analyses
Most specimens included in the analysis had all three markers successfully sequenced. To obtain more geographic variation in the dataset, 15 Cryptops specimens (mostly from
Phylogenetic analysis was conducted using both parsimony and maximum likelihood as optimality criteria. Parsimony analysis was done with TNT v. 1.5 (
Uncorrected p-distances of aligned COI, 16S and 18S data were calculated with MEGA v. 7.0.21 (
Results
Order Scolopendromorpha Pocock, 1895
Family Cryptopidae Kohlrausch, 1881
Genus Cryptops Leach, 1814
Cryptops (Cryptops) anomalans
Material examined
Romania: SE Romania: Lalomiţa County, Călugărească Forest, 18.II.2016, leg. and det. S. Baba, 1 subad. ex. (
Cryptops (Cryptops) speleorexsp. nov.
Previous records
Material examined
Holotype
: Romania: Constanța County, Mangalia, Movile Cave (Peștera Movile), Lake Hall, June, 2014, leg. S. Sarbu, 1 ex. (
Diagnosis
A species morphologically similar to Cryptops anomalans, but differing from it by the much elongated antennae and legs, generally less setose forcipules and body, coxopleures with more than 300 coxal pores (vs. less than 100 in anomalans), ultimate leg with 13–17 saw teeth on tibia (usually 7–10, occasionally 12 in anomalans), and larger and elongated spiracles (see Table
Differences in morphological characters between Cryptops anomalans and C. speleorex sp. nov.
| Morphological character | Cryptops anomalans | Cryptops speleorex sp. nov. |
|---|---|---|
| Body size (mm) | 25–50 | >46–52 |
| Antennae length | Until posterior end of T3 | Until mid of T5 |
| Antennal article 7 L/W (mm) | 0.5 × 0.25 | 1.0 × 0.5 |
| Antennae: spines on basal articles | Present, numerous | Lacking or just a few |
| Ultimate leg length | 7.65 mm | 13.25 mm |
| Ultimate leg pretarsus (mm) | 0.25 | 1 |
| Ultimate leg saw teeth on tibia and tarsus 1 | Tibia: 7–12 (usually 7–10); Tarsus: 3–5 | Tibia: 13–17; Tarsus: 5–6 |
| Legs | Short, compact, pretarsus short | Strongly elongated, pretarsus long |
| Spiracles | Ovoid, small to medium sized (Fig. |
Strongly elongated, large (Fig. |
| Forcipular trochanteroprefemur | With spines medially (4–6) | Without spines, just stout setae |
| Coxopleural pore field | Approx. 2/3 of coxapleura; composed of less than 100 pores (86–90) | Approx. 4/5 of coxopleura; composed of more than 310 pores (317–320) |
Description (holotype)
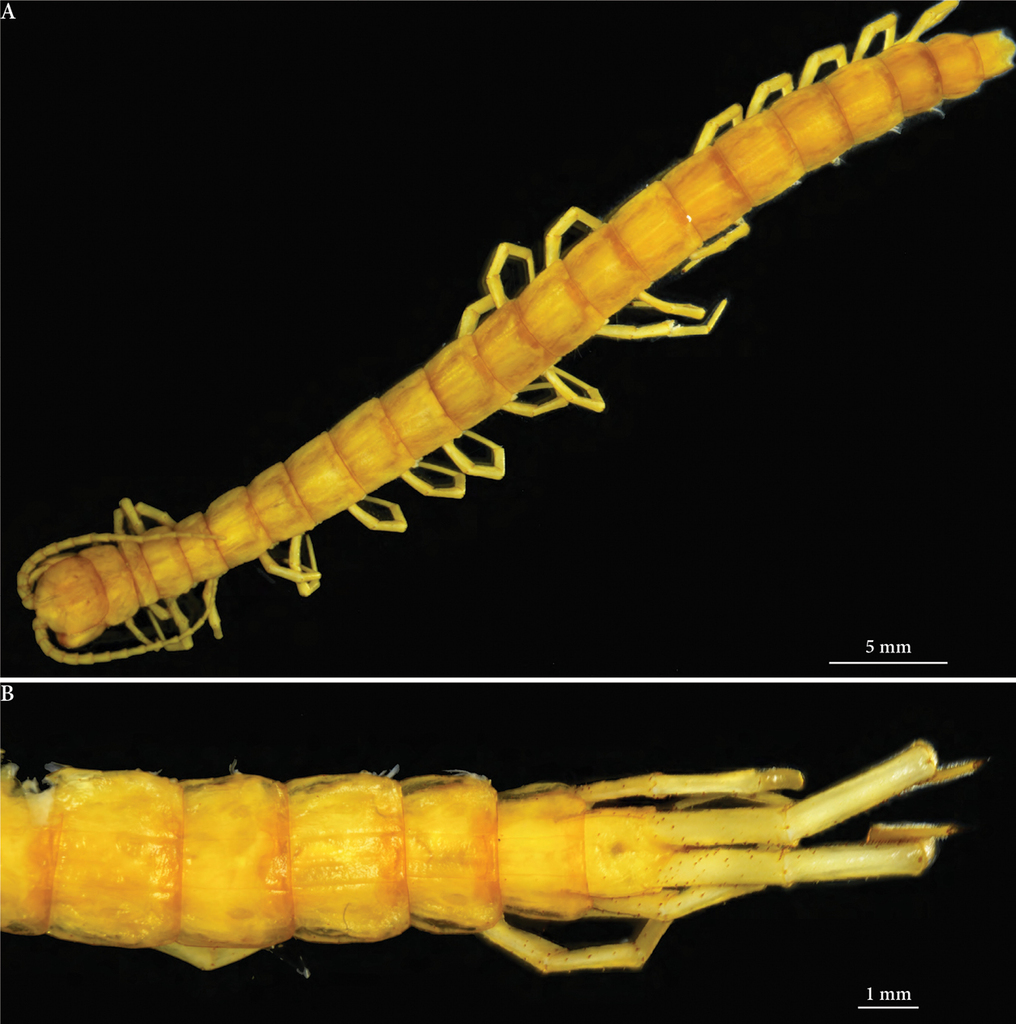
Length (anterior margin of head plate to posterior margin of telson) approx. 52 mm (46 mm in an adult paratype) (Figs
Antenna
relatively long, extending to the middle of tergite 5 when folded backward (Figs
Clypeus
with 2 setae; prelabral setae in one row of 21–22; 4 short setae between clypeus and prelabral row, irregularly or more evenly scattered. Labral mid piece with a short, but well-developed tooth; side pieces rounded (Fig.
Forcipular segment
anterior margin of coxosternite convex on each side, with a weak median diastema, fringed by 2 marginal setae on each side. Surface of coxosternite (Fig.
Maxilla 2
with a well-developed pretarsus; dorsal brush white, dense, situated on the distalmost part of article 3 of telopodite. Proximal side of first maxillary telopodite covered by 10–15 setae (Fig.
Tergites
Tergite 1 with a complete anterior transverse suture and cruciform sutures (Figs
Sternites
1–2 and 19–21 without transverse and median sutures; S 3–18 with median longitudinal and curved transverse sutures, more prominent from sternite 5 onward (Fig.
Spiracles
strongly elongated on T3, reducing in size towards the posterior end of the body; slit-like (Fig.
Coxopleural pore field
elliptical, covering 4/5 of surface, with more than 310 coxal pores (317–320), extending nearly to posterior margin of coxopleuron (Fig.
Legs
generally long; leg 10: prefemur 1.47 mm long, femur 1.59 mm, tibia 1.76 mm, tarsus 2.35 mm, pretarsus 0.7 mm. All tarsi single (Fig.
Ultimate leg
(Fig.
Etymology
The species epithet is a noun in apposition, meaning "king of the cave", referring to the species top position in the food chain of the Movile ecosystem.
Distribution
The species is hitherto known only from the aphotic zone of the Cave Movile in the southern part of Romanian Dobrogea.
Ecological remarks
Cryptops speleorex sp. nov. is the largest invertebrate species in Movile Cave. It has been observed feeding on terrestrial isopods (Trachelipus troglobius Tabacaru & Boghean, 1989, Armadillidium tabacarui Gruia, Iavorschi & Sarbu, 1994), smaller beetles, Diplura or spiders (
Phylogenetic analyses
Parsimony analysis resulted in a single most-parsimonious (MP) tree of length 1586 steps (Fig.
Regarding the placement of C. speleorex sp. nov. and the relationships among the C. anomalans specimens, the likelihood analysis (Fig.
When analyzed separately (only likelihood, tree not shown), the mitochondrial COI and 16S resolved C. speleorex sp. nov. as a distinct clade (BS = 100) within C. anomalans specimens, the tree topology regarding C. speleorex sp. nov./C. anomalans being identical to that of the parsimony tree. Not surprisingly, the level of variation in the nuclear 28S was low and the likelihood analysis based on it could not resolve the relationships among the C. anomalans/C.speleorex sp. nov. specimens (tree not shown).
Pairwise distances
Pairwise distances between the samples by each marker are shown in Tables
Estimates of evolutionary divergence between sequences. COI: The number of base differences per site from between sequences are shown. The analysis involved 30 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding.
| All positions containing gaps and missing data were eliminated. There were a total of 556 positions in the final dataset. | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | ||
| 1 | Scolopendra cingulata (IZ-131446) | |||||||||||||||||||||||||||||
| 2 | Cryptops anomalans UK (IZ-131458) | 0.248 | ||||||||||||||||||||||||||||
| 3 | C. anomalans Germany (KM491639) | 0.248 | 0.000 | |||||||||||||||||||||||||||
| 4 | C. anomalans Germany (KM491699) | 0.248 | 0.000 | 0.000 | ||||||||||||||||||||||||||
| 5 | C. anomalans Germany (KM491703) | 0.248 | 0.000 | 0.000 | 0.000 | |||||||||||||||||||||||||
| 6 | C. anomalans Germany (KM491706) | 0.248 | 0.000 | 0.000 | 0.000 | 0.000 | ||||||||||||||||||||||||
| 7 | C. anomalans Germany (KU497151) | 0.248 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |||||||||||||||||||||||
| 8 | C. anomalans Germany (KU497158) | 0.248 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||||||||||||||||||||||
| 9 | C. anomalans Germany (KU497159) | 0.248 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |||||||||||||||||||||
| 10 | C. anomalans SE Serbia cave (4) | 0.237 | 0.110 | 0.110 | 0.110 | 0.110 | 0.110 | 0.110 | 0.110 | 0.110 | ||||||||||||||||||||
| 11 | C. anomalans SW Serbia (1a) | 0.223 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.144 | |||||||||||||||||||
| 12 | C. anomalans SW Serbia (1b) | 0.223 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.144 | 0.000 | ||||||||||||||||||
| 13 | C. anomalans SW Serbia (7) | 0.225 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.149 | 0.005 | 0.005 | |||||||||||||||||
| 14 | Cryptops speleorex sp. nov. Movile cave, Romania (K3) | 0.246 | 0.155 | 0.155 | 0.155 | 0.155 | 0.155 | 0.155 | 0.155 | 0.155 | 0.153 | 0.121 | 0.121 | 0.126 | ||||||||||||||||
| 15 | Cryptops speleorex sp. nov. Movile cave, Romania (K4) | 0.225 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.142 | 0.095 | 0.095 | 0.101 | 0.085 | |||||||||||||||
| 16 | C. anomalans Bucharest Romania (57a) | 0.212 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.138 | 0.135 | 0.074 | 0.074 | 0.079 | 0.112 | 0.103 | ||||||||||||||
| 17 | C. anomalans SE Romania (54a) | 0.225 | 0.146 | 0.146 | 0.146 | 0.146 | 0.146 | 0.146 | 0.146 | 0.146 | 0.142 | 0.092 | 0.092 | 0.097 | 0.122 | 0.117 | 0.040 | |||||||||||||
| 18 | C. anomalans southern Serbia (2) | 0.243 | 0.129 | 0.129 | 0.129 | 0.129 | 0.129 | 0.129 | 0.129 | 0.129 | 0.138 | 0.103 | 0.103 | 0.108 | 0.104 | 0.097 | 0.088 | 0.099 | ||||||||||||
| 19 | C. anomalans Belgrade, Serbia (8) | 0.239 | 0.133 | 0.133 | 0.133 | 0.133 | 0.133 | 0.133 | 0.133 | 0.133 | 0.140 | 0.097 | 0.097 | 0.103 | 0.110 | 0.094 | 0.086 | 0.094 | 0.023 | |||||||||||
| 20 | C. anomalans Belgrade, Serbia (13) | 0.239 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.138 | 0.095 | 0.095 | 0.101 | 0.108 | 0.092 | 0.085 | 0.092 | 0.022 | 0.002 | ||||||||||
| 21 | C. anomalans Belgrade, Serbia (9) | 0.239 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.138 | 0.095 | 0.095 | 0.101 | 0.108 | 0.092 | 0.085 | 0.092 | 0.022 | 0.002 | 0.000 | |||||||||
| 22 | C. anomalans southern Serbia (3) | 0.239 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.131 | 0.138 | 0.104 | 0.104 | 0.110 | 0.112 | 0.092 | 0.099 | 0.106 | 0.032 | 0.027 | 0.025 | 0.025 | ||||||||
| 23 | C. anomalans southern Serbia (12) | 0.241 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.137 | 0.135 | 0.094 | 0.094 | 0.099 | 0.106 | 0.090 | 0.090 | 0.101 | 0.040 | 0.029 | 0.027 | 0.027 | 0.043 | |||||||
| 24 | C. hortensis UK (IZ-130582) | 0.243 | 0.203 | 0.203 | 0.203 | 0.203 | 0.203 | 0.203 | 0.203 | 0.203 | 0.182 | 0.174 | 0.174 | 0.178 | 0.173 | 0.189 | 0.180 | 0.187 | 0.198 | 0.200 | 0.198 | 0.198 | 0.191 | 0.191 | ||||||
| 25 | Cryptops sp. Austria (KM491620) | 0.228 | 0.178 | 0.178 | 0.178 | 0.178 | 0.178 | 0.178 | 0.178 | 0.178 | 0.167 | 0.182 | 0.182 | 0.185 | 0.174 | 0.167 | 0.176 | 0.183 | 0.180 | 0.173 | 0.173 | 0.173 | 0.185 | 0.182 | 0.169 | |||||
| 26 | Cryptops sp. Croatia (KU497153) | 0.230 | 0.182 | 0.182 | 0.182 | 0.182 | 0.182 | 0.182 | 0.182 | 0.182 | 0.180 | 0.200 | 0.200 | 0.203 | 0.201 | 0.191 | 0.203 | 0.209 | 0.198 | 0.196 | 0.194 | 0.194 | 0.194 | 0.192 | 0.156 | 0.156 | ||||
| 27 | C. parisi UK (IZ-130592) | 0.221 | 0.192 | 0.192 | 0.192 | 0.192 | 0.192 | 0.192 | 0.192 | 0.192 | 0.173 | 0.176 | 0.176 | 0.182 | 0.191 | 0.182 | 0.171 | 0.180 | 0.171 | 0.169 | 0.167 | 0.167 | 0.167 | 0.171 | 0.196 | 0.192 | 0.185 | |||
| 28 | C. croaticus (KU342049) | 0.239 | 0.201 | 0.201 | 0.201 | 0.201 | 0.201 | 0.201 | 0.201 | 0.201 | 0.185 | 0.173 | 0.173 | 0.176 | 0.192 | 0.178 | 0.174 | 0.180 | 0.194 | 0.194 | 0.192 | 0.192 | 0.196 | 0.196 | 0.169 | 0.192 | 0.192 | 0.137 | ||
| 29 | Cryptops sp. Slovenia (KU497143) | 0.255 | 0.192 | 0.192 | 0.192 | 0.192 | 0.192 | 0.192 | 0.192 | 0.192 | 0.185 | 0.189 | 0.189 | 0.192 | 0.201 | 0.203 | 0.189 | 0.203 | 0.201 | 0.205 | 0.207 | 0.207 | 0.201 | 0.196 | 0.173 | 0.207 | 0.180 | 0.187 | 0.165 | |
| 30 | Cryptops sp. Germany (KU342042) | 0.223 | 0.201 | 0.201 | 0.201 | 0.201 | 0.201 | 0.201 | 0.201 | 0.201 | 0.187 | 0.192 | 0.192 | 0.196 | 0.210 | 0.194 | 0.178 | 0.192 | 0.198 | 0.194 | 0.194 | 0.194 | 0.205 | 0.198 | 0.210 | 0.185 | 0.187 | 0.196 | 0.203 | 0.216 |
Estimates of evolutionary divergence between sequences. 16S: The number of base differences per site from between sequences are shown. The analysis involved 17 nucleotide sequences. All positions containing gaps and missing data were eliminated.
| There were a total of 392 positions in the final dataset. | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | ||
| 1 | Scolopendra cingulata (IZ-131446) | ||||||||||||||||
| 2 | Cryptops anomalans UK (IZ-131458) | 0.390 | |||||||||||||||
| 3 | C. anomalans SW Serbia (1a) | 0.372 | 0.092 | ||||||||||||||
| 4 | C. anomalans SW Serbia (1b) | 0.372 | 0.092 | 0.000 | |||||||||||||
| 5 | C. anomalans SW Serbia (7) | 0.372 | 0.094 | 0.003 | 0.003 | ||||||||||||
| 6 | C. anomalans southern Serbia (3) | 0.372 | 0.082 | 0.036 | 0.036 | 0.038 | |||||||||||
| 7 | C. anomalans Belgrade, Serbia (8) | 0.367 | 0.092 | 0.041 | 0.041 | 0.041 | 0.018 | ||||||||||
| 8 | C. anomalans Belgrade, Serbia (13) | 0.372 | 0.089 | 0.043 | 0.043 | 0.043 | 0.020 | 0.008 | |||||||||
| 9 | C. anomalans Belgrade, Serbia (9) | 0.372 | 0.087 | 0.041 | 0.041 | 0.041 | 0.010 | 0.008 | 0.010 | ||||||||
| 10 | C. anomalans southern Serbia (12) | 0.365 | 0.084 | 0.033 | 0.033 | 0.036 | 0.018 | 0.031 | 0.033 | 0.023 | |||||||
| 11 | C. anomalans SE Romania (54a) | 0.372 | 0.092 | 0.059 | 0.059 | 0.059 | 0.048 | 0.054 | 0.051 | 0.048 | 0.051 | ||||||
| 12 | C. anomalans Bucharest Romania (57a) | 0.372 | 0.099 | 0.066 | 0.066 | 0.066 | 0.054 | 0.059 | 0.056 | 0.054 | 0.059 | 0.013 | |||||
| 13 | Cryptops speleorex sp. nov. Movile cave, Romania (K4) | 0.365 | 0.099 | 0.082 | 0.082 | 0.082 | 0.066 | 0.074 | 0.071 | 0.069 | 0.071 | 0.066 | 0.069 | ||||
| 14 | Cryptops speleorex sp. nov. Movile cave, Romania (K3) | 0.383 | 0.112 | 0.087 | 0.087 | 0.084 | 0.071 | 0.077 | 0.074 | 0.069 | 0.082 | 0.079 | 0.082 | 0.066 | |||
| 15 | C. anomalans SE Serbia cave (4) | 0.385 | 0.084 | 0.094 | 0.094 | 0.097 | 0.077 | 0.087 | 0.084 | 0.082 | 0.079 | 0.097 | 0.107 | 0.107 | 0.125 | ||
| 16 | C. parisi UK (IZ-130592) | 0.355 | 0.217 | 0.209 | 0.209 | 0.212 | 0.214 | 0.227 | 0.224 | 0.222 | 0.219 | 0.224 | 0.227 | 0.232 | 0.235 | 0.232 | |
| 17 | C. hortensis UK (IZ-130582) | 0.360 | 0.230 | 0.217 | 0.217 | 0.219 | 0.222 | 0.235 | 0.230 | 0.232 | 0.224 | 0.235 | 0.245 | 0.230 | 0.219 | 0.230 | 0.260 |
Estimates of evolutionary divergence between sequences. 28S: The number of base differences per site from between sequences are shown. The analysis involved 18 nucleotide sequences.
| All positions containing gaps and missing data were eliminated. There were a total of 316 positions in the final dataset. | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | ||
| 1 | Scolopendra cingulata (IZ-131446) | |||||||||||||||||
| 2 | Cryptops anomalans UK (IZ-131458) | 0.187 | ||||||||||||||||
| 3 | C. anomalans Bucharest Romania (57a) | 0.187 | 0.000 | |||||||||||||||
| 4 | C. anomalans southern Serbia (2) | 0.190 | 0.003 | 0.003 | ||||||||||||||
| 5 | C. anomalans southern Serbia (3) | 0.190 | 0.003 | 0.003 | 0.000 | |||||||||||||
| 6 | C. anomalans Belgrade, Serbia (8) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | ||||||||||||
| 7 | C. anomalans Belgrade, Serbia (9) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | 0.000 | |||||||||||
| 8 | C. anomalans southern Serbia (12) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | ||||||||||
| 9 | C. anomalans Belgrade, Serbia (13) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |||||||||
| 10 | Cryptops speleorex sp. nov. Movile cave, Romania (K3) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||||||||
| 11 | Cryptops speleorex sp. nov. Movile cave, Romania (K4) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |||||||
| 12 | C. anomalans SE Romania (54a) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||||||
| 13 | C. anomalans SW Serbia (1a) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |||||
| 14 | C. anomalans SW Serbia (1b) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||||
| 15 | C. anomalans SW Serbia (7) | 0.190 | 0.003 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |||
| 16 | C. anomalans SE Serbia cave (4) | 0.184 | 0.006 | 0.006 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | ||
| 17 | C. parisi UK (IZ-130592) | 0.196 | 0.054 | 0.054 | 0.057 | 0.057 | 0.057 | 0.057 | 0.057 | 0.057 | 0.057 | 0.057 | 0.057 | 0.057 | 0.057 | 0.057 | 0.057 | |
| 18 | C. hortensis UK (IZ-130582) | 0.190 | 0.063 | 0.063 | 0.066 | 0.066 | 0.066 | 0.066 | 0.066 | 0.066 | 0.066 | 0.066 | 0.066 | 0.066 | 0.066 | 0.066 | 0.070 | 0.079 |
Key for identification of cave-specialized (troglomorphic/troglophilic) Cryptops
| 1 | Forcipular coxosternal margin with blunt, rounded or slightly flattened, hyaline lobes; tarsungulum very short | C. (Paracryptops) indicus |
| – | Forcipular coxosternal margin without hyaline lobes; tarsungulum moderate or long | 3 |
| 3 | Trigonal sutures present on the posterior part of sternites. Tarsus of most legs bipartite |
Cryptops (Trigonocryptops)
|
| – | Sternal trigonal sutures absent. Tarsus of most legs a single article | Cryptops (Cryptops) |
| 5 | Ultimate legs with saw teeth present from prefemur to tarsus 2, saw teeth formula: 28-30-14-17-17 | C. spelaeoraptor Ázara & Ferreira, 2014 |
| – | Ultimate legs with saw teeth present on tibia and tarsus 1 only | 7 |
| 7 | T1 with transverse suture only | 9 |
| – | T1 with transfer and other sutures | 11 |
| 9 | Head without paramedian sutures; length: 19 mm, antennae short, 3+3 saw teeth on tibia and tarsus of ultimate legs | C. beroni |
| – | Head with incomplete paramedian sutures on the posterior half and the anteriormost quarter of the cephalic plate; length: 28–29 mm; antennae long, 4+9 saw teeth on tibia and tarsus 1 of ultimate leg | C. illyricus |
| 11 | T1 with inverted Y-shaped sutures | C. legagus Edgecombe, Akkari, Netherlands, Du Preez, 2020 |
| – | T1 with transverse and/or paramedian sutures | 13 |
| 13 | T1 with transverse suture and two paramedian sutures; prefemur and femur of ultimate legs with dorsodistal spinous process; small species, ca 15 mm, cave in India | C. kempi |
| – | T1 with transverse suture and U-shaped or cruciform suture; prefemur and femur of ultimate legs without dorsodistal spinous process; caves in Europe | 15 |
| 15 | T1 with transverse and cruciform sutures; head with 2 complete paramedian sutures, large species | Cryptops speleorex sp. nov. |
| – | T1 with transverse suture and characteristic U-shaped suture attached to it; head with incomplete paramedian sutures | 17 |
| 17 | Labrum tridentate | 19 |
| – | Labrum unidentate | 21 |
| 19 | Antennae short, head plate with incomplete anterior and posterior paramedian sutures; saw teeth on tibia and tarsus in combination 13+6 | C. dianae |
| – | Antennae long, head plate with posterior paramedian sutures only | C. umbricus umbricus |
| 21 | Head with two incomplete posterior paramedian sutures only; anterior margin of forcipular coxosternite strongly convex and covered by spiniform setae, cave in France | C. umbricus lewisi |
| – | Head with two incomplete short posterior paramedian sutures only; anterior margin of forcipular coxosternite slightly rounded and barely protuberant; spiniform setae missing, cave on Tenerife | C. vulcanicus |
Discussion
Scolopendromorphs are strictly terrestrial and most species are found in forest leaf litter, decomposed wood, under bark of dead trees, in the soil, under stones or in caves in the temperate and tropical areas of the world. Few species are well adapted to eremic environments (
An annotated list of the troglobitic/troglophilic Cryptops species in the world.
| Species | Distribution | Category | References |
|---|---|---|---|
| Cryptops (Cryptops) beroni Matic & Stavropoulos, 1988 | Greece: Crete, Acrotiri, Cave Katholiko | Troglobite? |
|
| Cryptops (Trigonocryptops) camoowealensis Edgecombe, 2006 | Australia: Queensland, Camooweal area, Five O’Clock Cave | Troglobite |
|
| Cryptops (Trigonocryptops) cavernicolus Negrea & Fundora Martinez, 1977 | Cuba | Troglobite |
|
| Cryptops (Cryptops) dianae Matic & Stavropoulos, 1990 | Greece: Thassos Island, cave Dracotrypa | unknown | Matic and Stavropoulos (1990) |
| Cryptops (Trigonocryptops) hephaestus Ázara & Ferreira, 2013 | Brazil: known from three iron ore caves of the “Quadrilátero Ferrífero” (Iron Quadrangle) in Minas Gerais in Mariana and Itabirito municipalities | Troglophile |
|
| Cryptops (Cryptops) illyricus Verhoeff, 1933 | Caves only?; Slovenia and Croatia | Verhoeff 1933 | |
| Cryptops (Trigonocryptops) iporangensis Ázara & Ferreira, 2013 | Brazil: known from four caves (Ressurgência das Areias de Água Quente, Gruta Monjolinho, Caverna Alambari de Baixo, Caverna Santana) in Iporanga, São Paulo | Troglobite |
|
| Cryptops (Paracryptops) indicus (Silvestri, 1924) | India: Assam, Garo Hills, Siju Cave | Troglophile | (Silvestri 1924) |
| Cryptops (Cryptops) kempi Silvestri, 1924 | India: Assam, Garo Hills, Siju Cave | Troglophile | (Silvestri 1924) |
| Cryptops (Cryptops) legagus Edgecombe, Akkari, Netherlands, Du Preez, 2020 | Botswana: Diviner’s Cave (Koanaka Hills) and Dimapo Cave (Gcwihaba Hills) | Epigean/Troglophile? |
|
| Cryptops (Trigonocryptops) longicornis (Ribaut, 1915) | Caves in Spain | Troglobite | Ribaut (1915) |
| Cryptops (Cryptops) speleorex sp. nov. | Romania: Mangalia, Movile Cave | Troglobite | This paper (see also |
| Cryptops (Trigonocryptops) roeplainsensis Edgecombe, 2005 | Australia: known from three caves (Nurina Cave 6N-46, Burnabbie Cave, cave 6N-1327), Roe Plains | Troglobite |
|
| Cryptops (Cryptops) spelaeoraptor Ázara & Ferreira, 2014 | Brazil: Bahia, Campo Formoso, only known from the type locality, Toca do Gonçalo Cave |
|
|
| Cryptops (Trigonocryptops) troglobius Matic, Negrea & Fundora Martinez, 1977 | Cuba | Troglobite |
|
| Cryptops (Cryptops) umbricus umbricus Verhoeff, 1931 | Caves in France and Italy but also found outside caves | Troglophile |
|
| Syn. Cryptops jeanneli Matic, 1960 | |||
| Cryptops umbricus ischianus Verhoeff, 1942 | |||
| Cryptops (Cryptops) umbricus lewisi Iorio, 2010 | France: Alpes-Maritimes, Gourdon, Aven du Fourchu Cave | Troglobite |
|
| Cryptops (Cryptops) vulcanicus Zapparoli, 1990 | Spain: Tenerife Island, Cueva Felipe Reventón | Troglobite |
|
Several morphological characters traditionally used in centipedes taxonomy could be subject to intraspecific variation related to postembryonic development, animal life stage and ecology (
Intraspecific distance between the two sequenced Cryptops speleorex sp. nov. specimens is relatively high in comparison to the detected interspecific variation (Tables
Taxonomic and evolutionary implications of C. speleorex sp. nov
The type locality of C. anomalans is unknown and therefore it is impossible to conclude which part (if any) of the studied population is the actual C. anomalans described by
Acknowledgements
We are especially grateful to Serban M. Sarbu (Adjunct Faculty, California State University Chico) for calling our attention to this interesting material and for committing samples from Movile Cave for study. Stefan Baba (“Emil Racoviță” Institute of Speleology & Faculty of Biology, University of Bucharest, Romania), Dragan Antić and Dalibor Stojanović (both from University of Belgrade – Faculty of Biology) committed further specimens of C. speleorex and C. anomalans from Serbia and Romania for study. The study was partially funded by project #KP-06-H21/1-17.12.2018 of the National Science Fund, Ministry of Education and Science of the Republic of Bulgaria to PS and by Helsinki Entomological Society to VV. We thank G.D. Edgecombe, C. Martínez-Muñoz, A. Schileyko and Ivan H. Tuf for their constructive comments that greatly benefited the manuscript.
References
- Akkari N, Komerički A, Weigand AM, Edgecombe GD, Stoev P (2017) A new cave centipede from Croatia, Eupolybothrus liburnicus sp. n., with notes on the subgenusSchizopolybothrus Verhoeff, 1934 (Chilopoda, Lithobiomorpha, Lithobiidae). ZooKeys 687: 11–43. https://doi.org/10.3897/zookeys.687.13844
- Ázara LN, Ferreira RL (2013) The first troglobitic Cryptops (Trigonocryptops) (Chilopoda: Scolopendromorpha) from South America and the description of a non-troglobitic species from Brazil. Zootaxa 3709(5): 432–444. https://doi.org/10.11646/zootaxa.3709.5.2
- Ázara LN, Ferreira RL (2014) Cryptops (Cryptops) spelaeoraptor n. sp. a remarkable troglobitic species (Chilopoda: Scolopendromorpha) from Brazil. Zootaxa 3826(1): 291–300. https://doi.org/10.11646/zootaxa.3826.1.10
- Bonato L, Edgecombe G, Lewis J, Minelli A, Pereira L, Shelley R, Zapparoli M (2010) A common terminology for the external anatomy of centipedes (Chilopoda). ZooKeys 69: 17–51. https://doi.org/10.3897/zookeys.69.737
- Bonato L, Chagas Jr A, Edgecombe GD, Lewis JGE, Minelli A, Pereira LA, Shelley RM, Stoev P, Zapparoli M (2016) ChiloBase 2.0 – A World Catalogue of Centipedes (Chilopoda). http://chilobase.biologia.unipd.it
- Chagas Jr A, Bichuette ME (2018) Synopsis of centipedes in Brazilian caves (Arthropoda, Myriapoda, Chilopoda), a hidden diversity to be protected. ZooKeys 737: 13–56. https://doi.org/10.3897/zookeys.737.20307
- Crabill RE (1962) Concerning chilopod types in the British Museum (Natural History). Part I. Chilopoda: Geophilomorpha: Scolopendromorpha. Annals and Magazine of Natural History 13(5): 505–510. https://doi.org/10.1080/00222936208651277
- Edgecombe G (2005) A troglomorphic species of the centipede Cryptops (Trigonocryptops) (Chilopoda: Scolopendromorpha) from Western Australia. Records of the Western Australian Museum 22: 315–323. https://doi.org/10.18195/issn.0312-3162.22(4).2005.315-323
- Edgecombe G (2006) A troglobitic cryptopid centipede (Chilopoda: Scolopendromorpha) from western Queensland. Records of the Western Australian Museum 23: 193–198. https://doi.org/10.18195/issn.0312-3162.23(2).2006.193-198
- Edgecombe GD, Akkari N, Netherlands EC, Du Preez G (2020) A troglobitic species of the centipede Cryptops (Chilopoda, Scolopendromorpha) from northwestern Botswana. ZooKeys 977: 25–40. https://doi.org/10.3897/zookeys.977.57088
- Farris JS, Albert VA, Källersjö M, Lipscomb D, Kluge AG (1996) Parsimony jackknifing outperforms neighbor-joining. Cladistics 12: 99–124. https://doi.org/10.1111/j.1096-0031.1996.tb00196.x
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek RC (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299.
- Giurginca A, Vănoaica L, Šustr V, Tajovský K (2020) A new species of the genus Archiboreoiulus Brolemann, 1921 (Diplopoda, Julida) from Movile Cave (Southern Dobrogea, Romania). Zootaxa 4802(3): 463–476. https://doi.org/10.11646/zootaxa.4802.3.4
- Goloboff PA (1999) Analyzing large data sets in reasonable times: solutions for composite optima. Cladistics 15: 415–428. https://doi.org/10.1111/j.1096-0031.1999.tb00278.x
- Goloboff PA, Catalano SA (2016) TNT version 1.5, including a full implementation of phylogenetic morphometrics. Cladistics 32: 221–238. https://doi.org/10.1111/cla.12160
- Iorio E (2010) Description d'une nouvelle sous-espèce de Cryptops umbricus Verhoeff, 1931 (Chilopoda, Scolopendromorpha, Cryptopidae). Bulletin de la Société linnéenne de Bordeaux 144 (N.S. ) 37(4): 471–481.
- Iorio E, Geoffroy J-J (2007) Étude comparative de quatre espèces du genre Cryptops Leach, 1814 (Chilopoda, Scolopendromorpha, Cryptopidae) en France. Le Bulletin d’Arthropoda 31: 29–35.
- Iorio E, Geoffroy J-J (2008) Les scolopendromorphes de France (Chilopoda, Scolopendromorpha): identification et distribution géographique des espèces. Riviera scientifique 91: 73–90.
- Iorio E, Minelli A (2005) Un Chilopode confirmé pour la faune de France: Cryptops umbricus Verhoeff, 1931 (Scolopendromorpha, Cryptopidae). Bulletin mensuel de la Société linnéenne de Lyon 74(4): 150–157. https://doi.org/10.3406/linly.2005.13592
- Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. https://doi.org/10.1093/bib/bbx108
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. https://doi.org/10.1093/molbev/msw054
- Kuraku S, Zmasek CM, Nishimura O, Katoh K (2013) aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Research 41: W22–W28. https://doi.org/10.1093/nar/gkt389
- Krapelin K (1903) Revision der Scolopendriden. Jahrbuch der Hamburgischen Wissenschaftlichen Anstalten (2)20: 1–276.
- Lewis JGE (1982) The scolopendrid centipedes of the Oxford University 1932 Sarawak Expedition. Journal of Natural History 16: 389–397. https://doi.org/10.1080/00222938200770321
- Maddison WP, Maddison DR (2019) Mesquite: a modular system for evolutionary analysis. Version 3.61. http://www.mesquiteproject.org
- Matic Z (1960) Beiträge zur Kenntnis der blinden Lithobius-Arten (Chilopda-Myriopoda) Europas. Zoologischer Anzeiger 164: 443–448.
- Matic Z, Stavropoulos G (1988) Contributions à la connaissance des chilopodes de Grece. Biologia Gallo-Hellenica 14: 33–46.
- Matic Z, Negrea Ş, Fundora Martinez C (1977) Recherches sur les Chilopodes hypogés de Cuba. II. In: Résultats des expéditions biospéologiques cubano-roumaines à Cuba, vol. 2. Editura Academiei R.S.R., Bucureşti, 277–301.
- Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8. https://doi.org/10.1109/GCE.2010.5676129
- Minelli A, Golovatch SI (2013) Myriapods. In: Levin SA (Ed.) Encyclopedia of Biodiversity, 2nd edn. , Volume 5. Waltham, MA, Academic Press, 421–432. https://doi.org/10.1016/B978-0-12-384719-5.00208-2
- Morgan-Richards M, Bulgarella M, Sivyer L, Dowle EJ, Hale M, McKean NE, Trewick SA (2017) Explaining large mitochondrial sequence differences within a population sample. Royal Society Open Science 4(11): e170730. https://doi.org/10.1098/rsos.170730
- Negrea Ş (1993) Sur une population troglobionte de Cryptops anomalans Newport, 1844 (Chilopoda, Scolopendromorpha) trouvée dans la grotte “Pestera de la Movile” (Dobrogea, Roumanie). Travaux de l’Institut de Spéologie “Émile Racovitza” 32: 87–94.
- Negrea Ş (1994) Chilopodes (Chilopoda) cavernicoles de Roumanie connus jusqu’à present. Travaux du Muséum National d’Histoire Naturelle “Grigore Antipa” 34: 265–283.
- Negrea Ş (1997) Nouvelles données sur les Chilopodes souterrains et endogés de la zone karstique de Mangalia (Dobrogea, Roumanie). Travaux du Muséum National d’Histoire Naturelle “Grigore Antipa” 39: 45–51.
- Negrea Ş (2004) On the Chilopoda from south-eastern Dobrogean karstic area (Romania). 3. The material collected using deep traps placed in drillings and artificial microcaves. Travaux du Muséum National d’Histoire Naturelle “Grigore Antipa” 47: 111–128.
- Negrea Ş, Minelli A (1994) Chilopoda. In: Juberthie C, Decu V (Eds) Encyclopaedia Biospeologica, tome I. Imprimerie Fabbro, Saint Girons (France), 249–254.
- Newport G (1844) A List of the species of Myriapoda, Order Chilopoda, contained in the Cabinets of the British Museum, with synoptic descriptions of forty-seven new Species. Annals and Magazine of Natural History 1, 13: 94–101. https://doi.org/10.1080/03745484409442576
- Phillips JW, Chung AYC, Edgecombe GD, Ellwood MDF (2020) Bird’s nest ferns promote resource sharing by centipedes. Biotropica 52: 335–344. https://doi.org/10.1111/btp.12713
- Sarbu S, Lascu C, Brad T (2019) Dobrogea: Movile Cave. In: Ponta GML, Onac BP (Eds) Cave and Karst Systems of Romania. Springer, Cham, 429–436. https://doi.org/10.1007/978-3-319-90747-5_48
- Spelda J, Reip H, Oliveira Biener U, Melzer R (2011) Barcoding Fauna Bavarica: Myriapoda – a contribution to DNA sequence-based identifications of centipedes and millipedes (Chilopoda, Diplopoda). ZooKeys 156: 123–139. https://doi.org/10.3897/zookeys.156.2176
- Siriwut W, Edgecombe GD, Sutcharit C, Tongkerd P, Panha S (2016) A taxonomic review of the centipede genus Scolopendra Linnaeus, 1758 (Scolopendromorpha, Scolopendridae) in mainland Southeast Asia, with description of a new species from Laos. ZooKeys 590: 1–124. https://doi.org/10.3897/zookeys.590.7950
- Stamatakis A (2014) RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
- Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57: 758–771. https://doi.org/10.1080/10635150802429642
- Vaidya G, Lohman DJ, Meier R (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x
- Vahtera V, Edgecombe GD, Giribet G (2012) Evolution of blindness in scolopendromorph centipedes (Chilopoda: Scolopendromorpha): Insights from an expanded sampling of molecular data. Cladistics 28(1): 4–20. https://doi.org/10.1111/j.1096-0031.2011.00361.x
- Vahtera V, Edgecombe GD, Giribet G (2013) Phylogenetics of scolopendromorph centipedes: Can denser taxon sampling improve an artificial classification? Invertebrate Systematics 27(5): 578–602. https://doi.org/10.1071/IS13035
- Verhoeff KW (1931) Über europäische Cryptops-Arten. Zoologische Jahrbücher, Abteilung für Systematik 62: 263–288.
- Wesener T, Voigtländer K, Decker P, Oeyen JP, Spelda J (2016) Barcoding of Central European Cryptops centipedes reveals large interspecific distances with ghost lineages and new species records from Germany and Austria (Chilopoda, Scolopendromorpha). ZooKeys 564: 21–46. https://doi.org/10.3897/zookeys.564.7535
- Xiong B, Kocher TD (1991) Comparison of mitochondrial DNA sequences of seven morphospecies of black flies (Diptera: Simuliidae). Genome 34: 306–311. https://doi.org/10.1139/g91-050
- Zapparoli M (1990) Cryptops vulcanicus n. sp., a new species from a lava tube of the Canary Islands (Chilopoda, Scolopendromorpha). Vieraea 19: 153–160.
- Zapparoli M (2002) A catalogue of the centipedes from Greece (Chilopoda). Fragmenta Entomologica 34: 1–146.