Monograph |
|
Corresponding author: Michael J. Sharkey ( msharkey@uky.edu ) Academic editor: Lyubomir Penev
© 2021 Michael J. Sharkey, Daniel H. Janzen, Winnie Hallwachs, Eric G. Chapman, M. Alex Smith, Tanya Dapkey, Allison Brown, Sujeevan Ratnasingham, Suresh Naik, Ramya Manjunath, Kate Perez, Megan Milton, Paul Hebert, Scott R. Shaw, Rebecca N. Kittel, M. Alma Solis, Mark A. Metz, Paul Z. Goldstein, John W. Brown, Donald L. J. Quicke, C. van Achterberg, Brian V. Brown, John M. Burns.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Sharkey MJ, Janzen DH, Hallwachs W, Chapman EG, Smith MA, Dapkey T, Brown A, Ratnasingham S, Naik S, Manjunath R, Perez K, Milton M, Hebert P, Shaw SR, Kittel RN, Solis MA, Metz MA, Goldstein PZ, Brown JW, Quicke DLJ, van Achterberg C, Brown BV, Burns JM (2021) Minimalist revision and description of 403 new species in 11 subfamilies of Costa Rican braconid parasitoid wasps, including host records for 219 species. ZooKeys 1013: 1-665. https://doi.org/10.3897/zookeys.1013.55600
|
Abstract
Three new genera are described: Michener (Proteropinae), Bioalfa (Rogadinae), and Hermosomastax (Rogadinae). Keys are given for the New World genera of the following braconid subfamilies: Agathidinae, Braconinae, Cheloninae, Homolobinae, Hormiinae, Ichneutinae, Macrocentrinae, Orgilinae, Proteropinae, Rhysipolinae, and Rogadinae. In these subfamilies 416 species are described or redescribed. Most of the species have been reared and all but 13 are new to science. A consensus sequence of the COI barcodes possessed by each species is employed to diagnose the species, and this approach is justified in the introduction. Most descriptions consist of a lateral or dorsal image of the holotype, a diagnostic COI consensus barcode, the Barcode Index Number (BIN) code with a link to the Barcode of Life Database (BOLD), and the holotype specimen information required by the International Code of Zoological Nomenclature. The following species are treated and those lacking authorship are newly described here with authorship attributable to Sharkey except for the new species of Macrocentrinae which are by Sharkey & van Achterberg: AGATHIDINAE: Aerophilus paulmarshi, Mesocoelus davidsmithi, Neothlipsis bobkulai, Plesiocoelus vanachterbergi, Pneumagathis erythrogastra (Cameron, 1905), Therophilus bobwhartoni, T. donaldquickei, T. gracewoodae, T. maetoi, T. montywoodi, T. penteadodiasae, Zacremnops brianbrowni, Z. coatlicue Sharkey, 1990, Zacremnops cressoni (Cameron, 1887), Z. ekchuah Sharkey, 1990, Z. josefernandezi, Zelomorpha sarahmeierottoae. BRACONINAE: Bracon alejandromarini, B. alejandromasisi, B. alexamasisae, B. andresmarini, B. andrewwalshi, B. anniapicadoae, B. anniemoriceae, B. barryhammeli, B. bernardoespinozai, B. carlossanabriai, B. chanchini, B. christophervallei, B. erasmocoronadoi, B. eugeniephillipsae, B. federicomatarritai, B. frankjoycei, B. gerardovegai, B. germanvegai, B. isidrochaconi, B. jimlewisi, B. josejaramilloi, B. juanjoseoviedoi, B. juliodiazi, B. luzmariaromeroae, B. manuelzumbadoi, B. marialuisariasae, B. mariamartachavarriae, B. mariorivasi, B. melissaespinozae, B. nelsonzamorai, B. nicklaphami, B. ninamasisae, B. oliverwalshi, B. paulamarinae, B. rafamoralesi, B. robertofernandezi, B. rogerblancoi, B. ronaldzunigai, B. sigifredomarini, B. tihisiaboshartae, B. wilberthbrizuelai, Digonogastra montylloydi, D. montywoodi, D. motohasegawai, D. natwheelwrighti, D. nickgrishini. CHELONINAE: Adelius adrianguadamuzi, A. gauldi Shimbori & Shaw, 2019, A. janzeni Shimbori & Shaw, 2019, Ascogaster gloriasihezarae, A. grettelvegae, A. guillermopereirai, A. gustavoecheverrii, A. katyvandusenae, A. luisdiegogomezi, Chelonus alejandrozaldivari, C. gustavogutierrezi, C. gustavoinduni, C. harryramirezi, C. hartmanguidoi, C. hazelcambroneroae, C. iangauldi, C. isidrochaconi, C. janecheverriae, C. jeffmilleri, C. jennyphillipsae, C. jeremydewaardi, C. jessiehillae, C. jesusugaldei, C. jimlewisi, C. jimmilleri, C. jimwhitfieldi, C. johanvalerioi, C. johnburnsi, C. johnnoyesi, C. jorgebaltodanoi, C. jorgehernandezi, C. josealfredohernandezi, C. josefernandeztrianai, C. josehernandezcortesi, C. josemanuelperezi, C. josephinerodriguezae, C. juanmatai, C. junkoshimurae, C. kateperezae, C. luciariosae, C. luzmariaromeroae, C. manuelpereirai, C. manuelzumbadoi, C. marianopereirai, C. maribellealvarezae, C. markmetzi, C. markshawi, C. martajimenezae, C. mayrabonillae, C. meganmiltonae, C. melaniamunozae, C. michaelstroudi, C. michellevanderbankae, C. mingfangi, C. minorcarmonai, C. monikaspringerae, C. moniquegilbertae, C. motohasegawai, C. nataliaivanovae, C. nelsonzamorai, C. normwoodleyi, C. osvaldoespinozai, C. pamelacastilloae, C. paulgoldsteini, C. paulhansoni, C. paulheberti, C. petronariosae, C. ramyamanjunathae, C. randallgarciai, C. rebeccakittelae, C. robertoespinozai, C. robertofernandezi, C. rocioecheverriae, C. rodrigogamezi, C. ronaldzunigai, C. rosibelelizondoae, C. rostermoragai, C. ruthfrancoae, C. scottmilleri, C. scottshawi, C. sergioriosi, C. sigifredomarini, C. stevearonsoni, C. stevestroudi, C. sujeevanratnasinghami, C. sureshnaiki, C. torbjornekremi, C. yeimycedenoae, Leptodrepana alexisae, L. erasmocoronadoi, L. felipechavarriai, L. freddyquesadai, L. gilbertfuentesi, L. manuelriosi, Phanerotoma almasolisae, P. alvaroherrerai, P. anacordobae, P. anamariamongeae, P. andydeansi, P. angelagonzalezae, P. angelsolisi, P. barryhammeli, P. bernardoespinozai, P. calixtomoragai, P. carolinacanoae, P. christerhanssoni, P. christhompsoni, P. davesmithi, P. davidduthiei, P. dirksteinkei, P. donquickei, P. duniagarciae, P. duvalierbricenoi, P. eddysanchezi, P. eldarayae, P. eliethcantillanoae, P. jenopappi, Pseudophanerotoma alanflemingi, Ps. albanjimenezi, Ps. alejandromarini, Ps. alexsmithi, Ps. allisonbrownae, Ps. bobrobbinsi. HOMOLOBINAE: Exasticolus jennyphillipsae, E. randallgarciai, E. robertofernandezi, E. sigifredomarini, E. tomlewinsoni. HORMIINAE: Hormius anamariamongeae, H. angelsolisi, H. anniapicadoae, H. arthurchapmani, H. barryhammeli, H. carmenretanae, H. carloswalkeri, H. cesarsuarezi, H. danbrooksi, H. eddysanchezi, H. erikframstadi, H. georgedavisi, H. grettelvegae, H. gustavoinduni, H. hartmanguidoi, H. hectoraritai, H. hesiquiobenitezi, H. irenecanasae, H. isidrochaconi, H. jaygallegosi, H. jimbeachi, H. jimlewisi, H. joelcracrafti, H. johanvalerioi, H. johnburleyi, H. joncoddingtoni, H. jorgecarvajali, H. juanmatai, H. manuelzumbadoi, H. mercedesfosterae, H. modonnellyae, H. nelsonzamorai, H. pamelacastilloae, H. raycypessi, H. ritacolwellae, H. robcolwelli, H. rogerblancosegurai, H. ronaldzunigai, H. russchapmani, H. virginiaferrisae, H. warrenbrighami, H. willsflowersi. ICHNEUTINAE: Oligoneurus kriskrishtalkai, O. jorgejimenezi, Paroligoneurus elainehoaglandae, P. julianhumphriesi, P. mikeiviei. MACROCENTRINAE: Austrozele jorgecampabadali, A. jorgesoberoni, Dolichozele gravitarsis (Muesebeck, 1938), D. josefernandeztrianai, D. josephinerodriguezae, Hymenochaonia kalevikulli, H. kateperezae, H. katherinebaillieae, H. katherineellisonae, H. katyvandusenae, H. kazumifukunagae, H. keithlangdoni, H. keithwillmotti, H. kenjinishidai, H. kimberleysheldonae, H. krisnorvigae, H. lilianamadrigalae, H. lizlangleyae, Macrocentrus fredsingeri, M. geoffbarnardi, M. gregburtoni, M. gretchendailyae, M. grettelvegae, M. gustavogutierrezi, M. hannahjamesae, M. harisridhari, M. hillaryrosnerae, M. hiroshikidonoi, M. iangauldi, M. jennyphillipsae, M. jesseausubeli, M. jessemaysharkae, M. jimwhitfieldi, M. johnbrowni, M. johnburnsi, M. jonathanfranzeni, M. jonathanrosenbergi, M. jorgebaltodanoi, M. lucianocapelli. ORGILINAE: Orgilus amyrossmanae, O. carrolyoonae, O. christhompsoni, O. christinemcmahonae, O. dianalipscombae, O. ebbenielsoni, O. elizabethpennisiae, O. evertlindquisti, O. genestoermeri, O. jamesriegeri, O. jeanmillerae, O. jeffmilleri, O. jerrypowelli, O. jimtiedjei, O. johnlundbergi, O. johnpipolyi, O. jorgellorentei, O. larryspearsi, O. marlinricei, O. mellissaespinozae, O. mikesmithi, O. normplatnicki, O. peterrauchi, O. richardprimacki, O. sandraberriosae, O. sarahmirandae, O. scottmilleri, O. scottmorii, Stantonia billalleni, S. brookejarvisae, S. donwilsoni, S. erikabjorstromae, S. garywolfi, S. henrikekmani, S. luismirandai, S. miriamzunzae, S. quentinwheeleri, S. robinkazmierae, S. ruthtifferae. PROTEROPINAE: Hebichneutes tricolor Sharkey & Wharton, 1994, Proterops iangauldi, P. vickifunkae, Michener charlesi. RHYSIPOLINAE: Pseudorhysipolis luisfonsecai, P. mailyngonzalezae Rhysipolis julioquirosi. ROGADINAE: Aleiodes adrianaradulovae, A. adrianforsythi, A. agnespeelleae, A. alaneaglei, A. alanflemingi, A. alanhalevii, A. alejandromasisi, A. alessandracallejae, A. alexsmithi, A. alfonsopescadori, A. alisundermieri, A. almasolisae, A. alvarougaldei, A. alvaroumanai, A. angelsolisi, A. annhowdenae, A. bobandersoni, A. carolinagodoyae, A. charlieobrieni, A. davefurthi, A. donwhiteheadi, A. doylemckeyi, A. frankhovorei, A. henryhowdeni, A. inga Shimbori & Shaw, 2020, A. johnchemsaki, A. johnkingsolveri, A. gonodontovorus Shimbori & Shaw, 2020, A. manuelzumbadoi, A. mayrabonillae, A. michelledsouzae, A. mikeiviei, A. normwoodleyi, A. pammitchellae, A. pauljohnsoni, A. rosewarnerae, A. steveashei, A. terryerwini, A. willsflowersi, Bioalfa pedroleoni, B. alvarougaldei, B. rodrigogamezi, Choreborogas andydeansi, C. eladiocastroi, C. felipechavarriai, C. frankjoycei, Clinocentrus andywarreni, Cl. angelsolisi, Cystomastax alexhausmanni, Cy. angelagonzalezae, Cy. ayaigarashiae, Hermosomastax clavifemorus Quicke sp. nov., Heterogamus donstonei, Pseudoyelicones bernsweeneyi, Stiropius bencrairi, S. berndkerni, S. edgargutierrezi, S. edwilsoni, S. ehakernae, Triraphis billfreelandi, T. billmclarneyi, T. billripplei, T. bobandersoni, T. bobrobbinsi, T. bradzlotnicki, T. brianbrowni, T. brianlaueri, T. briannestjacquesae, T. camilocamargoi, T. carlosherrerai, T. carolinepalmerae, T. charlesmorrisi, T. chigiybinellae, T. christerhanssoni, T. christhompsoni, T. conniebarlowae, T. craigsimonsi, T. defectus Valerio, 2015, T. danielhubi, T. davidduthiei, T. davidwahli, T. federicomatarritai, T. ferrisjabri, T. mariobozai, T. martindohrni, T. matssegnestami, T. mehrdadhajibabaei, T. ollieflinti, T. tildalauerae, Yelicones dirksteinkei, Y. markmetzi, Y. monserrathvargasae, Y. tricolor Quicke, 1996. Y. woldai Quicke, 1996.
The following new combinations are proposed: Neothlipsis smithi (Ashmead), new combination for Microdus smithi Ashmead, 1894; Neothlipsis pygmaeus (Enderlein), new combination for Microdus pygmaeus Enderlein, 1920; Neothlipsis unicinctus (Ashmead), new combination for Microdus unicinctus Ashmead, 1894; Therophilus anomalus (Bortoni and Penteado-Dias) new combination for Plesiocoelus anomalus Bortoni and Penteado-Dias, 2015; Aerophilus areolatus (Bortoni and Penteado-Dias) new combination for Plesiocoelus areolatus Bortoni and Penteado-Dias, 2015; Pneumagathis erythrogastra (Cameron) new combination for Agathis erythrogastra Cameron, 1905. Dolichozele citreitarsis (Enderlein), new combination for Paniscozele citreitarsis Enderlein, 1920. Dolichozele fuscivertex (Enderlein) new combination for Paniscozele fuscivertex Enderlein, 1920. Finally, Bassus brooksi Sharkey, 1998 is synonymized with Agathis erythrogastra Cameron, 1905; Paniscozele griseipes Enderlein, 1920 is synonymized with Dolichozele koebelei Viereck, 1911; Paniscozele carinifrons Enderlein, 1920 is synonymized with Dolichozele fuscivertex (Enderlein, 1920); and Paniscozele nigricauda Enderlein,1920 is synonymized with Dolichozele quaestor (Fabricius, 1804). (originally described as Ophion quaestor Fabricius, 1804).
Keywords
Accelerated taxonomy, Agathidinae, BIN code, BioAlfa, Braconidae, Braconinae, caterpillar, Cheloninae, COI barcode, conservation, DNA barcode, Homolobinae, Hormiinae, Hymenoptera, Ichneumonoidea, Ichneutinae, Lepidoptera, Macrocentrinae, parasitoid host associations, Proteropinae, Rogadinae, Rhysipolinae, tri-trophic interaction, tropical
Chapter 1: Introduction and methods
Introduction
It is the purpose of this article to further refine methods to overcome the taxonomic impediment of ichneumonoid biodiversity. The current treatment deals with eleven subfamilies of Braconidae, i.e., Agathidinae, Braconinae, Cheloninae, Homolobinae, Hormiinae, Ichneutinae, Macrocentrinae, Orgilinae, Proteropinae, Rhysipolinae, and Rogadinae. Although not a thorough review of all species of these subfamilies, we here document a large number of Costa Rican species and provide names and unique identifiers for future keys, biodiversity analyses, conservation (e.g.,
In the following paragraphs we justify our method and address criticisms of the
Criticisms
1. “The method ignores previously described species”
This is not true, not for the
2. “Mitochondrial trees often disagree with nuclear species trees, especially in taxa where Wolbachia may be altering mtDNA introgression” and “congruence between nuclear and mitochondrial signal should be tested to better reinforce the species units identified”
This mostly appears to be pointless because we are not trying to recapitulate phylogeny. BOLD automatically screens out sequence uploads for Wolbachia (and other contaminants). Indeed, when barcode sequence libraries were surveyed for Wolbachia, evidence was found in only 0.16% of cases (
3. “purely DNA-based descriptions will… make the identification of millions of historical specimens impossible”
As we have made clear in the
4. “it will impair this science [taxonomy] in developing countries which house most of the undiscovered portion of biodiversity, due to high costs and lack of staff and technology”
This is a point that one of the reviewers of our present manuscript also mentioned. Firstly, high costs, lack of public interest, and lack of staff, to say nothing of lack of biopolitical enthusiasm or budgets, have caused the taxonomic impediment worldwide, not just in developing countries. In Canada, for example, a Canadian Council of Academics report from a decade ago found that previously deep and respected taxonomic resources suffered from decades of lack of investment that resulted in insufficient capacity to describe Canadian biodiversity (
5. [it will] “drastically affect other related fields of study and, importantly, conservation”
The authors did not expand on this criticism, but the opposite is true. For example, barcoded species will allow for the identification of metabarcoded specimens. This is a tool that is being used to determine the rareness and distribution of a species and potentially allow for their protection. Our method allows for the rapid naming of species and it is very unlikely that unnamed species will ever be protected, and they will have no chance of being integrated into the socioeconomics of a tropical country, a process that is vital for the conservation of tropical biodiversity (
6. “there must be several photographs available, not a single lateral photograph of a single specimen”
Of course many photographs are desirable as are fully integrated revisions. However, if we are to rid ourselves of the taxonomic impediment, time is a critical factor. Our simple images are meant simply as a voucher for a comparison with newly barcoded specimens, as well as to offer many gestalt traits. For example, we have a number of species in our treatment of Costa Rican fauna below that appear to have conspecific specimens on BOLD that were collected by others in other countries, e.g., Belize and Argentina, i.e., they are in the same BIN. In most of these cases the details on BOLD are “private” (unfortunately owing to other taxonomists’ possessiveness) and so we cannot easily examine the specimens to include them in our revision. However, the proprietors of these specimens can look at our image and conclude either, “yes that looks a lot like my specimen and is probably the same species or not”, and all of our specimens are available for examination from the CBG or from their final deposit in the Canadian National Collection in Ottawa.
7. “some text highlighting important diagnostic features is valuable… [and] the actual description of the species may be written within minutes when material and expertise are already available”
This is false. If morphological descriptions and diagnostics could be written in minutes we would include them, and the taxonomic impediment would be just a pebble in the taxasphere’s road. Even the revision of Alabagrus by
8. “Simply assigning all BINs taxonomic names.. would indeed complete the inventory of life on Earth extremely quickly (at precisely the same pace as the rate of barcoding) – that we do not dispute. But it would also remove the quantitative and qualitative difference between these preliminary identifiers (based on a single DNA marker) and full taxonomic recognition (based on a more comprehensive diagnosis, ideally supported by multiple lines of evidence including genetic data) that lend taxonomy its value”
When the authors and the taxasphere have the budgets, the enthusiasm, the time, and the biopolitical permissions to achieve this for the 20+ million tropical species of terrestrial Eukaryota, it will be wonderful. As mentioned earlier, our method is not meant to be the final treatment for a higher taxon. We admire and we practice the integrative approach, and all authors who are taxonomists in this publication regularly publish integrative revisions; this is our standard. But every integration starts with pieces to integrate. The barcode and its associated administrative accounting, along with the specimen itself, and whatever higher taxon information is available, is what we can offer to tropical conservation through sustainable biodevelopment (e.g., Janzen et al. 2020a). We are now offering an alternative method as a taxonomic first pass for megadiverse understudied taxa, the goal being to overcome the taxonomic impediment, thereby allowing the full power of tropical biodiversity value to become truly part of tropical societies. The critics also misrepresent our method. We do not simply assign taxonomic names to BINs. Firstly, ca. 10% of BINs contain multiple species. More rarely, BINs that are presently on BOLD today may coalesce, resulting in one of the BINS disappearing, just as happens when a species is found to bear several classical scientific names (incidentally, often revealed by barcoding). A careful examination of the content of the BINS and their nearest neighbors is necessary. Working with the specimens reared by Dan Janzen, Winnie Hallwachs et al. in the Costa Rican ACG mega-inventory (
9. “It would supplant taxonomists with technicians, who need to know nothing of the biology of the units with which they are dealing”
We wish this were true and imagine the power it would and perhaps will give the general public. Certainly, more people will play a larger part in a barcoding approach. If the criticism were true, we could overcome the taxonomic impediment much more quickly, to say nothing of exporting bioliteracy possibilities to the majority of the world (
Solutions
1. “real revolutions are undoubtedly coming, especially from the fields of machine learning and integrative species delimitation (
The ‘good news’ reported by the critics does nothing, and will do nothing, to attack the core of the taxonomic impediment. The visual identification system introduced by
2. “a true paradigm shift in taxonomy will come only when there is a revolution in the level of financial investment in taxonomy and the natural history museums that house the described and undescribed reference material of life on Earth”
The taxasphere has been hoping for this for many decades and it will likely never happen until we show the funding agencies that we can make a dent in the taxonomic impediment without costing billions of dollars, to say nothing of showing the overall public that there is reason to bother. To our knowledge, in the USA (and we presume in many other parts of the world) there are no taxonomists that are federally funded for alpha taxonomic research unless they are also addressing interesting scientific, commercial, or public questions. Trivial funding is available for higher level phylogenetic research, although on a very limited scale. We believe that NSF (USA) and other agencies may fund large scale revisions if the revisor, employing a barcode approach, is treating thousands of species and attacking the taxonomic impediment. Unfortunately, current economic crises are causing many of the countries with appropriate resources to be more interested in funding research that will largely benefit themselves rather than the tropical countries so rich in rapidly disappearing biodiversity.
We continue now by justifying our diagnostic approach. COI barcodes are indispensable for diagnosing the species treated herein, as well as signaling where seemingly trivial morphological variation actually indicates a species boundary, and they are an invaluable tool for studies of most other hyperdiverse arthropod taxa as well. For our taxa, COI barcodes, though not infallible, are a magnitude more effective than morphology, and the evidence presented below demonstrates this. Dozens of additional examples can be found in the following studies:
To understand how morphologically similar the species of the aforementioned complex are, compare the images of A. scottshawi and A. genemonroei (Figs
In the Alabagrus revision (
COI data are not always diagnostic. In the revision of Lytopylus (Braconidae, Agathidinae) (
As a final illustration of how ineffective morphology is to differentiate braconid species, we will paraphrase a report published by
Hopefully, we have demonstrated the utility of COI barcode data for species circumscription. Now we address the hurdle of species descriptions. Critics claim that the description of a new taxon must include more than an image and COI sequence data, insisting that keys, morphological diagnoses, detailed figures, and the examination of the holotypes of all congeneric species known from the same realm are necessary. Our answer to these criticisms is that there are too many species and too little time. For many species-rich higher taxa, detailed place-based neotropical inventories such as that of ACG (e.g.,
In the following paragraphs we will demonstrate the magnitude of the task of describing the world fauna of one hymenopteran superfamily and demonstrate that the present approach to taxonomy is woefully inadequate regardless of the number of taxonomists employed. The Ichneumonoidea contains the two most species-rich families of Hymenoptera, i.e., Braconidae and Ichneumonidae. As of 2016, the superfamily comprised ca. 44,000 valid species (
Extrapolating again from
Given the number of the species treatments needed to tackle even a small portion of this diversity, classical taxonomic monographic treatments are not the answer. Most of the criticisms offered against our approach stifle or skate over the fundamental purpose of the method. More images, checking types of dubious likelihood of being conspecific with any of the species treated, and producing morphological keys and diagnoses all take time, too much time, and far greater financial resources are required. Worse, they can only be used by the ever-dwindling number of people that society will support to be “experts” in the field, and even when such experts do exist, they and their literature are not available to the global society at large that needs to have reason to know and understand. But there are other more practical reasons not to include some of these elements which we will get to shortly.
With a COI barcode approach, it will be relatively easy to check the identity of a new specimen of braconid from Costa Rica, and in these new stages of method evolution, we are strategically fortunate that > 95% are undescribed and will remain so unless treated differently than in a traditional manner. To identify any specimen, one simply needs to obtain the COI barcode from the specimen and check for a match in BOLD with a barcode or BIN, then compare the images and other information in the BOLD database to confirm or refute the identification. If there is no match, that specimen is then a starting point for further identification or description, and it has its collateral and barcode data already databased with its specimen voucher code. This may not yet be an effective approach today for most braconid taxa from Costa Rica, but it will be effective for all of the genera that are given a preliminary treatment, such as those presented in the chapters that follow. If a Costa Rican specimen is absent from BOLD after the publication of this and further preliminary revisions, it is straightforward for anyone to describe the species and make it available to all through the web. Processing at the Centre for Biodiversity Genomics (CBG) and the output in BOLD allows its placement in a taxonomic hierarchy; e.g., family, subfamily, and usually genus, the latter almost 95% accurate in braconids. Such description can simply follow an example from any of the following chapters. This is why some refer to COI barcoding as the democratization of taxonomy, and perhaps why it frightens some of our more conservative taxonomic practitioners, and especially those who normally work with just a few (today) important species far from the hyperdiverse tropics.
Compare this to the standard approach of identifying a braconid from Costa Rica. One would start by identifying the specimen to subfamily and genus, impossible tasks for most people, even with good keys, and a difficult task for specialists. In the case of Costa Rican braconids, this is also a rather tedious process for a specialist due to the number of new genera being encountered and generated annually. To make this example a little more understandable to the reader, we will not make a reference to the publications required to do this, rather we will let the reader verify the difficulty for themselves by trying to find the references themselves to attempt the subfamilial and generic identification of any braconid specimen from any Neotropical country. Once a generic name is in hand, the identifier will need to proceed to the species level with the knowledge that fewer than 10% of neotropical species are described. Typically, this means obtaining numerous publications, many of which are protected by copyright and expensive to purchase, even if available on the web, and then comparing the specimen in hand with diverse keys and descriptions, in assorted languages (e.g., German for Costa Rican Opiinae). Furthermore, many of the abundant English texts are based on a vocabulary that often does not have unambiguous translations to Spanish or Portuguese. When a tentative identification is made, an examination of the type specimen will almost certainly be needed for confirmation. Most readers understand the impossible barrier that this presents, especially for non-taxonomists from developing countries. Even if a morphological match with a holotype is obtained, the likelihood of cryptic species will make this morphological comparison far from certain.
Morphological keys for masses of hyperdiverse, poorly known species are all but useless, and their exclusion from the following chapters is not an oversight. This is why. For the genera treated herein, we estimate that we have barcoded at most 10–20% of the Costa Rican fauna. Most of the specimens come from caterpillar rearings in Área de Conservación Guanacaste (the size of the footprint of New York City and its suburbs, www.acguanacaste.ac.cr), and for practical reasons, fruit-feeding, stem-boring, and leaf-mining caterpillars have not yet been reared, nor those from canopy-restricted caterpillars. Added to this are specimens from approximately 16 Malaise trap-years of samples from six localities scattered across dry forest, rain forest, and cloud forest (e.g.,
We have often heard the criticism that a barcoding approach to species circumscription will result in duel taxonomic systems, one morphological and the other DNA-based. There is some truth to this statement if an effort is not made to blend the old with the new, and our suggestion is to discontinue revisions of complex faunas based only on morphology, as well as stop imagining that a visual comparison with ancient holotypes allows accurate decisions as to which of multiple species in a cryptic complex actually matches the holotype. Furthermore, once the species have been tagged with interim names, be they codes or human-readable scientific names, they can be studied and grouped morphologically at will.
Our arguments follow. In the generic treatments below every effort has been made to search the literature for species described by specialists who have spent a lifetime doing braconid taxonomy. In some cases, this has resulted in the discovery of published names for our barcoded specimens, e.g., Adelius gauldi and Adelius janzeni. A recent publication by
As a final comment, the strategy employed herein may not be optimal for all taxa and in all regions of the globe. For example, even in Costa Rica, there are taxa for which there are already many described species (e.g., some families of Lepidoptera), and in these groups our approach may result in the description of an unacceptable number of synonyms, so caution has been employed. For example, the tortricid fauna of Costa Rica includes ca. 250 described species, the vast majority of which have not been barcoded. In this group, describing the 300‒400 BINs that currently have not been identified, would almost certainly result in an unacceptable number of synonyms. Also, because the genitalia of Lepidoptera harbor many of the most compelling morphological features for species discrimination and circumscription, considerably greater effort would be required to examine the morphology in these families than in Braconidae. On the other hand, the comparison of barcodes at $10/specimen is vastly cheaper in specialist’s time than is genitalic study of thousands of specimens and every effort should be made to barcode holotypes.
If there is any hope of overcoming the taxonomic impediment it will be achieved with the approach outlined here or something similar to it. The shortcomings of the method are obvious in that it only provides a first pass at the comprehensiveness that we all desire. First passes, such as those presented in the following chapters, will provide the foundations that can be augmented by subsequent generations of revisors as time, money, and determination allow.
Materials and methods
Specimens and generic placement
Approximately half of the species newly described here, and most of the specimens, were collected by rearing wild-caught host caterpillars in ACG in northwestern Costa Rica (
Four sites in ACG are the subject of most of the Malaise-trapped specimens described here. Bosque San Emilio (BSE) is a 100-year old, 10‒20 m tall, secondary successional Pacific dry forest at ca. 300 m elevation in Sector Santa Rosa. Estación San Gerardo (ESG) is 30‒80-year old mid-elevation Caribbean rain forest at ca. 600 m elevation. Derrumbe (Cloud Forest) is a somewhat fragmented old-growth cloud forest at ca. 1400 m elevation near the top of 1500 m Volcán Cacao, a member of Cordillera Guanacaste that separates BSE from ESG. These three sites had one trap apiece that were operated continually, emptied once a week, for a year, and positioned beneath a broken portion of the canopy rather than in full sun or full heavy shade. The fourth site is a complex of seven Malaise traps in an area of ca. 5 km2 of forest with a 1.5 km2 bare-earth-rock geothermal drilling platform (trap numbers PL12-1-7) in the middle of the area; this forest is mostly composed of old growth at ca. 800 m elevation. The year of sampling at this site ranged from traps nearly insolated at the platform edge to 150 m distant into the shady forest understory. This forest lies at the intersection of ACG dry forest and rainforest at 800 m but slightly below the lower margin of cloud forest that starts at ca. 1000 m. All wild-caught caterpillars from which braconids were reared were found in some variant of the four ecosystems (dry, rain, cloud and interface) and within 20 km of one or more of the Malaise traps. In their first year (sometime between 2011 and 2014, these traps collected ca. 28,000 species of arthropods (excluding spiders, collembola and mites) as based on their BINs (Janzen et al. 2020).
Holotypes of all newly described species are deposited in the insect collection of the Canadian National Collection of Insects, Ottawa (
Identification of specimens to the subfamily level can be achieved using the key by
Morphological terms largely follow Sharkey and Wharton (1997), and definitions in English may be found at the Hymenoptera Anatomy Ontology Portal (http://portal.hymao.org/projects/32/public/ontology/).
Some host species are still awaiting full identification and are given interim names. For example, Antaeotricha Janzen233 is identified to the genus Antaeotricha by classical morphology-based criteria and to Janzen233 by barcode and ecological information. However, no formal scientific species name is available until a barcode-match is obtained with an existing holotype or until it is described as new, which may be decades. Equally, Antaeotricha radicalisEPR03 is also an interim name based on what the species LOOKS like, but is not, by its barcode, and note that the species epithet is not italicized and contains a number: it is not a scientific name, but temporarily retains the information that this species was called simply its look-alike A. radicalis before barcoding and associating it with other ecological data. Also, a name such as gelJanzen01 Janzen407 signifies a caterpillar in the family Gelechiidae for which even a generic name is not obtainable at present. In the future, the barcode, the temporary name, name, and BIN will remain searchable in the Janzen and Hallwachs database as well as in BOLD, GenBank, and any other public sequence repository. Continually updated copies of the Janzen and Hallwachs master database are deposited in the National Museum of Natural History, Washington, DC (
Focus-stacked images of specimens were taken using a JVC digital camera mounted on a Leica microscope and compiled with the program Automontage. Non-distortional image post-photography was done in Adobe Photoshop.
DNA extraction and sequencing
Molecular work was carried out at the CBG using their standard protocols. A leg of each specimen was destructively sampled for DNA extraction using a glass fiber protocol (
Databases
Voucher codes are presented for all holotype specimens (and other barcoded individuals) treated herein; and all host caterpillars are individually vouchered to their individual records (yy-SRNP-xxxxx). Codes beginning with DHPARxxxxx are for the parasite (or hyperparasite) specimens reared from the caterpillar. The SRNP voucher codes are from the Janzen and Hallwachs’ database (http://janzen.sas.upenn.edu/caterpillars/database.lasso). Specimen voucher codes beginning with BIOUG are from the BOLD database (http://www.boldsystems.org), and are specimens obtained from ACG Malaise traps. The abundant collateral information obtainable from these two databases complements the species treatments in important ways. A brief introduction to what to look for and how the databases supplement the species treatments follows.
As mentioned above, the braconid specimens reared in the ACG have a DHJPAR code and their host caterpillar has a SRNP code. These codes can be entered on the home page of the Janzen and Hallwachs database (Fig.
Anacrucis nephrodes (Tortricidae) last instar caterpillar parasitized by Chelonus michellevanderbankae, specimen DHJPAR0029134. Note white wasp larvae in the abdomen, visible through the translucent cuticle. The caterpillar has been exposed for the photograph by opening its leaf and silk nest, littered with its own fecal pellets.
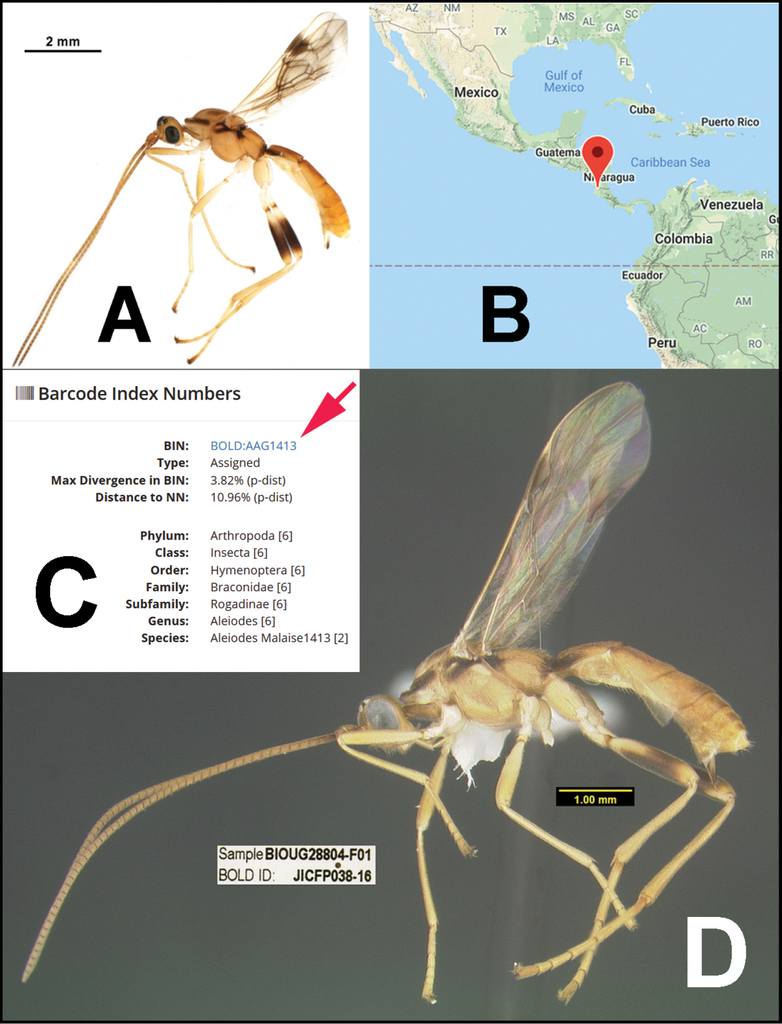
The BOLD website has a wealth of information related to the species that are treated in the chapters below. For example, selecting the link associated with the alphanumeric voucher code of the holotype of Aleiodes adrianaradulovae, BIOUG28804-F01, (http://boldsystems.org/index.php/Public_RecordView?processid=JICFP038-16) brings one to the specimen’s page on BOLD. Some of the contents of that page are shown in Fig.
Data for Aleiodes adrianaradulovae specimen BIOUG28804-F01 A image of specimen BIOUG28804-F01as it appears on BOLD B distribution map on BOLD (which can be enlarged for more detailed locality) C BIN code and other data on BOLD D image of specimen BIOUG28804-F01 as it appears in this publication, image taken after point-mounting. Note its interim species epithet in BOLD, which was updated to the “real” code-compliant species epithet adrianaradulovae when the name was made public, i.e., this publication.
On the BIN page (Fig.
Partial contents of BOLD:AAG1413.
The “Nearest Neighbor Details” section provides the details on the closest BIN, including its member count and the mean and maximum genetic distances among the members. It also includes the “Process ID” used in the CBG (not to be confused with the specimen voucher code) for the closest member of that BIN as well as the taxonomy for that BIN. In the ‘Specimen Images” section, there are four members of the BIN that have images. Clicking on the thumbnail images will bring them into the large image field above them.
Also included in the BIN page (Fig.
Partial contents of BOLD:AAG1413.
NJ tree of BIN BOLD:AAG1413.
In Fig.
From the list of the public records as shown in Fig.
List of specimens in BIN BOLD:AAG1413.
A focus of BOLD Systems is to support collaboration and research networks, and as such, the support team at BOLD helps to connect interested parties with the owners of the specimens with data in BOLD so that they might inquire for further details or gain access to physical specimens, samples, or DNA extracts. Requests for these particular physical objects can be directed to the CBG but for ACG the request should go to DHJ and WH, who will distribute accordingly.
Sequence analysis and species determination
Sequences (barcodes) of the Costa Rican specimens were assigned to multi-specimen operational taxonomic units called Barcode Index Numbers (BINs) (
Morphology and host information were compared to the BIN assignments that “package” the members of the NJ tree into putative species. Most specimen groups (most BINs) were supported by all data sources and treated as one species. However, as mentioned previously, in the treatment of Macrocentrus that follows here, seven species are lumped into the same BIN (BOLD:ACK7466). This is commonplace in other taxonomic groups; there are many cases where a single BIN has been shown unambiguously to be two or more species by their biology and a deeper genome probe (e.g.,
Species diagnoses
Our species treatments are comprised of a link to the BIN on BOLD, a consensus COI barcode, and a lateral or dorsal image of the specimen. Added to this are certain details such as the type depository and holotype locality information. There are a few exceptions to this generality. In several cases more than one species fall in the same BIN and in these cases a morphological key or morphological diagnoses are given to separate these. To facilitate an ongoing revision of the Neotropical Macrocentrinae we have also included morphological diagnoses and more comprehensive imaging for this taxon.
To comply with the Code of Zoological Nomenclature, a word-based diagnosis or definition is necessary for each new species proposed. In the past this was usually in the form of a morphological key or a suite of morphological characters that distinguish the species from others being treated. In our case consensus barcodes of the COI barcode region were employed. These can be seen as a suite of character states that are universally shared by all members of a species and unique to the species. Consensus barcodes were generated using custom software written by one of us (BVB) implemented at phorid.net/DNAbarcode. Fasta files of barcodes for all specimens of a species were downloaded from BOLD and aligned in AliView (
Braconid specimens from the following New World countries appear to be relatively well-sampled in BOLD: Canada, USA, Belize, Argentina, French Guiana, and Mexico. There is a small number of cases where specimens from these countries fall in the same BIN as one of our Costa Rican species. More sampling between these disparate localities, and more genomic and/or morphological and behavioral data will help resolve these species-level cases, which are beyond the scope of this paper. Only rarely are there images of these extra-Costa Rican specimens that suggest that they are not conspecific. These are discussed in more detail under their respective species treatments. To reiterate and confirm: we fully understand that barcodes are not a panacea that will solve all species-limit questions, we simply suggest that they are the optimal available first step.
We attach 10 supplementary chapters for the 11 subfamilies treated here representing ten COI NJ trees, all downloaded from BOLD in November 2020. Each is a snapshot of the dynamic project database at http://djanzen.sas.upenn.edu, select fields of which are routinely updated to BOLD for public use and taxonomic treatment as in this work. The project database is forever dynamic owing to the need for constant updating of the names of species in the ever-shifting taxonomy of plants, hosts and parasites, as new publication and biological relationships emerge. We use the NJ trees as quick visual summaries of sample sizes per species, degrees of specialization of the parasites, flagging of species new to the project, indications of taxonomic puzzles, variability in barcode length, BIN composition (the equivalent of sorting morphological look-alikes into unit trays), and data-checking. Typically, what we consider to be conspecifics are grouped together in their own terminal clade. However, specimens with less than about 550 base pairs are potentially suspect as to their placement in a NJ tree. The shorter the barcode, the more likely it is to be misplaced on the tree, requiring taxonomic placement by other traits, such as morphology or host data or both. It is not uncommon for unrelated specimens with short sequences to group together; this is because the sequences that would group them with their conspecifics are not present and only conserved base pairs, common to many species, are present. Defectively short barcodes are not honored with a BIN code, even though they may group with BINed conspecifics. The arrangement of presumed conspecifics within a BIN into shallowly separated monophyletic groups has high potential for suggesting the presence of a species complex within the BIN, a moderately frequent occurrence (10%). Resolution of such species complexes requires ecological correlates, morphological correlates, and/or a deep genomic probe (e.g.,
Chapter 2: Agathidinae
The key to genera by
Key to the New World genera of Agathidinae
Aerophilus Szépligeti, 1902
Aerophilus is worldwide in distribution with hundreds (described plus undescribed) of species in the New World. Members are parasitoids of a wide range of caterpillar families. The genus is well represented in both the Nearctic and Neotropical realms.
Aerophilus paulmarshi , sp. nov.
Diagnostics
BOLD:ADC6602. Consensus barcode. TATTTTATATTTTATTTTTGGAATTTGATCAGGAATTTTAGGTTTATCAATAAGAATAATTATTCGAATAGAATTAAGATTAGGGGGTAATTTAATTGGTAAT---GATCAAATTTATAATAGAATTGTTTCTGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGTTTTGGAAATTGATTAGTTCCTTTAATATTAGGGGGACCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTATTAATATTAATTTTAAGATCTTTAATTAATATTGGAGTTGGTACAGGATGAACAGTTTATCCTCCTTTATCAATAAATATGAGTCATAGAGGAATATCAGTAGATTTAGCTATTTTTTCTTTACATATTGCAGGAATTTCTTCTATTATAGGAGCAATAAATTTTATTACAACTATTATAAATATATGAATAATCAATGTTAAAATTGATAAAATACCTTTATTAGTATGATCAATTTTTATTTCTGCTATTTTATTATTATTATCATTACCAGTATTAGCTGGAGCTATTACAATATTATTAACTGATCGAAATTTAAATACAAGATTTTTTGATCCATCAGGAGGAGGAGATCC----------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.6138, 820 meters, Malaise trap, 6/iii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29687-E06.
Paratypes
None.
Etymology
Named in honor of Paul Marsh, eminent braconidologist.
Note
In the key provided by
Mesocoelus Schulz, 1911
Members are widespread but rarely collected in the neotropics. The extremely reduced venation of Mesocoelus is rare among agathidines, shared only with the Old World genus Aneurobracon (Agathidinae). The two described species are both from the Caribbean (holotypes from St. Vincent and Cuba). Reported hosts are Chilocampyla psidiella and Acrocercops sp., leaf-miners in the family Gracillariidae. No new host records for the genus are reported here.
Mesocoelus davidsmithi , sp. nov.
Diagnostics
BOLD:AAV4319. Consensus barcode. AATTTTATATTTTATTTTTGGAGTATGGGCAGGAATTTTAGGGTTATCAATAAGATTAATTATTCGTATAGAATTAAGAGTTATTGGAAATTTTATTGGTAATGATCAGATTTATAATAGGATTGTTACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGGTTTGGTAATTGATTAGTTCCATTAATAGTAGGAGGACCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTATTATTATTAATTTTAAGGTCAATAGTTAATGTTGGGGTAGGAACTGGGTGAACAGTTTACCCTCCTTTATCTTTAAATATAAGGCATAGAGGGATTTCTGTTGATTTGGCTATTTTTTCTTTACACATTGCAGGAGTTTCTTCAATTATAGGAGCTATAAACTTTATTACTACTATTTTAAATATATGAATCATTAATATTAAAATTGATAAAATACCTTTATTAGTATGATCTATTTTAATTACTGCAATTTTATTATTATTATCATTACCTGTATTAGCCGGAGCAATTACTATATTGTTAACTGATCGTAATTTAAATACAAGATTTTTTGACCCGACAGGAGGAGGGGATCCTATTTTATATCAACATTTATTT. Of the two described species, M. davidsmithi is closest to M. laeviceps in that they both lack distinct notauli. To differentiate these two, M. davidsmithi has 27 flagellomeres, whereas M. laeviceps has between 19 and 22.
Holotype ♀
Heredia, 6 km ENE Vara Blanca, 10.183, -84.12, 2000 meters, 09/iv/2002, Malaise trap. Depository:
Host data. None.
Holotype voucher code. H1731.
Paratypes
None.
Neothlipsis Sharkey, 2011
Neothlipsis is widespread in the Nearctic and northern Neotropical region. There may be ca. 70 species including undescribed species. The sole host record is Samea multiplicalis (Guenée) (Crambidae).
Neothlipsis bobkulai , sp. nov.
Diagnostics
BOLD:ADB1901. Consensus barcode. TCTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTTTAATATTAGGAGGTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTATTAATATTGATTTCTAGATCTATAATTAATATTGGTGTTGGTACAGGTTGAACTGTTTATCCTCCTTTATCTTTAAATTTAAGACATAGTGGAATTTCTGTAGATTTAGCTATTTTTTCTTTACATATTGCAGGAATTTCTTCTATTATGGGAGCAATAAATTTTATTACAACTATTTTAAATATATGAATAATAAATATTAAAATTGATAAAATACCATTATTAATTTGATCTATTTTTATTACAGCTATTTTATTATTATTATCTTTACCAGTATTAGCTGGGGCAATTACAATATTATTAACAGATCGAAATTTAAATACTAGATTTTTTGATCCTACAGGAGGGGGAGATCCA. Similar to Neothlipsis taeniativentris (Enderlein, 1920) but differing slightly in color, with the pale yellow on the head of N. taeniativentris more extensive.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.6138, 820 meters, Malaise trap, 5/xii/2013. Depository:
Host data. None.
Holotype voucher code. BIOUG29112-G02.
Paratypes
BIOUG29646-F07, BIOUG29620-B06, BIOUG29646-E01.
Plesiocoelus van Achterberg, 1990
Members are widespread but rarely collected in the neotropics. It is unlikely that there are more than ten species including undescribed species The holotype of the only described species from the New World, P. bassiformis, is from Ecuador. van
Bortoni and Penteado-Dias (2015) described two new species under Plesiocoelus, however, they both belong to other genera: Therophilus anomalus (Bortoni and Penteado-Dias) new combination for Plesiocoelus anomalus Bortoni & Penteado-Dias, 2015; Aerophilus areolatus (Bortoni and Penteado-Dias) new combination for Plesiocoelus areolatus Bortoni & Penteado-Dias, 2015. See the key to genera above that incorporates these aberrant species with reduced wing venation.
Plesiocoelus vanachterbergi , sp. nov.
Diagnostics
BOLD:ABX6701. Consensus barcode. AATTTTATATTTTATTTTCGGAATTTGATCAGGAATTTTGGGRTTATCAATAAGWTTGGTTATTCGAATGGAATTAAGAATTACTAGAAATTTTATTGGTAATGATCAAATTTATAATAGTATTGTT-CTRCTCATGCTTTTATTATAATTTTTTTTATGGTAATACCTATTATGATTGGGGGATTYGGAAATTGATTAATTCCTTTAATATTAGGAGGTCCTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCATCTTTATTATTATTAATTTCTAGCTCAATTATTAATGTTGGAGTAGGAACAGGATGAACAGTTTATCCTCCTTTATCATTAAATTTAAGTCATAGTGGAATTTCAGTAGATTTAGCTATTTTTTCTTTACATATTGCTGGAATTTCATCAATTATAGGAGCAATAAATTTTATTACT-CTATTTTAAATATATGRATAATTAAAATTAATGTTGATAAAATAACTTTATTGATTTGRTCAATTTTAATTACAGCAATTTTATTATTATTATCATTACCGGTWTTAGCAGGAGCAATTACAATATTATTAACAGATCGAAATTTAAATACTAGATTTTTTGATCCTTCAGGAGGGGGAGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Jacobo, 10.94076, -85.3177, 461 meters, caterpillar collection date: 28/vii/2013, wasp eclosion date: 8/viii/2013. Depository:
Host data. Parasitoid of Brenthia Janzen02 (Choreutidae) feeding on leaves of Rhynchosia calycosa (Fabaceae).
Caterpillar and holotype voucher codes. 13-SRNP-69969, DHJPAR0053046.
Paratypes
Hosts = Brenthia Janzen02, Brenthia Janzen05, Brenthia Janzen14, Brenthia Janzen15. DHJPAR0049867, DHJPAR0048215, DHJPAR0050114, DHJPAR0053040, DHJPAR0053044, DHJPAR0053045, DHJPAR0053033, DHJPAR0045360. Depository:
Pneumagathis Sharkey, 2015
Members of Pneumagathis are restricted to North and Central America. There appear to be only two species, P. erythrogastra (Neotropical) and P. spiracularis (Nearctic), both of which are parasitoids of Hesperiidae.
Pneumagathis erythrogastra , comb. nov.
Agathis erythrogastra Cameron, 1905.
Bassus brooksi Sharkey, 1998, syn. nov.
Pneumagathis brooksi (Sharkey, 1998), in Chapman and Sharkey (2015).
Diagnostics
BOLD:AAV3035. Consensus barcode. GATTTTATATTTTATTTTTGGAATTTGRTCRGGRATATTRGGTTTATCAATAAGTTTAATTATTCGAATAGAATTAAGAATTACAGGAAATTTTATTGGTAATGATCAAATTTATAATTCAATTGTT-CTGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGATTYGGAAATTGRTTAGTYCCTTTGATATTAGGAGGRCCYGATATAGCTTTTCCDCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATATTATTAATATTAAGTTCAATTATTAATATTGGRGTAGGTACTGGATGAACAGTTTATCCTCCTTTATCTTTAAATATAAGTCATAGAGGWATATCAGTTGATTTAGCTATTTTTTCTTTACATATTGCTGGAATTTCTTCTATTATAGGAGCAATAAATTTTATTACMACAATTTTAAAYATATGAATTATTAATATTAAAATTGATAAAATACCTTTATTAGTRTGATCAATTTTTATTACTGCAATTTTATTATTATTATCTTTACCAGTTTTAGCTGGAGCTATTACTATATTATTGACTGAYCGAAATTTAAATACAAGATTWTTTGATCCTTCAGGAGGGGGRGATCCAATTTTATATCAACATTTRTTT------------------------------------------------------------------.
Notes
As reported in
Therophilus Wesmael, 1837
Therophilus is a cosmopolitan genus with many hundreds of species (mostly undescribed). It is relatively rare in the neotropics. Published hosts include caterpillars of a wide range of lepidopteran families (
Therophilus penteadodiasae , sp. nov.
Diagnostics
BOLD:ABZ8830. Consensus barcode. TATTTTATATTTTATTTTTGGGATTTGATCGGGAATTTTAGGGTTATCAATAAGATTATTAATTCGAATAGAGTTAAGTATTGGTGGTAATTTTATTGGTAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATRATTGGTGGTTTTGGTAATTGATTAATTCCTTTAATATTAGGAGGTCCAGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAACATTATTAATTTTAAGTTCTTTAATTAATATTGGAGTTGGAACTGGATGAACAGTCTATCCTCCTTTATCGTTAAATATAAGACACAGAGGAATATCTGTTGATTTAGCAATTTTTTCTTTACATATTGCTGGTGTTTCTTCAATTATAGGGGCAATAAATTTTATTACAACTATTTTAAATATATGAATTGTAAATATTAAAATTGATAAAATATCTTTATTGGTTTGATCAATTTTAATTACAGCGATTTTATTATTGTTATCTTTACCAGTATTGGCTGGTGCAATTACTATATTATTAACAGATCGAAATTTAAATACAAGATTTTTTGATCCTTCAGGAGGAGGGGATCCAATTTTATATCAGCATTTATTT.
Holotype ♀
Guanacaste, Sector El Hacha, Casa Hacha Vieja, 10.98050, -85.54429, 290 meters, 290 meters, caterpillar collection date: 24/ii/2014, wasp eclosion date: 17/iii/2014. Depository:
Host data. Parasitoid of Antaeotricha Janzen146 (Depressariidae) feeding on Lonchocarpus oliganthus (Fabaceae).
Caterpillar and holotype voucher codes. 14-SRNP-55508, DHJPAR0055292.
Paratypes
Hosts = Antaeotricha Janzen146 and Antaeotricha Janzen366 (Depressariidae). DHJPAR0055209, DHJPAR0055544.
Etymology
The species is named in honor of Angelica Penteado-Dias, eminent Brazilian braconidologist.
Therophilus bobwhartoni , sp. nov.
Diagnostics
BOLD:AAY4688. Consensus barcode. AATTTTATATTTTATTTTTGGAATTTGGTCGGGTATTTTGGGATTATCGATAAGTTTATTAATTCGGATGGAATTAAGAGTAGGGGGTAATTTTATTGGAAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCATTTATTATAATTTTTTTTATGGTTATACCAATTATAATTGGGGGATTTGGTAATTGATTAATTCCTTTAATGTTAGGGGGTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCCTCATTAATATTATTAATTTTGAGGTCATTAATTAATATTGGAGTTGGGACTGGRTGAACAGTTTACCCTCCTTTATCATTAAATATAAGTCATAGGGGGATATCTGTTGATTTAGCGATTTTTTCTTTGCATATGGCAGGAGTTTCTTCAATTATGGGGGCAATAAATTTTATTACTACAATTTTAAATATATGAATTATAAATATTAAAATTGATAAGATACCTTTATTAGTTTGATCGATTTTAATTACGGCAATTTTGTTATTATTATCATTACCTGTGTTAGCTGGAGCTATTACTATATTATTAACTGACCGAAATTTAAATACAAGATTTTTTGATCCTTCAGGTGGAGGGGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Cacao, Sendero Nayo, 10.92446, -85.46953, 1090 meters, caterpillar collection date: 05/ii/2007, wasp eclosion date: 18/iii/2007. Depository:
Host data. Parasitoid of elachJanzen01 Janzen161 (Depressariidae) feeding on Viburnum costaricanum.
Caterpillar and holotype voucher codes. 07-SRNP-35262, DHJPAR0062169.
Paratype
Host = elachJanzen01 Janzen161: DHJPAR0041864.
Therophilus donaldquickei , sp. nov.
Diagnostics
BOLD:ADM6790. Consensus barcode. GGGAATGATCAAGTTTATAATAGAATTGTTACTGCGCATGCATTTATTATAATTTTTTTTATGGTGATACCAATTATAATTGGTGGGTTTGGAAATTGATTAGTTCCTTTAATGTTGGGAGGACCTGATATGGCTTTCCCACGAATAAATAATATAAGTTTTTGGTTATTAATTCCATCATTAATATTATTAATTTTGAGTTCTTTAATTAATATTGGGGTTGGAACAGGGTGGACAGTTTATCCTCCATTGTCATTAAATATAAGTCATAGAGGTATATCAGTTGATTTAGCTATTTTTTCTTTACATATAGCCGGTGTTTCTTCAATTATAGGAGCTATAAATTTTATTACTACAATTTTAAATATATGAATAATAACAATTAAAATTGATAAAATACCATTGTTAGTTTGATCTATTTTAATTACAGCAATTTTATTATTGTTATCTTTACCTGTGTTGGCGGGGGCTATTACTATATTATTAACAGATCGAAATTTAAATACAAGATTTTTTGATCCATCAGGTGGTGGTGACCCAATTTTATATCAACATTTATTTTGATTTTTTGGTCATCCTGAAGTTTATATTTTAATTTTACCGGGATTTGGGATTATTTCTCATATT.
Holotype ♀
Heredia, 16 km SSE La Virgen, 1050‒1150 meters, 21/iii/2001, Malaise trap. Depository:
Host data. None.
Holotype voucher code. H155.
Paratypes
None.
Therophilus gracewoodae , sp. nov.
Diagnostics
BOLD:ADD0457. Consensus barcode. TATTTTATATTTTATTTTTGGAATTTGGTCGGGTATTTTGGGATTATCAATAAGTTTATTAATTCGTATAGAATTAAGAATTGGGGGTAACTTAATTGGTAACGATCAAATTTATAATAGAATTGTTACTGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGTTTTGGTAATTGATTGATTCCTTTAATATTGGGGGGGCCTGATATAGCTTTCCCCCGGATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAATATTATTAATTTTGAGTTCATTAATTAATATTGGAGTTGGAACAGGATGAACAGTTTATCCTCCTTTATCATTAAATATAAGTCATAGAGGTATATCAGTTGATTTAGCTATTTTTTCTTTACATATAGCTGGTATTTCATCAATTATGGGGGCAATAAATTTTATTACTACAATTTTTAATATATGAATTATAATAATTAAAATTGATAAAATACCATTATTAGTTTGATCTATTTTAATTACAGCAATTTTATTATTATTATCATTACCTGTATTGGCTGGGGCTATTACAATATTATTAACAGATCGAAATTTAAATACAAGATTTTTTGATCCTTCAGGGGGGGGGGA.
Holotype ♀
Guanacaste, Sector Cacao, Sendero Arenales, 10.92471, -85.46738, 1,080 meters, caterpillar collection date: 09/iv/2010, wasp eclosion date: 10/v/2010. Depository:
Host data. Parasitoid of Rhodoneura terminalis (Thyrididae) feeding on Hampea appendiculata (Malvaceae)
Caterpillar and holotype voucher codes. 10-SRNP-35134, DHJPAR0039519.
Paratypes
None.
Therophilus maetoi , sp. nov.
Diagnostics
BOLD:AAU5369. Consensus barcode. ATTTTTATATTTTATTTTTGGAATTTGATCAGGAATTTTAGGATTATCAATAAGTTTATTAGTTCGAATGGAATTAAGAATTRGTGGTAATTTTATTGGKAATGATCAAATTTATAATAGTATTGTAACTGCACATGCATTTATTATAATTTTTTTTATAGTTATGCCAATYATAATTGGAGGRTTTGGTAATTGGTTAGTACCTTTAATATTAGGAGGTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGGTTTTGATTATTAATYCCTTCATTAATATTATTAATTTTRAGATCATTAATTAATATTGGAGTTGGWACWGGTTGAACAGTATAYCCRCCRTTATCWTTAAATATAAGACATAGGGGTATATCTGTTGATTTRGCTATTTTTTCTTTACATATTGCTGGTATTTCATCTATTATAGGGGCAATAAATTTTATTACAACTATTTTAAATATATGAATTATTAATATTAAAATTGATAAAATRCCTTTATTAGTTTGATCAATTTTAATTACTGCAATTTTATTATTATTATCATTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACAGATCGAAATTTAAATACAAGATTTTTTGATCCTTCAGGAGGTGGTGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Heredia, 6 km ENE Vara Blanca, 10.183, -84.12, 2000 meters, 21/ii/2002, Malaise trap. Depository:
Host data. None.
Holotype voucher code. H1168.
Paratypes
H1148, H1150, H1168.
Therophilus montywoodi , sp. nov.
Diagnostics
BOLD:ADD0519. Consensus barcode. AATTTTATATTTTATTTTTGGGGTTTGATCAGGAATTGTAGGTTTATCAATAAGATTATTAATTCGAATGGAATTAAGAATTGGTGGTAATTTTATTGGTAATGATCAAATTTATAATAGAATTGTTACAGCTCATGCATTTGTAATAATTTTTTTTATAGTTATACCAATTATGATTGGGGGTTTTGGAAATTGATTAGTTCCTTTAATATTGGGGGGYCCAGATATAGCTTTCCCTCGAATAAATAATATAAGATTCTGGTTATTAATTCCTTCATTAATATTATTAATTTTAAGTTCATTAGTTAATATTGGAGTTGGAACAGGGTGAACAGTTTATCCTCCTTTATCTTTAAATATAAGTCATAGAGGTATATCGGTTGATTTAGCTATTTTTTCTTTACATATTGCAGGGGTTTCATCAATTATAGGGGCTATAAATTTTATTACTACAATTTTAAATATATGAATTATGAATATTAAAATTGATAAAATACCTTTATTAGTGTGATCAATTTTAATTACAGCAATTTTATTATTATTATCATTACCAGTGTTAGCAGGGGCAATTACAATATTATTAACAGATCGTAATTTGAATACAAGATTTTTTGATCCTTCTGGTGGGGGGGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Finca San Gabriel, 10.87766, -85.39343, 645 meters, caterpillar collection date: 31/vii/2009, wasp eclosion date: 26/viii/2009. Depository:
Host data. Parasitoid of Strepsicrates Brown25 (Tortricidae) feeding on Psidium guajava (introduced) (Myrtaceae).
Caterpillar and holotype voucher codes. 09-SRNP-4023, DHJPAR0040068.
Paratypes
Host = Strepsicrates Brown25: DHJPAR0038338. Depository:
Zacremnops Sharkey & Wharton, 1985
Members are widespread in the neotropics.
Zacremnops brianbrowni , sp. nov.
Diagnosis
The COI barcode has only 293 bases and therefore was not given a BIN. This species can be distinguished from all other species of Zacremnops by red coloration restricted to the metapleuron (i.e., not on the propodeum) in combination with the melanic hind tarsus.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Sendero Juntas, 10.90661, -85.28784, 400 meters, caterpillar collection date: 11/viii/2010, wasp eclosion date: 3/ix/2010. Depository:
Host data. Parasitoid of Microthyris prolongalis (Crambidae) feeding on Ipomoea batatas (Convolvulaceae).
Caterpillar and holotype voucher codes. 10-SRNP-42859, DHJPAR0041189.
Paratypes
None.
Zacremnops coatlicue
Diagnostics
No barcode available. This species can be distinguished from all other species of Zacremnops by restriction of the red coloration to the metapleuron and propodeum, in combination with the melanic hind tarsus
Host data. All parasitoids of Syllepte belialis (Crambidae).
Material examined
Zacremnops cressoni
Diagnosis
BOLD: AAV3186. Z. cressoni is the only known species of Zacremnops that has yellow hind tarsi. What appears to be this species is widespread from southern USA to northern Colombia and Venezuela, including the Caribbean.
Host data . The sole reared specimen parasitized Microthyris prolongalis (Crambidae) feeding on Ipomoea batatas (Convolvulaceae).
Zacremnops ekchuah
Diagnosis
There are no COI barcodes available for any of the six reared specimens; however, Z. ekchuah is the only species of Zacremnops that is entirely black. What appears to be this species is widespread in Mexico with southern records in Costa Rican Caribbean mid-elevation rain forest.
Host data. Specimens were reared from spilomeline cambids, i.e., Microthyris prolongalis, Microthyris prolongalisDHJ02, Pantographa suffusalis, and Syllepte amandoDHJ02 (Crambidae).
Zamicrodus Viereck, 1912
Members of this genus are distributed widely across the neotropics. The two described species are from South America. The first host record (Hesperiidae) for the genus is reported here. The genus has not been revised, though there are likely to be fewer than ten species.
Zamicrodus josefernandezi , sp. nov.
Diagnostics
BOLD:ACC5208. Consensus barcode. AATTTTATATTTTATTTTTGGTGTTTGATCAGGAATTTTAGGTTTATCTATAAGAATAATTATTCGAATAGAATTAAGAATTACAAGTAATTTTATTGGAAATGATCAAATTTATAATTCTATTGTCCTGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGTGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGGACCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTATTATTATTAATTATAAGATCTATTATTAATATTGGAGTAGGACTGGATGAACAGTTTATCCTCCTTTATCATTAAATTTAAGTCATAGAGGTATTTCAGTAGATTTAGCTATTTTTTCTTTACATATTGCAGGAATTTCTTCTATTATAGGAGCTATAAATTTTATTACAACAATTTTAAATATATGAATTATTAATATTAAAATTGATAAAATACCCTTATTAGTTTGATCAATTTTTATTACAGCAATTTTATTATTATTATCATTACCAGTTTTAGCTGGAGCTATTACTATATTATTACTGATCGAAATTTAAATACAAGATTTTTTGATCCTTCTGGAGGAGGAGACCCAATTTTATATCAACATTTATTT. The two described species differ from Z. josefernandezi in that their mesosomata are partly to completely pale yellow-orange, whereas that of Z. josefernandezi is completely black.
Holotype ♀
Guanacaste, Sector Pitilla, Medrano, 11.01602, -85.38053, 380 meters, caterpillar collection date: 9/ii/2012, wasp eclosion date: 8/iii/2012. Depository:
Host data. Parasitoid of Quasimellana sethos (Hesperiidae) feeding on Rhipidocladum racemiflorum (Poaceae).
Caterpillar and holotype voucher codes. 12-SRNP-70305, DHJPAR0048733.
09-SRNP-4023, DHJPAR0048733.
Paratypes
Host, Quasimellana sethos: DHJPAR0057459, DHJPAR0048728.
Zelomorpha Ashmead, 1900
The Costa Rican species of Zelomorpha were revised by
Zelomorpha sarahmeierottoae , sp. nov.
Diagnostics
BOLD:ACG5511. Consensus barcode. TGTTTTATATTTTTTATTTGGTATATGAAGGGGAATTTTAGGATTAAGATTGAGTTTATTGGTTCGATTTGAATTAGGATTAAGAGGAAATTTAATTGGAAGTGATCAAATTTATAATAGGATAGTACTTATCATGCATTAATTATGATTTTTTTTATAGTTATACCTATTATGATTGGAGGATTTGGTAATTGGTTGGTTCCTTTATTGTTAGGAAGACCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTATTACCTTCATTAATATTATTAATTTTAAGTTCTTTTGTTAATATTGGAGCAGGTACAGGTTGGACTATTTATCCTCCATTATCATTAAATTTTAGACATAGGGGTATGTCGGTTGATTTAATAATTTTTGCTTTGCATATTGCTGGTGTTTCATCAATTATAGGTGCAATTAATTTTATTACTACTATTTTAAATATATGAATAATAAATATTAATATAGATAAAATATCTTTATTTGTTTGATCTATTTTATTAACAGCTATTTTATTATTATTATCTTTGCCAGTTTTAGCTGGAGCTATTACTATATTATTAAGAGATCGAAATTTAAATTCAAGATTTTTTGATCCTACTGGGGGAGGGGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, Malaise trap, 04/vii/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG05883-E01.
Paratypes
None.
Chapter 3: Braconinae
Braconines are primarily idiobiont parasitoids of Coleoptera and Lepidoptera, but many other insect orders have been used. Below we treat only Costa Rican species. The ten reared species described below are all gregarious parasitoids of caterpillars. Hosts of the Malaise-trapped species could be larval Lepidoptera or the larvae of a number of other insect orders, the most likely being Coleoptera. The key by
Key to the New World genera of Braconinae
The shape of the antennal scape is important for identification of genera. For determination of the relative lengths of the dorsal and ventral sides of the scape, as in couplets 3‒5, 11, 12, and 16, the antennae should be positioned as those of couplet 3, i.e., horizontally.
Bracon Fabricius, 1804
Bracon is an enormous, polyphyletic, cosmopolitan genus with thousands of undescribed species. They are idiobiont ectoparasitoids, attacking hosts in many insect orders, but primarily Coleoptera and Lepidoptera.
Bracon alejandromarini , sp. nov.
Diagnostics
BOLD:ADH4980. Consensus barcode. GGAGTTTTATATTTTTTATTCGGTATATGAGCTGGTATAATTGGTTTATCTATAAGTTTAATTATTCGTTTAGAATTAGGTAYACCAGGAAGTATACTAGGGAATGACCAAATTTATAATAGAATAGTGACTGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGTTTAAATAATATAAGGTTTTGGTTATTAATTCCTTCATTAACTTTATTATTATTAAGAAGAATTTTAAATATTGGTGTAGGAACAGGATGAACTATATATCCTCCTTTATCTTCAAGTTTAGGCCATAGAGGTATATCAGTTGATTTGGCAATTTTTTCTTTACATTTAGCTGGGGCATCTTCAATTATAGGGGCAATAAATTTTATTACTACTATTTTAAATATACATTTAATAATAATAAAATTAGATCAATTAACTTTATTAATTTGATCTATTTTTATCACAACTATTTTATTATTATTATCTTTACCAGTATTAGCAGGGGCAATTACTATATTATTGACAGAT.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 12/iii/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG32921-A02.
Paratype
BIOUG32997-A06. Depository:
Etymology
Bracon alejandromarini is named to honor Alejandro Marin for his efforts to shadow his father, Sigifredo Marin, as an apprentice to the overall managerial aspects of the projects of the Guanacaste Dry Forest Conservation Fund in support of Área de Conservación Guanacaste, and for his enthusiastic willingness to apply his professional medical knowledge to GDFCF staff in Costa Rica.
Bracon alejandromasisi , sp. nov.
Diagnostics
BOLD:AAA5367. Consensus barcode. TGTATTATATTTTTTATTTGGAATATGAGCYGGAATAATTGGTTTATCAATAAGTTTAATTATTCGTTTAGAATTAGGRATACCAGGTAGTTTAYTAGGTAATGATCAAATTTATAATAGTATAGTTACAGCKCATGCTTTTATTATAATTTTTTTTATAGTTATACCAGTAATATTAGGAGGWTTTGGTAATTGATTAGTTCCTTTAATATTAGGTGCTCCTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATTTTATTATTATTAAGAAGAATTTTAAATGTTGGTGTAGGRACAGGCTGAACTATTTATCCTCCTTTATCTTCTATAATAGGTCATAGAGGWATATCTGTRGATTTATCTATTTTYTCTTTACATTTAGCTGGTATTTCTTCTATTATAGGATCGATTAATTTTATTACAACAATTTTAAATATACATTTATTAATATTAAAATTAGATCAATTAACTTTATTTATTTGATCAATTTTTATTACAACTATTTTATTATTATTATCTTTACCTGTATTAGCAGGAGCTATTACTATAYTATTAACTGATCGAAATTTWAATACTTCATTTTTTGATTTTTCTGGAGGTGGGGATCCAATTYTATTTCAACATTTATTT. Bracon alejandromasisi and B. tihisiaboshartae occupy the same BIN and have the same consensus barcode. Bracon tihisiaboshartae can be differentiated from B. alejandromasisi by the color of the metasomal terga: entirely yellow in B. tihisiaboshartae and partly black in B. alejandromasisi
Holotype ♀
Guanacaste, Sector Cacao, Sendero Nayo, 10.92446, -85.46953, 1090 meters, caterpillar collection date: 22/x/2013, wasp eclosion date: 09/xi/2013, number eclosed 17. Depository:
Host data. Gregarious parasitoid of Consul electra (Nymphalidae) feeding on leaves of Piper psilorhachis (Piperaceae).
Caterpillar and holotype voucher codes. 13-SRNP-36214, DHJPAR0054650.
Note
Bracon alejandromasisi and Bracon tihisiaboshartae occupy the same BIN but are clearly two species, see comments for B. tihisiaboshartae.
Paratypes
Hosts = Consul electra and Consul cecrops (Nymphalidae) feeding on four species of Piper (Piperaceae). 14 specimens, same data as holotype and DHJPAR0029029, DHJPAR0029036, DHJPAR0029031, DHJPAR0034263. Depository:
Etymology
Bracon alejandromasisi is named in honor of Alejandro Masis’ persistent high-quality biopoliticking on behalf of ACG in San José, Costa Rica, and especially in Guanacaste Province.
Bracon alexamasisae , sp. nov.
Diagnostics
BOLD:ADF2876. Consensus barcode. GTTTTATATTTTTTATTTGGTATATGATCTGGAATATTAGGTTTATCAATAAGGTTAATTATTCGTTTAGAATTAGGTATACCAGGGAGATTATTGGGTAATGATCAAATTTATAATAGTATAGTTACTGCTCATGCTTTTGTAATAATTTTTTTTATGGTTATACCTGTAATAATTGGAGGTTTTGGAAATTGATTATTGCCTTTAATATTAGGAGCTCCAGATATAGCTTTCCCTCGTCTTAATAATATAAGATTTTGGTTATTAATTCCTTCTTTATTTTTATTACTTATAAGAAGAGTTTTAAATGTTGGTGTAGGTACAGGTTGAACAGTTTATCCACCTTTATCTTCTTCTATGGGTCATAGAGGTTTATCTGTTGATTTAGCTATTTTTTCTTTACATATTGCTGGTATTTCTTCTATTTTGGGTGCTATTAATTTTATTACAACTATTTTAAATATACATTTGTATACTTTAAAATTAGATCAGATAACTTTATTAATTTGATCAGTTTTTATTACAGTAATTTTATTATTATTATCTTTACCAGTTTTAGCTGGTGCTATTACTATATTGTTAACTGAT.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 6/xi/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG31518-E04.
Paratypes
None.
Etymology
Bracon alexamasisae is named to honor Alexa Masis for her cheerful participation in the lives and dedication of the Masis + Boshart family as it has carried out a vital role in the growth and management of the Guanacaste Dry Forest Conservation Fund and Área de Conservación Guanacaste.
Bracon andresmarini , sp. nov.
Diagnostics
BOLD:ADD6766. Consensus barcode. ATATTATATTTTTTATTTGGATTATGAGCTGGTATATTAGGATTATCTATAAGTTTAATTATTCGTTTAGAATTAGGTATACCTGGTAGATTATTAGGTAATGATCAAATTTATAATAGAATAGTTACTGCCCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAGTAATATTAGGAGGATTTGGAAATTGATTAATTCCGTTAATATTAGGTGCACCAGATATAGCTTTCCCTCGTTTAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATTTTATTAATATTAAGAAGAATTTTAAATATTGGAGTAGGCACAGGATGAACTATATATCCTCCTTTATCATCAAATTTAGGCCACAGAGGTATATCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCCGGAGTATCCTCAATTATAGGCTCAATAAATTTTATTACAACTATTTTAAATATACATTTAGTTATAATAAAATTAGATCAACTAACTTTATTAATTTGATCAATTTTTATTACAACTATTTTATTATTATTATCTTTACCTGTTTTAGCAGGA.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-2, 10.7634, -85.335, 824 meters, Malaise trap, 23/i/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG30922-D05.
Paratypes
None.
Etymology
Bracon andresmarini is named to honor Andres Marin for his cheerful participation in the lives and business future of the Marin and Romero families as they have carried out its vital role in the growth and management of the Guanacaste Dry Forest Conservation Fund and Área de Conservación Guanacaste.
Bracon andrewwalshi , sp. nov.
Diagnostics
BOLD:AAY4684. Consensus barcode. AGTTTTATATTTTTTATTTGGTATATGAGCTGGTATAGTAGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAGGTATACCTGGAAGTTTATTAGGTAATGACCAAATTTATAATAGAATAGTTACAGCTCATGCTTTTGTAATAATTTTTTTTATAGTYATACCAGTTATATTAGGTGGATTTGGTAATTGATTAATCCCTTTAATATTAGGAGCCCCTGATATAGCTTTCCCTCGAATAAATAATATAAGTTTTTGGTTGTTAATTCCTTCATTAATTTTATTATTATTAAGAAGAATTTTAAATGTGGGTGTAGGAACAGGATGAACTATATACCCACCTTTATCTTCAAGATTAGGGCATAGAGGTTTATCTGTTGATTTAGCTATTTTTTCTCTACATTTAGCAGGAGTTTCTTCAATTATAGGTTCAATAAATTTTATTACAACTATTTTAAATATACATCTATTAATATTAAAATTAGATCAATTAACTTTATTAGTTTGATCAATTTTTATTACTACTATTTTATTATTGTTATCATTACCTGTTTTAGCAGGAGCTATCACTATATTATTAACTGATCGTAATTTAAATACTTCATTTTTTGATTTTTCAGGAGGTGGGGACCCAATTTTATTCCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Rincon Rain Forest, Zompopera, 10.8884, -85.259, 440 meters, caterpillar collection date: 19/iv/2018, wasp eclosion date: 2/v/2018. Depository:
Host data. Pirascca tyriotes (Riodinidae) feeding on Miconia argentea (Melastomataceae).
Caterpillar and holotype voucher codes. 18-SRNP-40675, DHJPAR0062680.
Paratypes
None.
Bracon anniapicadoae , sp. nov.
Diagnostics
BOLD:ACL5572. Consensus barcode. AATTTTATATTTTTTATTTGGCATATGATCAGGTATAATTGGATTATCTATAAGTTTAATTATTCGATTAGAATTAAGAATACCTGGTAGATTATTAGGTAATGATCAAATTTATAATAGTATAGTAACAGCTCATGCTTTAATAATAATTTTTTTTATAGTTATACCAATTATATTAGGAGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGAGCCCCTGATATAGCTTTCCCACGATTAAATAATATAAGTTTTTGATTATTAATTCCTTCTTTAATTTTATTATTATTAAGAAGAATTTTAAATGTAGGGGTAGGAACAGGTTGAACTATATATCCACCTTTATCATCAAATATGGGACATAGAGGTTTATCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCCATTATAGGATCAATTAATTTTATTTCAACTATTTTAAATATACATTTAAAAATATTAAAATTAGATCAATTAACATTATTAATTTGATCAATTTTTATTACAACTATTTTATTATTATTATCATTACCTGTTTTAGCAGGAGCAATTACTATATTATTAACTGATCGAAATTTAAATACTTCTTTTTTTGATTTTTCAGGAGGAGGAGACCCAATTTTATTCCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 3/ix/2012. Depository:
Host data. None,
Holotype voucher code. BIOUG08904-B08..
Paratypes
BIOUG09441-F08, BIOUG09739-C05, BIOUG28764-A07, BIOUG28769-G07, BIOUG29282-F10. Depository:
Etymology
Bracon anniapicadoae is named to honor Annia Picado for her years of dedicated fly and microlepidopteran processing for the development of the former INBio arthropod collection, now in the Museo Nacional de Costa Rica, and now her furthering the BioAlfa program to DNA barcode all of Costa Rican eukaryote biodiversity.
Bracon anniemoriceae , sp. nov.
Diagnostics
BOLD:ADA8088. Consensus barcode. ATTTTATATTTTTTATTTGGTATATGGGCTGGTATAGTTGGTTTATCTATAAGTTTAATTATTCGTTTAGAATTGGGGATACCTGGTAGTTTATTAGGTAATGATCAAATTTATAATAGAATAGTTACAGCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAGTTATATTAGGGGGRTTTGGTAATTGATTAATTCCTTTAATATTAGGGTCGCCTGATATAGCATTTCCTCGTTTAAATAATATAAGATTTTGRTTATTAGTTCCTTCATTAATTTTATTATTATTAAGAAGAATTTTAAATGTAGGAGTAGGAACTGGGTGGACAATATATCCCCCTTTATCTTCAAGTTTAGGTCATAGAGGYTTATCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGGGTTTCTTCAATTATAGGTTCAATAAATTTTATTACTACTATTCTTAATATGCATTTATTAATATTAAAATTAGATCARTTGAGTTTATTGATTTGATCAATTTTTATTACTACTATTTTATTATTATTATCTTTACCTGTTTTAGCAGGTGCTATTACTATATTATTAACAGATCGTAATTTAAATACT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-2, 10.7634, -85.335, 824 meters, Malaise trap, 13/ii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG28680-H11.
Paratypes
BIOUG28680-G08, BIOUG28770-H05. Depository:
Bracon barryhammeli , sp. nov.
Diagnostics
BOLD:ACZ9844. Consensus barcode. TTTTTATTTGGAATTTGATCAGGTTTATTAGGGTTATCAATAAGTTTAATTATTCGTTTAGAATTAGGGACACCTAGAAGTTTAATAATAAATGATCAAATTTATAATAGAATAGTAACATCCCATGCTTTTATTATAATTTTTTTTATAGTAATACCTGTAATATTAGGAGGATTTGGAAATTGACTATTACCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTCATTATACCTTCTTTATTTTTGTTATTAATAAGAAGAATTCTGAATGTAGGGGTTGGGACTGGGTGAACTATATACCCTCCATTATCTAGTTCTTTAGGACATAACGGATTATCGGTAGATTTAGCTATTTTTGCTTTACATATAGCTGGAATATCCTCTATTTTAGGATCAATTAATTTTATTACAACTATTTTTAATATACAAATATTAAATTTAAAATTAGATCAATTAACTTTATTTATTTGATCAATTCTTATTACTACTTTTTTATTATTATTATCTTTACCTGTTTTAGCAGGAGCTATTACCATATTACTTACAGATCGT---------------------------------------------------.
Holotype ♂
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, Malaise trap, 3/viii/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG28152-C12.
Paratypes
None.
Etymology
Bracon barryhammeli is named to honor Barry Hammel for his many years of dedicated curatorial and taxonomic efforts for the development of the former INBio plant collection, now in the Museo Nacional de Costa Rica, and now his furthering the BioAlfa program to DNA barcode all of Costa Rican eukaryote biodiversity, with an emphasis on all Costa Rica’s plant species.
Bracon bernardoespinozai , sp. nov.
Diagnostics
BOLD:ACG5363. Consensus barcode. TGTTTTATATTTTTTATTTGGGATTTGAGCAGGAATAATTGGATTATCAATAAGATTAATTATTCGGTTAGAATTA---GGAATACCAGGAAATTTATTAAATAATGATCAAATTTATAATAGTATGGTTACTTCTCATGCTTTTGTTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGGGCTCCTGATATAGCATTCCCCCGTCTAAATAATATAAGATTTTGGTTAATTATTCCTTCAATAATTTTATTATTATTAAGAAGAGTTGTAAATGTAGGTGTAGGTACAGGATGAACAATTTATCCACCTTTATCTTCTAATTTAGGTCACAGAGGGGTTTCAGTAGATATAGCAATTTTTTCTTTACATTTAGCTGGGGTTTCTTCTATTTTAGGGGCAATTAATTTTATTACAACAATTTTAAATATACATTTAAACATTATAAAATTAGATCAATTAACTTTATTAATCTGGTCAATTTTTATTACAACAATTTTATTGCTTTTATCTTTACCAGTATTAGCAGGTGCAATTACTATATTATTAACAGATCGAAATTTAAATACATCTTTTTT-------------------------------------------------------------------.
Holotype ♂
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 30/vii/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG05883-A04.
Paratypes
None.
Etymology
Bracon bernardoespinozai recognizes Bernardo Espinoza for his many years of dedicated curatorial and taxonomic efforts for the enhancement of the former INBio arthropod collection, now in the Museo Nacional de Costa Rica, and now his furthering the BioAlfa program to DNA barcode all of Costa Rican eukaryote biodiversity with an emphasis on Arctiinae.
Bracon carlossanabriai , sp. nov.
Diagnostics
BOLD:ADA3386. Consensus barcode. GTTTTATATTTTTTATTTGGGATATGAGCTGGTATAGTAGGTTTATCTATAAGATTAATCATTCGTTTAGAATTAGGTATACCTGGTAGTTTACTAGGTAATGATCAAATTTATAATAGAATAGTTACAGCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAGTAATAATTGGTGGATTTGGGAATTGATTAATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTTCCACGTTTAAATAATATAAGATTTTGGTTATTAATTCCTTCATTAATTTTATTATTATTAAGAAGAATTTTAAATGTAGGTGTAGGTACTGGTTGAACAATATACCCTCCATTATCTTCAAGATTAGGACATAGGGGTTTATCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCAGGGGTTTCTTCAATTATAGGAGCAATAAATTTTATTACAACTATTTTAAATATACATTTATTAATATTAAAATTAGATCAATTAACTTTATTAATTTGATCTATTTTTATTACAACAATTTTATTATTATTATCTTTACCTGTTTTAGCAGGAGCTATTACTATA---------------------------------------------------------.
Holotype ♂
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, Malaise trap, 14/iv/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG28246-E07.
Paratypes
None.
Etymology
Bracon carlossanabriai is named to honor Carlos Sanabria for his many years of dedicated efforts on behalf of Costa Rica’s Phytosanitary Service (Servicio Fitosanitario del Estado, or SFE), which in turn supports Costa Rica’s new BioAlfa program to DNA barcode the country.
Bracon chanchini , sp. nov.
Diagnostics
BOLD:ACR7966. Consensus barcode. ATTTTATATTTTTTATTTGGAATATGAGCTGGAATATTAGGTATATCTATAAGTTTAATTATTCGATTAGAATTAGGTATACCAGGTAGTTTATTGGGTAATGATCAAATTTATAACAGTATAGTTACTGCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGGTTTGGGAATTGATTATTACCTTTAATATTAGGAGCCCCTGATATAGCATTCCCTCGTTTGAATAATATAAGGTTTTGATTAATTATCCCTTCTTTAATTTTATTATTAATAAGAAGAATTTTAAATGTAGGTGTTGGAACTGGATGAACAGTATACCCTCCTTTATCTTCTTCTTTAGGACATGGAGGATTATCTATAGATTTAGCTATTTTTTCTTTACATATGGCTGGAATTTCATCTATTTTAGGTGCAATTAATTTTATTACAACTATTTTAAATATGCATTTATTTATTTTGAAGTTGGATCAGTTAACTTTATTAATTTGATCTATTTTTATTACAGTAATTTTATTATTATTATCTTTACCAGTTTTAGCTGGAGCAATCACTATATTATTAACTGAT---------------------------------------------.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 11/vi/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG17967-B03.
Paratypes
None.
Bracon christophervallei , sp. nov.
Diagnostics
BOLD:ADB0826. Consensus barcode. GTTTTATATTTTTTATTTGGTATATGAGCTGGGATAATTGGTTTATCAATAAGTTTAATTATTCGTTTAGAATTAGGCATACCAGGATCTTTATTAAGAAATGATCAAATTTATAATAGAATAGTTACAGCTCATGCTTTTGTTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGTTCTCCAGATATAGCTTTCCCTCGTTTAAATAATATGAGATTTTGATTAATTATTCCTGCAATAATTTTATTATTATTAAGGAGAATTTTAAATGTAGGTGTAGGTACTGGTTGAACAATATACCCACCTTTATCTTCTTCATTAGGTCATAGAGGAATTTCAGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGAGTTTCATCTATTTTAGGTTCAATTAATTTTATTACTACCATTTTAAATATACATTTAAATATTTTAAAGATAGATCAATTAACTTTATTAGTTTGATCAATTTTTATTACAACAATTTTATTACTTTTATCTTTACCTGTTTTAGCAGGT---------------------------------------------------------.
Holotype ♂
Guanacaste, Sector Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, Malaise trap, 20/iii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29096-E05.
Paratypes
None.
Bracon erasmocoronadoi , sp. nov.
Diagnostics
BOLD:ADH0668. Consensus barcode. TTTTTATTTGGAATATGAGCCGGAATATTAGGAATATCTATAAGTTTAATTATTCGGTTAGAATTAGGGATACCAGGTAGTTTATTGGGTAATGATCAAATTTATAATAGAATAGTTACTGCTCATGCTTTTGTAATGATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTACTACCTTTAATATTAGGTGCTCCTGATATAGCATTTCCTCGTTTAAATAATATAAGATTTTGATTAATTATTCCTTCTTTAATTTTATTATTAATAAGAAGAATTTTAAATGTAGGTGTTGGAACTGGTTGAACAGTTTACCCTCCCTTATCTTCTTCTTTAGGTCATAGAGGGCTATCTGTAGATTTAGCTATTTTTTCTTTACATATAGCTGGTATCTCCTCTATTTTAGGAGCAATTAATTTTATCACAACTATTTTAAATATACATTTATTTATTTTAAAATTAGATCAATTGACTTTATTAATTTGATCAATTTTTATCACAGTAATTTTATTACTATTATCTTTACCAGTTTTAGCTGGA-------------------------------------------------------------------.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 26/ii/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG32860-H09.
Paratypes
None.
Bracon eugeniephillipsae , sp. nov.
Diagnostics
BOLD:ADA9990. Consensus barcode. GTTTTATATTTTTTATTTGGTTTATGATCAGGAATTTTAGGTCTATCAATAAGTTTAATTATTCGATTAGAATTAGGTATACCAGGCAGGATATTAGGTAATGATCAAATTTATAATAGTATTGTAACTGCTCATGCTTTTGTTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGGTTATTGCCTTTAATATTAGGAGCTCCTGATATGGCATTYCCTCGTTTAAATAATATAAGATTTTGATTAATTTTCCCTTCTTTAATTTTATTATTAATAAGTAGGATTTTAAATGTAGGAGCAGGTACAGGTTGGACAGTTTATCCTCCTTTATCTTCTTCATTAGGACATAGAGGTTTATCAGTTGATTTAGCTATTTTTTCTTTACATATAGCTGGTGTATCTTCAATTTTAGGAGCAATTAATTTTATCACTACAATTTTAAATATGCATTTAAATACTTTAAARTTAGATCAATTAACTTTAATAATTTGATCAATTTTTATTACTGTAATTTTATTATTGTTATCTTTACCAGTTTTAGCAGGGGCTATTACTATATTATTAACTGATCGA.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, Malaise trap, 6/ii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG28680-G05.
Paratypes
BIOUG28680-A07, BIOUG28680-E08, BIOUG29219-D01.
Etymology
Bracon eugeniephillipsae is named to honor Eugenie (Jenny) Phillips for her many years of dedicated curatorial and taxonomic efforts for the development of the former INBio arthropod collection, now in the Museo Nacional de Costa Rica, and now her administrative development of BioAlfa, the program to DNA barcode all of Costa Rican eukaryote biodiversity.
Bracon federicomatarritai , sp. nov.
Diagnostics
BOLD:ACM9419. Consensus barcode. TGTCTTATATTTTTTATTTGGTTTATGAGCTGGAATATTAGGTTTATCTATAAGTTTAATTATTCGATTAGAATTAGGTATACCTGGTAGATTATTAGGTAATGACCAAATTTATAATAGTATAGTTACAGCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAGTAATATTAGGAGGATTTGGAAATTGGTTAATTCCTTTAATATTAGGGGCCCCAGATATAGCTTTCCCACGTTTAAATAACATAAGATTTTGATTACTAATTCCTTCATTAATTTTATTATTATTAAGAAGAATTTTAAATATTGGGGTTGGTACAGGTTGGACAATATACCCCCCATTATCATCAAATTTAGGACATAGAGGGATATCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATAGGGTCAATAAATTTTATTACAACTATTTTAAATATACATTTAATTACAATAAAACTAGATCAACTAACTTTATTAGTTTGATCAATTTTTATTACAACTATTTTACTATTATTATCTCTACCTGTTTTAGCAGGGGCTATTACTATACTTTTAACAGATCGTAATTTAAATACTTCTTTTTTCGATTTTTCAGGAGGAGGGGACCCTATTTTATTCCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Sendero Anonas, 10.90527, -85.27881, 405 meters, caterpillar collection date: 18/iii/2014, wasp eclosion date: 30/iii/2014, 2 wasps eclosed from 2 cocoons. Depository:
Host data. Gregarious parasitoid of Tebenna Janzen02 (Choreutidae) feeding on Ficus citrifolia (Moraceae).
Caterpillar and holotype voucher codes. 14-SRNP-41359, DHJPAR0055286.
Paratype: One specimen same data as holotype. Depository:
Bracon frankjoycei , sp. nov.
Diagnostics
BOLD:ACJ2744. Consensus barcode. TATTTTATATTTTTTATTTGGTATATGAGCAGGTATAGTAGGTTTATCTATAAGATTAATTATTCGTTTAGAATTAGGTATACCT---GGGAGTTTATTAGGTAATGATCAAATTTATAATAGTATAGTTACAGCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCTATTATATTAGGTGGGTTTGGAAATTGGTTAATTCCTTTAATATTAGGAGCCCCTGATATAGCTTTCCCTCGATTAAATAATATGAGATTTTGATTATTAATCCCTTCATTAATTTTATTATTATTAAGAAGAATTTTAAATGTTGGTGTAGGTACTGGGTGAACAATATACCCTCCATTATCTTCAAGATTAGGCCATAGAGGTTTATCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTGTTTCTTCAATTATAGGGTCAATAAATTTTATTACTACAATTTTAAATATACATTTATTAATGTTAAAAATAGATCAATTAACTTTATTAATTTGATCAATTTTTATTACTACTATTTTATTATTATTATCTTTACCAGTATTAGCAGGGGCTATTACAATATTATTAACTGATCGTAACTTAAATACTTCTTTTTTTGACTTTTCTGGCGGAGGGG--------------------------.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 9/iv/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG07560-C12.
Paratypes
None.
Bracon gerardovegai , sp. nov.
Diagnostics
BOLD:ACW3155. Consensus barcode. TTATTAGGGATTTGATCTGGAATTTTAGGATTATCTATAAGATTAATTATTCGGTTAGAACTAGGAACTTCAGGATTTTTATTAGGTAATGATCAAATTTATAATAGATTAGTAACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATACTAGGAGGATTTGGAAATTGATTAATCCCTTTAATATTGGGGGCTCCTGATATAGCATTCCCTCGAATAAATAATATAAGGTTTTGACTTCTTATTCCTTCATTAATATTATTAATTTTAAGAAGAATCTTAAATGTAGGAGTTGGCACAGGGTGAACAATATATCCTCCTTTATCTTCTTCTATAGGTCATAGAGGATTATCTACAGATTTAGCAATTTTTTCTTTACATTTAGCAGGAGCATCCTCAATTTTAGGAGCTATTAATTTTATTACAACAATTTTTAATATAAAATTAAATTCAATAAAATTAGATCAATTAACTTTATTAATTTGATCTATTTTAATTACTACAATTTTATTATTATTATCTTTACCTGTGTTAGCTGGAGCTATTACTATATTATTAACGGAT.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, Malaise trap, 2/xii/2013. Depository:
Host data. None.
Holotype voucher code. BIOUG22966-G03.
Paratype
BIOUG27938-F06. Depository:
Bracon germanvegai , sp. nov.
Diagnostics
BOLD:ADL8270. Consensus barcode. TTATTAGGGATTTGATCTGGAATTTTAGGATTATCTATAAGATTAATTATTCGGTTAGAACTAGGAACTTCAGGATTTTTATTAGGTAATGATCAAATTTATAATAGATTAGTAACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATACTAGGAGGATTTGGAAATTGATTAATCCCTTTAATATTGGGGGCTCCTGATATAGCATTCCCTCGAATAAATAATATAAGGTTTTGACTTCTTATTCCTTCATTAATATTATTAATTTTAAGAAGAATCTTAAATGTAGGAGTTGGCACAGGGTGAACAATATATCCTCCTTTATCTTCTTCTATAGGTCATAGAGGATTATCTACAGATTTAGCAATTTTTTCTTTACATTTAGCAGGAGCATCCTCAATTTTAGGAGCTATTAATTTTATTACAACAATTTTTAATATAAAATTAAATTCAATAAAATTAGATCAATTAACTTTATTAATTTGATCTATTTTAATTACTACAATTTTATTATTATTATCTTTACCTGTGTTAGCTGGAGCTATTACTATATTATTAACGGAT.
Holotype ♂
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 4/vi/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG36580-A08.
Paratypes
None.
Bracon isidrochaconi , sp. nov.
Diagnostics
BOLD:ADA7614. Consensus barcode. GTTTTATATTTTTTATTTGGAATATGAGCTGGTATAGTTGGTTTATCAATAAGAATAATTATTCGTTTAGAATTAGGTATACCTGGATTTTTATTAATAAATGATCAAATTTATAATAGAATAGTAACTGCTCATGCTTTTGTTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGGAATTGATTAGTTCCTTTAATATTAGGTTCTCCAGATATAGCTTTCCCTCGTTTAAATAATATAAGATTTTGATTAATTATCCCAGCAATAATTTTATTATTATTAAGAAGAATTTTAAATGTTGGTGTAGGTACAGGTTGAACAATTTACCCTCCTTTATCTTCTTCTTTAGGACATAGAGGAATTTCAGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCATCTATTTTAGGTTCAATTAATTTTATTACAACAATTTTAAATATACATTTAAATGTATTAAAGATAGATCAATTAACTTTATTTGTTTGGTCAATTTTTATTACTACAATTTTATTATTATTATCTTTACCTGTTTTAGCTGGAGCTATTACAATATTA------------------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, Malaise trap, 23/i/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG28721-H09.
Paratypes
None.
Etymology
Bracon isidrochaconi is named to honor Isidro Chacon for his many years of dedicated curatorial and taxonomic efforts for the advancement of the former INBio arthropod collection, now in the Museo Nacional de Costa Rica, and now his furthering the BioAlfa program to DNA barcode all of Costa Rican eukaryote biodiversity with an emphasis on Notodontidae, butterflies, Bombycoidea, and Noctuoidea.
Bracon jimlewisi , sp. nov.
Diagnostics
BOLD:ADL5861. Consensus barcode. TGTTTTATATTTTTTATTTGGGGTTTGATCTGGATTTTTAGGTTTATCTATAAGATTAATTATTCGTATAGAATTGAGTATACCAGGAAGATTATTAAGTAATGATCAAATTTATAATAGTTTAGTAACTGCTCATGCTTTTGTTATAATTTTTTTTATAGTTATACCAGTAATAATTGGAGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTCCCTCGATTAAATAATATAAGATTTTGGTTAATTATTCCTTCATTAATACTATTAATTTTAAGAAGAATTTTAAATGTTGGGGTAGGTACAGGTTGAACAATATACCCCCCTTTATCTTCTTCTTTAGGGCATAGAGGTAATTCAACTGATTTAGCTATTTTTTCTTTACATATAGCTGGAATTTCTTCAATTTTAGGGGCTATTAATTTTATTACAACTATTTTTAATATAAAATTATTTTTTTTAAAATTTGATCAATTAACTTTATTTATTTGATCAATTTTAATTACAACAATTTTATTATTGTTATCTTTACCAGTTTTAGCTGGGGCAATCACAATA---------------------------------------------.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 9/vii/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG36967-F06.
Paratypes
None.
Etymology
Bracon jimlewisi is named to honor Jim Lewis for his many years of dedicated volunteer curatorial and taxonomic efforts for the construction of the former INBio arthropod collection, now in the Museo Nacional de Costa Rica, and now his furthering the BioAlfa program to DNA barcode all of Costa Rican eukaryote biodiversity, with an emphasis on Hemiptera.
Bracon josejaramilloi , sp. nov.
Diagnostics
BOLD:ACK7911. Consensus barcode. TATTTTATATTTTTTATTTGGTATATGAGCTGGTATGTTAGGTTTATCAATAAGAATAATTATTCGGTTAGAATTAGGTATACCAGGAAGATTACTAGGTAATGATCAAATTTATAATAGTATAGTTACTGCTCATGCATTTGTTATAATTTTTTTTATAGTTATACCAATTATATTAGGAGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGATTAATTATTCCTTCTTTAGTTTTATTATTATTAAGAAGAATTTTAAATATTGGGGCAGGAACAGGATGAACTATATATCCTCCTTTATCTTCTCATTTAGGTCATAGAGGTATATCAGTTGATTTAGCTATTTTTTCTTTACATATAGCAGGGATTTCATCAATTTTAGGATCTATTAATTTTATTACAACAATTTTAAATATACATTTATTTACTTTAAAATTAGATCAATTAACTTTATTAGTTTGATCAATATTTATTACTACAATTTTATTATTATTATCTTTACCAGTTTTAGCAGGGGGTATTACAATATTATTAACAGACCGTAAYCTAAATACTTCATTTTTTGATTTTTCTGGAGGAGGAGATCCTATTTTATTCCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Pasmompa, 440 meters, 11.01926, -85.40997, caterpillar collection date: 21/ii/2012, wasp eclosion date: 08/iii/2012, 11 wasps eclosed from 15 cocoons. Depository:
Host data. Gregarious parasitoid (Fig.
Caterpillar and holotype voucher codes. 12-SRNP-30473, DHJPAR0049094.
Paratypes
Hosts = Lerema liris and Morys valerius (Hesperiidae). DHJPAR0049094, DHJPAR0028977, DHJPAR0058913.
Etymology
Bracon josejaramilloi is named in honor of José Jaramillo of Área de Conservación Guanacaste in recognition of his 30 years of managing ACG mechanics, telecommunication, computer, and internet processing for ACG and its web site.
Bracon josejaramilloi (12-SRNP-30473) ultimate instar wasp larvae below their host ultimate instar caterpillar Lerema liris (Hesperiidae); as a group they will spin a roof-in-common followed by their individual closely packed cocoons below that roof, adjacent to the cadaver of the caterpillar.
Bracon juanjoseoviedoi , sp. nov.
Diagnostics
BOLD:ADB0539. Consensus barcode. ATTTTATATTTTTTATTTGGTATATGAGCTGGAATATTAGGTTTATCAATAAGATTAATTATTCGATTAGAATTAGGAATACCAGGAAGATTATTAGGAAATGATCAAATTTATAATAGAATAGTGACAGCTCATGCATTTGTTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTATTACCTTTAATATTAGGGGCTCCTGATATAGCTTTTCCTCGTCTTAATAATATAAGATTTTGATTAATTATTCCGGCTTTAATTTTATTACTAATAAGAAGAATTTTAAATGTAGGTGTAGGTACTGGTTGAACAGTTTATCCTCCTTTATCTTCTTCTTTAGGTCATAGAGGTTTATCTGTTGATTTAGCTATTTTTTCTTTACATATTGCTGGTATTTCTTCAATTTTAGGGGCAATTAATTTTATTACAACAATTTTAAATATACATTTATATAAATTAAAATTAGATCAATTAACTTTATTAACTTGATCAATTTTTATTACAGTAATTCTTTTACTTTTATCTTTACCAGTTTTAGCTGGAGCTATTACTATACTTTTAACTGATCGA------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-2, 10.7634, -85.335, 824 meters, Malaise trap, 13/ii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG28680-H09.
Paratypes
None.
Etymology
Bracon juliodiazi , sp. nov.
Diagnostics
BOLD:ACG3370. Consensus barcode. TATTTTATATTTTTTATTCGGAATATGAGCAGGTATATTAGGATTATCTATAAGAATAATTATTCGATTAGAATTAGGRATACCTGGGACTTTATTAGGCAATGATCAAATTTATAATAGAATAGTGACAGCTCATGCATTTGTTATAATTTTTTTTATAGTTATACCAATTATACTAGGGGGTTTCGGTAATTGATTAATTCCTTTAATATTAGGAGCCCCAGACATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTAATTATTCCTTCTTTAATTTTATTATTATTAAGAAGAATTTTAAATGTAGGGGTAGGTACTGGATGAACTGTTTATCCTCCCTTATCTTCTTCTTTAGGTCATAGAGGCATATCTGTTGATTTAGCAATTTTTTCATTACATTTAGCAGGTATTTCTTCAATTTTAGGTTCAATTAATTTTATTACAACTATTTTAAATATAAAATTACATACAATAAAATTAGATCAATTAACTTTATTAATTTGATCAATTTTTATTACTACAATTTTATTATTATTATCTTTACCAGTTYTAGCGGGAGCTATTACAATATTATTAACTGATCGAAATTTAAATACTTCATTTTTTGATTTTTCTGGGGGAGGTGACCCTATTTTATTCCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 29/x/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG09074-C11.
Paratypes
BIOUG09074-B05, BIOUG09074-B02. Depository:
Bracon luzmariaromeroae , sp. nov.
Diagnostics
BOLD:ADF0526. Consensus barcode. AATTTTATATTTTTTATTTGGAATATGATCTGGAATAATTGGTTTATCTATAAGTTTAATTATTCGTTTAGAATTAGGAATACCTGGAAGATTATTGAGAAATGACCAAATTTATAATAGAATAGTTACAGCTCATGCTTTAGTAATAATTTTTTTTATAGTTATACCAATTATATTAGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGATTAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATTTTATTATTATTAAGAAGAATTTTAAATGTTGGTGTTGGAACAGGATGAACTTTATACCCTCCTTTATCTTCAAATTTAGGACATAATGGAGTATCTGTAGATTTATCTATTTTTTCTTTACATTTAGCTGGTATTTCATCAATTATAGGTTCATTAAATTTTATTACTACTATTTTAAATATACATTTATTAATATTAAAATTAGATCAATTAACTTTATTAATTTGATCAATTTTTATTACAACTATTTTATTATTATTATCTTTACCTGTTTTAGCAGGAGCTATTACTATATTATTAACTGATCGTAATTTAAATACTTCATTTTTTGATTTTTCAGGAGGTGGTGACCCAATTCTATTTCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Casa Keyner, 10.95644, -85.26611, 121 meters, caterpillar collection date: 13/viii/2016, wasp eclosion date: 23/viii/2016, six wasps eclosed from six cocoons. Depository:
Host data. Gregarious parasitoid of Vettius aurelius (Hesperiidae) feeding on mature leaves of Lasiacis sorghoidea (Poaceae).
Caterpillar and holotype voucher codes. 16-SRNP-46268, DHJPAR0060193.
Paratypes
Five specimens, same data as holotype but still coded with their caterpillar code 16-SRNP-46268 (DHJPAR codes not assigned). Depository:
Etymology
Bracon manuelzumbadoi , sp. nov.
Diagnostics
BOLD:ADH5864. Consensus barcode. GTGTTTTATATTTTTTGTTTGGTATATGGTCTGGGATATTAGGGATATCTATAAGATTAATTATTCGATTAGAATTAGGAATACCCGGTAGATTATTAGGGAATGATCAACTTTATAATAGTATGGTAACAGCTCATGCTTTTGTAATAATTTTTTTTATAGTAATACCTGTAATAATTGGGGGTTTTGGCAATTGATTATTACCTTTAATATTAGGGTCCCCTGATATAGCTTTCCCTCGTCTTAATAATATAAGATTTTGATTGATTATTCCTTCATTAATTTTGTTATTAATAAGAAGAATTTTGAATGTAGGTGTAGGTACTGGTTGAACTGTATATCCTCCTTTATCTTCTTCTTTAGGACATAGGGGGATATCTGTTGATTTAGCTATTTTTTCTTTACATATTGCTGGAGTATCCTCAATTTTAGGAGCAATTAATTTTATTAGAACTATTTTAAATATACATTTATTTATATTAAAATTGGATCAATTAACTTTGTTAATTTGATCAATTTTTATTACTGTTATTTTATTATTATTATCTTTACCAGTTTTAGCTGGTGCAATTACTATATTATTAACTGAT.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 2/iv/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG33120-A12.
Paratypes
None.
Etymology
Bracon manuelzumbadoi is named to honor Manuel Zumbado for his many years of dedicated curatorial and taxonomic efforts for the construction of the former INBio arthropod collection, now in the Museo Nacional de Costa Rica, and now his furthering the BioAlfa program to DNA barcode all of Costa Rican eukaryote biodiversity, with an emphasis on Diptera.
Bracon marialuisariasae , sp. nov.
Diagnostics
BOLD:ADA9695. Consensus barcode. GTTTTATATTTTTTGTTTGGGATATGATCTGGTATAATTGGTTTATCAATAAGTTTAATTATTCGTTTAGAATTGGGTATACCAGGTAGATTATTAGGCAATGATCAGATTTATAATAGTATAGTTACTGCTCATGCTTTTGTTATAATTTTTTTTATAGTTATGCCTGTAATATTAGGAGGGTTTGGGAATTGATTAATTCCTTTAATGTTGGGTGCTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGGTTTTGGTTATTAATTCCTTCATTGGTTTTATTATTACTAAGAAGAATTTTAAATGTTGGTGTTGGGACTGGGTGAACTATATACCCTCCTTTATCTTCAAGATTAGGTCATAGTGGTTTGTCAGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGGGTATCTTCAATTATAGGGGCAATTAATTTTATTACAACTATTTTAAATATACATTTATTTATATTAAAATTAGATCAATTAACTTTGTTAATTTGATCAATTTTTATCACAACTATTTTATTATTATTATCATTACCTGTTTTAGCTGGAGCTATTACTATGTTATTAACTGATCGTAAT---------------------------.
Holotype ♂
Guanacaste, Sector Pailas Dos, PL12-2, 10.7634, -85.335, 824 meters, Malaise trap, 23/i/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG28680-C06.
Paratypes
None.
Bracon mariamartachavarriae , sp. nov.
Diagnostics
BOLD:AAV6295. Consensus barcode. AATTTTATATTTCTTATTTGGYATATGAGCAGGTATAGTAGGYYTATCAATAAGTTTAATTATTCGATTAGAATTAGGKATACCTGGRTCATTATTAGGWAATGATCAAATTTATAATAGAATAGTKACAGCTCATGCTYTARTWATAATTTTTTTTATAGTTATACCAGTAATATTAGGAGGTTTTGGTAATTGATTAATTCCTTTAATATTAGGWGCTCCTGATATAGCYTTCCCTCGATTAAATAATATAAGTTTTTGRTTAYTAATYCCTTCATTAATTTTATTAATATTAAGAAGAATTTTAAATGTAGGTGTAGGTACAGGTTGAACTTTATAYCCTCCATTATCYTCAAATTTAGGACATAGAGGDTTATCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTGTATCTTCAATTATAGGTTCAATAAATTTTATTACTACTATTTTAAATATACATTTATTAATATTAAAATTAGATCAATTAACTTTATTAATTTGATCAATTTTTATTACAACTATTTTATTATTATTATCTTTACCYGTWTTAGCAGGWGCAATTACAATATTATTAACTGATCGAAATTTAAATACTTCATTTTTTGATTTTTCAGGWGGAGGAGAYCCAATTYTATTYCAACATTTATTT. Terminal flagellomeres black, concolorous with basal flagellomeres.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Selva, 10.92291, -85.31877, 410 meters, caterpillar collection date: 05/viii/2017, wasp eclosion date: 21/viii/2017; nine wasps eclosed from 11 cocoons. Depository:
Host data. Gregarious parasitoid of Drephalys Burns01 (Hesperiidae) feeding on mature leaves of Vochysia guatemalensis (Vochysiaceae).
Caterpillar and holotype voucher codes. 17-SRNP-80872, DHJPAR0061668.
Paratypes
Eight specimens, same data as holotype. Depository:
Other specimens
There are three specimens from French Guiana (e.g., ASNUR330-11) and one from Argentina (BIOUG13458-C11) in the same BIN, none of which we consider to be conspecific with the Costa Rican specimens. Images on BOLD show that the French Guiana specimens have the terminal flagellomeres yellow. There are no images for the Argentinian specimen, but its locality makes it unlikely to be conspecific.
Etymology
Bracon mariamartachavarriae is named in honor of Maria Marta Chavarria in recognition of her persistent efforts as co-coordinator of ACG research; intense education efforts to adults, teenagers, and schoolchildren about ACG biology both terrestrial and marine; maintenance and data management of ACG weather data; ACG biopolitics; and rescue of key ACG animals.
Bracon mariorivasi , sp. nov.
Diagnostics
BOLD:ADA3193. Consensus barcode. GTATTATATTTTTTATTTGGTATATGATCTGGCATTTTAGGTTTATCAATAAGTTTAATAATTCGATTGGAATTGGGGACGCCAGGTAGATTGTTAGGTAATGATCAAATTTATAATAGAATGGTGACAGCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAGTTATAGTTGGAGGTTTTGGAAATTGATTATTACCTTTAATATTAGGATCTCCAGATATAGCATTTCCTCGATTAAATAATATAAGGTTTTGACTAATTATTCCTTCTTTAATTCTTTTATTAATAAGAAGGATTTTAAATGTAGGAGTTGGTACGGGATGAACAGTTTATCCTCCTTTATCTTCTTCTTTAGGTCATAGAGGTTTATCTATAGATTTAGCTATTTTTTCTCTTCATATAGCAGGAATTTCTTCAATTTTAGGTGCTATTAATTTTATTTCTACAATTTTTAATATACATTTATATAATTTAAAATTAGATCAATTAGTTTTATTAATTTGGTCTATTTTTATTACTGCTGTTTTATTATTGTTGTCATTACCTGTTTTGGCAGGGGCTATTACAATACTTTTAACAGAT------------------------------------------.
Holotype ♂
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, Malaise trap, 17/ii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG28290-E07.
Paratypes
None.
Bracon melissaespinozae , sp. nov.
Diagnostics
BOLD:ACJ4722. Consensus barcode. AATTTTATATTTTTTATTTGGGATATGGTCTGGAATAATTGGTTTATCTATAAGTTTAATTATTCGATTAGAATTGAGAATACCTGGTAGATTATTAGGTAATGATCAAATTTATAATAGTATAGTAACAGCTCATGCTTTAATAATAATTTTTTTTATAGTTATACCAATTATATTAGGAGGTTTTGGGAATTGATTAATTCCTTTAATGTTAGGAGCTCCTGATATAGCTTTCCCACGTTTAAATAATATAAGTTTTTGGTTATTAATCCCCTCTTTAATTTTATTATTATTAAGAAGAATTTTAAATGTAGGGGTAGGAACAGGTTGAACTATATATCCACCTTTATCATCAAATATAGGACATAGAGGTTTATCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTATAGGATCAATTAATTTTATTTCAACTATTTTAAATATACATTTAAAAATATTAAAATTAGATCAATTAACATTATTAATTTGATCAATTTTTATTACAACTATTTTATTATTGTTATCATTACCTGTTTTAGCGGGAGCAATTACTATATTATTAACTGATCGAAATTTAAATACTTCTTTTTTTGATTTTTCAGGAGGAGGTGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Maria, Light Site Trail, Volcán Santa Maria, 10.75772, -85.30598, 840 meters, caterpillar collection date: 29/viii/2012, wasp eclosion date: 10/ix/2012, 16 wasps eclosed. Depository:
Host data. Gregarious parasitoid of unknown caterpillar in leaf nest on Smilax spinosa (Smilacaeae).
Caterpillar and holotype voucher codes. 12-SRNP-13169, DHJPAR0051204.
Paratypes
15 specimens, same data as holotype, still labelled with 12-SRNP-13169. Depository:
Etymology
Bracon melissaespinozae is named in honor of Melissa Espinoza’s proactive and growing abilities as a science reporter for all things ACG for the ACG web site (www.acguanacaste.ac.cr).
Bracon nelsonzamorai , sp. nov.
Diagnostics
BOLD:ADB4596. Consensus barcode. ATTTTATATTTTTTATTTGGGATATGAGCTGGGATAATTGGTTTATCAATAAGTTTAATTATTCGATTAGAATTAGGGATACCTGGAAGTTTATTAGGTAATGACCAAATTTATAATAGTATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTAATATTAGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGTTTAAATAATATAAGATTTTGATTATTAATCCCTTCTTTAATTTTATTAATTTTGAGAAGAATTTTAAATATTGGGGTAGGCACAGGCTGAACTATATACCCCCCTTTATCGTCTAGATTAGGTCATAGAGGAGTATCTGTTGATTTAGCAATTTTTTCATTACATTTAGCAGGAATCTCTTCTATTATAGGAGCAATAAATTTTATTACCACTATTTTAAATATACATTTATTAATATTAAAATTAGACCAATTAACCTTATTAATCTGATCAATTTTTATTACAACTATTTTATTATTACTATCTTTACCAGTATTAGCTGGAGCTATCACTATACTATTAACAGATCGAAACCTAAAC---------------------------------.
Holotype ♂
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 22/v/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29002-G10.
Paratypes
None.
Etymology
Bracon nelsonzamorai is named to honor Nelson Zamora for his many years of dedicated curatorial and taxonomic efforts for the growth of the former INBio plant collection, now in the Museo Nacional de Costa Rica, and his furthering the BioAlfa program to DNA barcode all of Costa Rican eukaryote biodiversity, with an emphasis on all of Costa Rica’s plant species.
Bracon nicklaphami , sp. nov.
Diagnostics
BOLD:ACP6341. Consensus barcode. AATTTTATATTTTTTATTTGGTATATGATCTGGTATAATTGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAGGTATACCTGGAAGATTATTAAGAAATGATCAAATTTATAATAGAATAGTCACAGCCCATGCTTTAGTAATGATTTTTTTTATAGTTATACCAATTATATTAGGAGGTTTTGGAAATTGGTTAATTCCTTTAATATTAGGTGCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATTTTGTTATTATTAAGAAGAATTTTAAATGTAGGTGTAGGAACAGGATGGACATTATATCCTCCTTTATCTTCAAATTTAGGTCATAGAGGTTTATCTGTGGATTTATCTATTTTTTCTATTCATTTAGCTGGAATTTCATCAATTATAGGTTCATTAAATTTTATTACTACAATTTTAAATATACATTTATTAATATTAAAATTAGATCAATTAACTTTATTAATTTGATCAATTTTTATTACAACTATTTTATTATTATTATCTTTACCTGTTTTGGCAGGAGCTATTACTATATTATTAACTGATCGTAATTTAAATACTTCATTTTTTGATTTTTCAGGAGGTGGGGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Palomo, 10.96187, -85.28045, 96 meters, caterpillar collection date: 24/iv/2014, wasp eclosion date: 4/v/2014. Depository:
Host data. Damas immacula (Hesperiidae) feeding on Geonoma congesta (Arecaceae).
Holotype voucher code. DHJPAR0055350.
Paratypes
None.
Bracon ninamasisae , sp. nov.
Diagnostics
BOLD:ADA4862. Consensus barcode. ATTTTATATTTTTTATTTGGAATATGATCAGGAATAATTGGTCTATCAATAAGATTAATTATTCGGTTAGAATTAAGAATACCTGGTAGATTATTAGGTAATGACCAAATTTATAATAGTATAGTAACAGCTCATGCTTTAATAATAATTTTTTTTATAGTTATACCAATTATATTAGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGTTTAAATAATATAAGTTTTTGACTATTAATTCCTTCTTTAATTTTATTATTATTAAGAAGAATTTTGAATATAGGAGTAGGTACAGGTTGAACTTTATACCCACCTTTATCTTTAAGAATTGGTCATAGAGGTTTATCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTATAGGATCAATTAATTTTATTTCAACTATTTTAAATATACATTTAAAATCATTAAAATTAGATCAATTAACATTATTAATTTGATCAATTTTTATTACAACTATTTTATTATTATTATCATTACCTGTTTTAGCAGGAGCAATTACTATA---------------------------------------------------------.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, Malaise trap, 9/xii/2013. Depository:
Host data. None.
Holotype voucher code. BIOUG28011-B01.
Paratypes
None.
Etymology
Bracon ninamasisae is named to honor Nina Masis for her cheerful participation in the lives and dedication of the Masis + Boshart family as it carried out its vital role in the growth and management of the Guanacaste Dry Forest Conservation Fund and Área de Conservación Guanacaste.
Bracon oliverwalshi , sp. nov.
Diagnostics
BOLD:ABV9910. Consensus barcode. AATTCTTTATTTTTTATTTGGTATATGAGCAGGAATATTAGGTTTATCAATAAGATTAATTATTCGTTTAGAACTAGGAATACCAGGTAGATTATTAGGGAATGACCAAATTTATAATAGAATAGTTACAGCTCATGCTTTTGTTATAATTTTTTTTATAGTTATACCAGTAATAGTTGGAGGGTTTGGGAATTGATTATTACCTTTAATATTAAGGGCCCCTGATATAGCTTTCCCACGTTTAAATAATATAAGATTTTGGTTATTAATTCCTTCTTTATTTTTATTATTAATAAGAAGAGTATTAAATGTAGGTGTTGGTACTGGATGAACAATATATCCTCCTTTGTCTTCTTCTTTAGGTCATAGGGGTATATCAGTTGATTTAGCTATTTTTTCTTTACATATTGCGGGTATTTCATCAATTTTAGGGGCTATAAATTTTATTTCAACTATTTTTAATATACATTTATTAACTTTAAAATTAGATCAATTAACTTTATTTATTTGATCAATTTTTATTACAACTTTGTTATTATTATTATCTTTACCAGTATTAGCTGGGGCTATTACCATATTATTAACTGATCGAAATTTAAATACTTCTTTTTTTGATTTTTCTGGTGGAGGTGACCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Cacao, Sendero Cima, 10.933, -85.457, 1460 meters, Malaise trap, 9/iii/2009. Depository:
Host data. None.
Holotype voucher code. DHJPAR0046065.
Paratypes
None.
Bracon paulamarinae , sp. nov.
Diagnostics
BOLD:ADB2848. Consensus barcode. GTTTTATATTTTTTATTTGGTATATGGGCAGGAATAGTAGGATTATCAATAAGATTAATTATTCGATTAGAATTAGGTATGCCTGGCAGATTACTGGGGAATGATCAAATTTATAATAGGATAGTAACAGCTCATGCTTTTGTAATAATTTTTTTTATAGTAATACCAGTTATATTAGGGGGGTTTGGCAATTGATTAATTCCTTTAATATTGGGTGCTCCAGATATGGCTTTCCCTCGATTAAATAATATAAGATTTTGGTTATTAATTCCTTCTTTAATTTTATTATTATTAAGAAGAATTTTAAATGTAGGGGTAGGAACTGGTTGAACAATATACCCTCCTTTATCTTCTAATTTAGGACATAGGGGATTGTCAGTTGATTTAGCTATTTTTTCTCTACATTTAGCTGGGGCATCTTCAATTATAGGGTCTATAAATTTTATTACTACAATTTTAAATATACATTTAATAATATTAAAATTAGATCAATTAACTTTATTAATTTGGTCAATTTTTATTACTACTATTTTATTATTATTATCTTTACCAGTTTTAGCTGGAGCTATTACTATATTATTAACT------------------------------------------.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 9/vii/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG28824-F05.
Paratypes
None.
Etymology
Bracon paulamarinae is named to honor Paula Marin for her cheerful participation in the lives and nutritional health of the Marin and Romero families as they have carried out its vital role in the growth and management of the Guanacaste Dry Forest Conservation Fund and Área de Conservación Guanacaste.
Bracon rafamoralesi , sp. nov.
Diagnostics
BOLD:ADB5058. Consensus barcode. ATTTTATATTTTTTATTTGGTATATGATCAGGTATAGTTGGTTTATCAATAAGGTTAATTATTCGATTAGAATTAGGTTTACCTGGGAGTTTATTAGGTAATGATCAAATTTATAATAGAATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTTATATTAGGGGGGTTTGGTAATTGATTAATTCCTTTAATATTAGGAGCCCCAGATATAGCTTTCCCTCGTTTAAATAATATAAGATTTTGGTTATTATTTCCTTCTTTAATTTTATTATTATTGAGAAGAATTTTAAATGTTGGTGTAGGCACTGGTTGAACAATATATCCTCCTTTATCATCTAGGTTAGGTCATAGAGGTTTATCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGGGTTTCTTCTATTTTAGGTTCAATAAATTTTATTACAACAATTTTAAACATGCATTTATTAATATTAAAGTTAGATCAATTGACTTTATTAATTTGGTCAATTTTTATTACAACTATTTTATTATTATTGTCTTTACCTGTTTTGGCTGGTGCTATTACTATATTATTAACAGAT------------------------------------------------.
Holotype ♂
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 19/xii/2013. Depository:
Host data. None.
Holotype voucher code. BIOUG29288-B07.
Paratypes
None.
Bracon robertofernandezi , sp. nov.
Diagnostics
BOLD:ADC8245. Consensus barcode. GTTTTATATTTTTTATTTGGGATATGAGCTGGCATGCTGGGGTTATCAATAAGATTGATTATTCGATTAGAATTAGGCATACCAGGAAGAATATTAGGTAATGATCAAATTTATAATAGTATAGTTACTGCGCATGCTTTTATTATAATTTTTTTTATGGTTATACCTATTATAATTGGTGGGTTTGGAAATTGATTACTACCATTAATATTAGGGGCCCCAGATATGGCATTTCCTCGATTAAATAATATAAGATTTTGATTAATTATTCCTGCTTTAATTATATTATTAATAAGTAGAATTTTAAATGTTGGTGTAGGAACTGGTTGAACTGTTTATCCTCCTTTATCTTCTTCATTGGGGCATAGGGGGATATCTGTTGATTTAGCTATTTTTTCATTACATATAGCTGGAATTTCTTCAATTTTAGGTGCAATTAATTTTATTACTACTATTTTTAATATACAACTATATATTTTAAAATTAGACCAATTAACTTTATTAATTTGATCAATTTTTATCACAGTTGTCTTGTTATTATTATCATTACCAGTTTTAGCAGGAGCTATTACTATATTATTAACTGATCGT------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-2, 10.7634, -85.335, 824 meters, Malaise trap, 30/x/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG30497-D09.
Paratypes
None.
Etymology
Bracon robertofernandezi is named to honor Roberto Fernandez for his previous efforts on behalf of environmental planning for ICE, the National Electric Company, and now his work for BioAlfa the program to DNA barcode all of Costa Rican eukaryote biodiversity.
Bracon rogerblancoi , sp. nov.
Diagnostics
BOLD:AAA5369. Consensus barcode. AATTTTATATTTTTTATTTGGTTTATGATCTGGTATAGTTGGATTATCTATAAGTTTAATTATTCGTTTAGAATTAGGTATACCTGGAAGATTATTAAGTAATGACCAAATTTATAATAGAATAGTTACAGCTCATGCTTTAGTAATAATTTTTTTTATAGTTATACCAATTATATTAGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGTGCTCCTGATATAGCTTTCCCTCGATTAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATTTTATTATTATTAAGAAGAATTTTAAATGTTGGAGTAGGAACAGGTTGAACTTTATACCCTCCTTTATCTTCAAATTTAGGTCATAATGGTATATCTGTAGATTTATCTATTTTTTCTTTACATTTAGCTGGTATTTCATCAATTATAGGTTCATTAAATTTTATTACTACTATTTTAAATATACATTTATTAACATTAAAATTAGATCAATTAACTTTATTAATTTGATCAATTTTTATTACAACTATTTTATTATTATTATCTTTACCTGTTTTAGCAGGRGCTATTACTATATTATTAACTGATCGTAATTTAAATACTTCATTTTTTGATTTTTCAGGAGGTGGAGACCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Palomo, 10.96187, -85.28045, 96 meters, caterpillar collection date: 05/vii/2012, wasp eclosion date: 17/vii/2012, 8 wasps eclosed. Depository:
Host data. Gregarious parasitoid of Lerema liris (Hesperiidae) feeding on leaves of Paspalum fasciculatum (Poaceae) (Fig.
Caterpillar and holotype voucher codes. 12-SRNP-68015, DHJPAR0049816.
Paratypes
Hosts = Synapte salenus, Parphorus storax, Morys lydeDHJ01, Lerema liris (all Hesperiidae, Hesperiinae). 11 specimens, same data as holotype and DHJPAR0029034, DHJPAR0052239, DHJPAR0051799, DHJPAR0029037, DHJPAR0051186, DHJPAR0029035, DHJPAR0049816, DHJPAR0058162, DHJPAR0058165, DHJPAR0058166. Depository:
Etymology
Bracon rogerblancoi is named to honor Sr. Roger Blanco, former parataxonomist, and Research Program Coordinator, and now the Subdirector of ACG, in recognition of three decades of intense support and facilitation of ACG germination and growth.
Bracon rogerblancoi (DHJPAR0029037) mass of white cocoons spun by the wasp larvae below their roof-in-common and end-to-end cocoons adjacent to the ultimate instar of a Morys lydeDHJ01 (Hesperiidae, Hesperiinae) caterpillar cadaver in its rolled grass leaf nest (06-SRNP-43580); prepupal larvae exposed by tearing away one wall of the leaf-silk caterpillar nest.
Bracon ronaldzunigai , sp. nov.
Diagnostics
BOLD:ACR7910. Consensus barcode. GTATTATATTTTTTATTTGGAATATGGGCAGGTATATTAGGTTTATCAATAAGTATAATTATTCGTTTAGAATTAGGYAYAGTTGGAAGTTTATTAATAAATGATCAAATTTATAATAGTATTGTTACTTCTCATGCTTTTGTAATAATTTTTTTTATAGTTATGCCTGTAATAGTTGGGGGTTTTGGAAATTGATTATTACCTTTAATGTTAGGATCTCCTGATATAGCTTTTCCTCGGTTAAATAATATAAGATTTTGATTACTTATTCCTTCTTTAATTATATTATTGTTTAGTAGAGTTTTAAATATTGGTGTGGGGACTGGGTGAACTATATATCCTCCTTTATCTTCTTTATTAGGTCATGGTGGATTGTCAGTAGATTTAGCTATTTTTTCTTTACATATTGCAGGAGTTTCATCAATTTTAGGGGCTATTAATTTTATTTCAACTATTTTAAATATATTTTTATATACTTTAAAATTAGATCAATTAACTTTGTTGATTTGATCAATTTTTATTACAGCTATTTTGTTATTATTATCTTTACCTGTTTTAGCAGGCGCTATTACAATACTATTAACAGATCGAAATTTAAAT.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, Malaise trap, 19/v/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG25142-A09.
Paratype
BIOUG18005-A01. Depository:
Etymology
Bracon ronaldzunigai is named to honor Ronald Zuniga for his many years of dedicated curatorial and taxonomic efforts for the advancement of the former INBio arthropod collection, now in the Museo Nacional de Costa Rica, and his furthering the BioAlfa program to DNA barcode all of Costa Rican eukaryote biodiversity, with an emphasis on Hymenoptera.
Bracon sigifredomarini , sp. nov.
Diagnostics
BOLD:ACG5126. Consensus barcode. TATTTTATATTTTTTATTTGGAATATGAGCTGGGATATTAGGTATATCTATAAGATTAATTATTCGTTTAGAATTAGGGGTACCAGGTAGCTTATTAGGAAATGATCAAATTTATAATAGGATAGTTACTGCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTATTACCTTTAATATTAGGGGCTCCTGATATAGCATTCCCTCGTTTAAATAATATAAGGTTTTGATTGATTATTCCTTCTTTAATTTTATTATTAATAAGAAGAATTTTAAATGTAGGAGTTGGTACTGGTTGAACAGTTTACCCTCCTTTATCTTCATCTTTAGGACATAGAGGGCTATCTGTAGATTTAGCAATTTTTTCTTTACATATAGCTGGTATTTCATCTATTTTAGGTGCAATTAATTTTATTACAACTATTTTAAATATACATTTATTTATTTTGAAATTAGATCAATTAACTTTATTAATTTGATCAATTTTTATTACAGTTATTTTATTATTATTATCTTTACCAGTTTTAGCTGGGGCAATTACTATATTATTAACTGACCGAAATTTAAATACATCTTTTTTTGATTTTTCTGGAGGAGGGGATCCAATTTTATTCCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 30/vii/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG05422-G01.
Paratypes
None.
Etymology
Bracon sigifredomarini is named to honor Sigifredo Marin for his decades of managerial soul and energy and effort on behalf of Parque Nacional Santa Rosa, Área de Conservación Guanacaste, and then the Guanacaste Dry Forest Conservation Fund projects in and around ACG.
Bracon tihisiaboshartae , sp. nov.
Diagnostics
BOLD:AAA5367. Consensus barcode. TGTATTATATTTTTTATTTGGAATATGAGCYGGAATAATTGGTTTATCAATAAGTTTAATTATTCGTTTAGAATTAGGRATACCAGGTAGTTTAYTAGGTAATGATCAAATTTATAATAGTATAGTTACAGCKCATGCTTTTATTATAATTTTTTTTATAGTTATACCAGTAATATTAGGAGGWTTTGGTAATTGATTAGTTCCTTTAATATTAGGTGCTCCTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATTTTATTATTATTAAGAAGAATTTTAAATGTTGGTGTAGGRACAGGCTGAACTATTTATCCTCCTTTATCTTCTATAATAGGTCATAGAGGWATATCTGTRGATTTATCTATTTTYTCTTTACATTTAGCTGGTATTTCTTCTATTATAGGATCGATTAATTTTATTACAACAATTTTAAATATACATTTATTAATATTAAAATTAGATCAATTAACTTTATTTATTTGATCAATTTTTATTACAACTATTTTATTATTATTATCTTTACCTGTATTAGCAGGAGCTATTACTATAYTATTAACTGATCGAAATTTWAATACTTCATTTTTTGATTTTTCTGGAGGTGGGGATCCAATTYTATTTCAACATTTATTT. Bracon tihisiaboshartae and B. alejandromasisi occupy the same BIN. Bracon tihisiaboshartae can be differentiated from B. alejandromasisi by the color of the metasomal terga: entirely yellow in B. tihisiaboshartae and partly black in B. alejandromasisi. The two can also be differentiated based on their host caterpillars and style of cocoons, see note below.
Holotype ♀
Guanacaste, Sector Mundo Nuevo, Quebrada Tibio Perla, 10.76261, -85.42979, 330 meters, caterpillar collection date: 17/xi/2013, wasp eclosion date: 1/xii/2013, 26 wasps eclosed. Depository:
Host data. Gregarious parasitoid of Memphis boisduvali (Nymphalidae) feeding on Mespilodaphne veraguensis (Lauraceae) (Fig.
Caterpillar and holotype voucher codes. 13-SRNP-57185, DHJPAR0054608.
Paratype
Host = Memphis pithyusa: DHJPAR0040072. Depository:
Note
Bracon alejandromasisi and Bracon tihisiaboshartae are placed in the same BIN, but their COI barcodes differ by ca. 1%, and each is reared from different genera of caterpillars (Consul, Memphis) feeding on very different plants (Piper, Croton). The cocoons of the former are dark brown and tightly fill the caterpillar nest (Fig.
Etymology
Bracon tihisiaboshartae is name in honor of Tihisia Boshart in specific recognition of her rapid and high-quality rendering of artwork for Hymenoptera and Lepidoptera patronyms for major Costa Rican decision-makers helping ACG.
Bracon wilberthbrizuelai , sp. nov.
Diagnostics
BOLD:ADB1539. Consensus barcode. ATTTTATACTTCTTATTCGGAATATGAGCTGGAATAATTGGTTTATCTATAAGATTAATTATTCGGTTAGAATTAGGTATACCTGGGAGATTATTAAAAAATGACCAAATTTATAATAGAATAGTCACAGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTAATATTAGGAGGGTTTGGAAATTGATTAACCCCTTTAATATTAGGTGCTCCTGATATAGCTTTCCCTCGATTAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATTTTATTATTATTAAGAAGAATTTTAAATGTTGGGGTAGGAACAGGTTGAACTATATACCCCCCATTATCATCAAATTTAGGACACAGAGGTTTATCAGTTGATTTAGCTATTTTTTCCTTACATTTAGCAGGAATTTCTTCTATTATAGGGGCAATAAATTTTATTTCTACTATTTTAAATATACATTTATTTACATTAAAAATAGATCAATTAACTTTATTTGTTTGATCTATTTTTATCACAACTATTTTATTACTATTATCTTTACCAGTTTTAGCTGGTGCTATCACTATATTATTAACAGAT------------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-2, 10.7634, -85.335, 824 meters, Malaise trap, 2/i/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29474-A02.
Paratypes
None.
Digonogastra Viereck, 1912
Digonogastra is a large and species rich genus restricted to the New World. Members attack larval Lepidoptera and Coleoptera. The generic assignment of many species of Digonogastra from Cyanopterous is difficult or impossible because the precise limits of the two are not well defined at present.
Digonogastra montylloydi , sp. nov.
Diagnostics
BOLD:ADD0356. Consensus barcode. ATTTTATATTTTTTATTTGGGATGTGATCTGGAATAGTAGGGTTATCAATAAGTTTAATTATTCGATTAGAATTAGGAGTTCCTGGAAGGTTATTAGGTAATGATCAAATTTATAATAGAATAGTTACTGCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATATTAGGTGGATTTGGAAATTGGTTAATTCCTTTAATATTAGGTGCTCCTGACATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCATCTTTATTAATATTATTGTTGAGAGGAATTTTAAATGTGGGTGTAGGTACTGGATGAACAATTTATCCTCCATTATCTTCATTTTTGGGACATGGAGGGATTTCTGTTGATTTAGCTATTTTTTCTTTACATTTGGCTGGTGTTTCTTCAATTATAGGATCAATTAATTTTATTACAACAATTTTAAATATACGATTATTTTTTTTGAAATTAGATCAATTAACTTTATTTATTTGATCAATTTTTATTACTACAATTTTATTATTATTATCTTTACCAGTTTTAGCTGGGGGAATTACTATATTATTAACAGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGAGGAGATCCAATTTTATTTCAACATTTA.
Holotype ♀
Guanacaste, Sector Pitilla, Estación Quica, 10.99697, -85.39666, 470 meters, caterpillar collection date 28/iv/2008, wasp eclosion date: 30/iv/2008. Depository:
Host data. Oiketicus kirbyi (Psychidae) feeding on Byrsonima crassifolia (Malpighiaceae). The gregarious wasp larvae spin their cocoons inside the bagworm silk and twig nest; the paratypes are sibs of the holotype.
Caterpillar and holotype voucher codes. 08-SRNP-70260, DHJPAR0028324.
Paratypes
Host = same as holotype: DHJPAR0028320, DHJPAR0028321, DHJPAR0028322, DHJPAR0028323. Depository:
Digonogastra montywoodi , sp. nov.
Diagnostics
BOLD:ADJ0327. Consensus barcode. TATATTATATTTTTTATTTGGTATATGAGCTGGAATAATTGGTTTGTCTATAAGATTAATTATTCGTTTAGAGTTAGGTATACCGGGTAGAATATTAAATAATGATCAAATTTATAATAGAATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATGGTTATACCTATAATAGTAGGTGGATTTGGGAATTGATTAACACCTTTAATATTAGGGGCTCCTGATATGGCTTTCCCACGAATAAATAATATAAGATTTTGGTTATTGGTTCCTTCAATTTTATTATTAATATTAAGAAGAATTATAAATATTGGAGTAGGTACTGGATGAACAATATATCCTCCTTTATCTTCTTTATTAGGACATAGTGGAATTTCAGTTGATTTAGCAATTTTTTCTTTACATTTAGCGGGGGTTTCTTCAATTATAGGTTCAATTAATTTTATTTCAACAATTTTAAATATACGTTTATTTTATTTAAAATTAGATCAATTAACTTTATTTATTTGATCAATTTTTATTACAACAATTTTGTTATTATTATCTTTACCTGTTTTGGCGGGGGGTATTACTATGTTATTAACTGATCGTAATTTAAATTCTACATTTTTTGATTTTTCTGGAGGAGGAGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pailas, Catarata Borinquen, 10.817721, -85.390465, 945 meters, 29/i/2017, light-trapped. Depository:
Host data. None.
Holotype voucher code. DHJPAR0061034.
Paratypes
None.
Other material
A specimen from Panama is in the same BIN and is likely conspecific.
Digonogastra motohasegawai , sp. nov.
Diagnostics
BOLD:ADJ4657. Consensus barcode. GATATTGTATTTTTTATTTGGTATATGGGCTGGGATAGTTGGTTTATCAATAAGATTAATTATTCGTTTAGAGTTGGGGATACCTGGAAGGATGTTAGGTAATGATCAAATTTATAATAGAATAGTGACGGCACATGCTTTTGTAATAATTTTTTTTATAGTTATACCAGTAATATTGGGGGGGTTTGGAAATTGGTTAATTCCATTAATGTTAGGGGCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGGTTTTGATTACTTATTCCTTCAATTTTATTATTATTATTAAGAAGAATTTTAAATATTGGRGTAGGTACAGGGTGAACTGTTTATCCTCCTTTATCTTCATCTTTAGGACATAGAGGGGTATCAGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTGTTTCTTCTATTATAGGGTCAATTAATTTTATTACAACAATTTTAAATATACGTTTATTTTTTTTAAAATTAGATCAATTAACTTTATTTATTTGATCAATTTTTATTACAACAATTTTATTGTTATTATCTTTACCTGTTTTGGCAGGGGGAATTACAATACTATTAACAGATCGAAATTTAAATACAACATTTTTTGATTTTTCTGGGGGAGGGGATCCTGTTTTGTTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pailas, Catarata Borinquen, 10.817721, -85.390465, 945 meters, 29/i/2017, light-trapped. Depository:
Host data. None.
Holotype voucher code. DHJPAR0061037.
Paratypes
DHJPAR0061039. Depository:
Digonogastra natwheelwrighti , sp. nov.
Diagnostics
BOLD:ADM7321. Consensus barcode. AATATTGTATTTTTTATTTGGTATGTGAGCTGGTATAGTTGGATTATCAATAAGGTTGATTATTCGTTTAGAATTAGGTATACCAGGGAGTTTATTAGGTAATGATCAGATTTATAATAGTATAGTAACTGCTCATGCTTTTATTATAATTTTTTTTATGGTTATACCTATTATATTAGGGGGATTTGGGAATTGATTAATTCCTTTAATATTGGGGGCCCCAGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTACTTATTCCTTCTATTTTGTTGTTATTATTAAGGAGATTTATAAATATTGGGGTTGGTACAGGATGGACAGTTTATCCTCCTTTATCTTCTTCTTTGGGGCATAGGGGTATTTCAGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATGGGGTCAATTAATTTTATTTCTACTATTTTAAATATACATTTATTTTTTTTAAAATTAGATCAGTTAACTTTGTTTATTTGATCAATTTTTATTACAACAATTTTATTGTTATTATCTTTACCTGTTTTAGCTGGGGGTATTACTATATTATTAACGGATCGTAATTTAAATACAACTTTTTTTGATTTTTCGGGGGGGGGAGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pailas, PDL#5, 10.7627, -85.334, 825 meters, 4/i/2014, light-trapped. Depository:
Host data. None.
Holotype voucher code. DHJPAR0062413.
Paratypes
None.
Digonogastra nickgrishini , sp. nov.
Diagnostics
BOLD:ADJ3490. Consensus barcode. GGTATTATATTTTTTATTTGGGATGTGATCAGGAATATTAGGTTTATCAATAAGAATAATTATTCGTTTAGAATTAGGGATACCAGGAAGAATATTAGGTAATGATCAAATTTATAATAGAATAGTTACTATTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATATTAGGTGGATTTGGTAATTGATTAATTCCATTAATATTAGGGGCACCTGATATGGCTTTCCCTCGAATAAATAATATAAGATTTTGATTAATTATTCCTTCTTTAATGTTATTATTATTAAGAAGGGTAATAAATGTTGGAGTAGGAACTGGTTGAACTATATATCCTCCTTTATCATCATTTTTAGGTCATGGAGGAATATCAGTTGATTTATCAATTTTTTCTTTACATTTGGCTGGAATTTCTTCAATTATGGGTGCAATTAATTTTATTACAACTATTTTAAATATACGTTTATTTTTTTTAAAGTTAGATCAGTTAACTTTATTTATTTGATCAATTTTTATTACTACAATTTTATTATTATTATCATTACCAGTTTTAGCTGGTGGAATTACAATATTATTAACTGATCGTAATTTAAATACTACATTTTTTGATTTTTCTGGAGGGGGGGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pailas, Catarata Borinquen, 10.817721, -85.390465, 945 meters, 29/i/2017, light-trapped. Depository:
Host data. None.
Holotype voucher code. DHJPAR0061042.
Paratypes
None.
Chapter 4: Cheloninae
Cheloninae have been viewed as solitary, koinobiont, egg-larval parasitoids, i.e., lay one egg in the egg of the host and delaying development until the host caterpillar is nearing maturity (van
All reared Cheloninae reported here are parasitoids of wild-caught leaf rolling and webbing “Microlepidoptera” caterpillars and none from “macrocaterpillars” even though there are 550+ species of leaf-sheltering or leaf-nest Hesperiidae caterpillars in the rearing inventory (
Dan Boyce (1936) observed that a species of Ascogaster pupated within the pupal chamber of the host, and that has become the universal truth applied to all chelonines (
In some cases where there are many records for a single species of reared Cheloninae, this is because the host caterpillars are often gregarious and live in a sloppy mass of silked-together leaves. Presumably, the female moth lays a large mass of 50‒200 eggs, and when a female chelonine wasp finds it, it has the opportunity to oviposit into many eggs. The consequence is that the eclosing wasps will be sibs (e.g., Pseudophanerotoma alexsmithi, Chelonus sujeevanratnasinghami, Chelonus scottshawi, Chelonus gustavogutierrezi).
The ACG rearing inventory has found that an identification trait of most chelonine cocoons is that they are squared at both apices, as shown in Fig.
All Adelius and most Ascogaster and Phanerotoma were captured with Malaise traps and rarely reared. In part, this is because they may be parastioids of caterpillars that are miners in leaf tissue, rather than exposed in a leaf-silk webbing. For practical reasons, leaf-mining caterpillars are rarely reared in the ACG, remaining for further development of the inventory. For the Cheloninae NJ tree, see Suppl. material
Cocoons of Chelonus motohasegawai in the remains of the host caterpillar pupal chamber spun between sides of a plastic rearing bag (DHJPAR0042920).
Prepupae of Chelonus nataliaivanovae (DHJPAR0035559) inside the pupal chamber of a single caterpillar of its host (Anacrusis turrialbae 09-SRNP-44104) after the leaf ceiling was lifted off. The head and thorax capsule of the caterpillar are inside the ring of prepupae in the cocoons that they have spun; a yellow meconial pellet is at the posterior end of each cocoon.
Key to New World genera of Cheloninae
Adelius Haliday, 1833
Adelius is worldwide in distribution. Hosts are leaf-mining Lepidoptera, e.g., Nepticulidae.
Adelius adrianguadamuzi , sp. nov.
Diagnostics
BOLD:ADF7277. Consensus barcode. ATTTTATATTTTATATTTGGTATTTGATCAGGTGGTTTAGGTTTATCACTAAGAATAATAATTCGTATAGAATTAAGAACAGTAGGAAGTTATTTGGGTAATGATCAAATTTATAATAGAATTGTAACTCTTCATGCTTTTGTGATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGTAATTGGTTAATTCCATTAATAGTAATAAGACCTGATATATCATTTCCTCGAATAAATAATATGAGATTTTGATTATTAATTCCTTCTTTATTTTTATTGATTATGAGAAGATTTGTAAATGTTGGTGTAGGTACAGGATGAACAGTTTATCCTCCTTTATCTTTAATTATAGGTCATGGTGGTGTAGCTGTAGATTTATCAATTTTTTCTTTACACTTAGCTGGGATTTCATCAATTATGGGTGCTATAAATTTTATTGTTACAATTTTTAATAGATCAATATTAATAAATATATTTAATAAAATTTCTTTATTTTGTTGATCTGTTTTTATTACAGCTTTTTTATTGCTATTATCTTTACCAGTATTAGCTGGGGCTATTACTATATTATTAACTGAT----------------------------. Similar to A. excelsus Bortoni and Shimbori, but differs slightly in color, and the hind tibia is much more swollen in A. adrianguadamuzi.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, 18‒25/x/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG31661-B07..
Paratypes
None.
Note
The holotype and all but one paratype of A. excelsus are from high elevation localities in Colombia. The paratype of A. excelsus from Costa Rica is likely to be a member of A. adrianguadamuzi in the opinion of MJS.
Adelius gauldi
Diagnostics
BOLD:ACJ2315. Image available on BOLD.
Host data. None.
Material examined
♀, Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 16‒23/iv/2012, Malaise trap. BIOUG07619-D09, BIOUG18398-G03.
Adelius janzeni
Diagnostics
BOLD:ACJ2270. Image available on BOLD.
Host data. None, Malaise trap.
Material examined
♀, Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 16‒23/iv/2012, Malaise trap. BIOUG07614-F11.
Ascogaster Wesmael, 1835
No described species of Ascogaster are documented from Costa Rica, and only two species are recorded from the Neotropics, i.e., Peru and Uruguay, though there are probably a few hundred species undescribed. Numerous lepidopteran families serve as hosts. Here we add records from Tortricidae and Depressariidae, neither of which is a new family record.
Ascogaster gloriasihezarae , sp. nov.
Diagnostics
BOLD:ADA4144. Consensus barcode. ATTTTATATTTTATTTTTGGAATTTGATCAGGTATATTTGGTTTATCTTTAAGTCTAATTATTCGAATAGAATTAAGGTCGGTTACAGCTTATTTAGGTAACGATCAAATTTATAACAGGTTAGTTACAATACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGTTTTGGAAATTGATTAGTTCCTTTAATATTAGGTGGTCCTGATATATCATTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCACTTTTTTTATTGATTATAAGAAGTTTTGTAAATGTAGGTGTGGGTACAGGATGAACAGTTTATCCTCCTTTATCATTAATTATTGGGCATGGTGGGGTATCAGTTGATATTAGAATTTTTTCTTTACATATAGCTGGAATATCTTCTGTAATAGGTGCATTAAATTTTATTGTAACAATTATAAATATATGAATTGGAGTAAAATATATAGATAAATTGTCTTTATTCACTTGATCAGTTTTTATTACAGCTATTTTATTATTACTATCCTTGCCTGTTTTAGCTGGTGCAATTACTATATTATTAACTGATCGAAATTTTAAT---------------------------------.
Holotype ♂
Alajuela, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, 16‒23/xii/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28068-C02.
Paratypes
None.
Ascogaster grettelvegae , sp. nov.
Diagnostics
BOLD:ABX8525. Consensus barcode. TATTTGGACTATCTTTAAGTTTAATTATTCGAATAGAATTAAGTTCTATTACTTCTTATTTAGGTAATGATCAGATTTATAATAGAGTAGTTACTCTACATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGCGGTTTTGGAAATTGGTTGGTTCCTTTAATGTTAGGAGGACCAGATATATCATTTCCTCGAATAAATAATTTAAGATTTTGATTGTTAGTACCTTCAATTATTTTATTAATTAATAGAAGTTTTATTAATGTAGGGGTAGGAACAGGATGAACTGTATATCCGCCTTTATCTCTTATAATTGGCCATAGAGGAGCTTCTGTAGATTTAAGTATTTTTTCTTTGCATTTAGCTGGAATATCCTCTATTATAGGGGCTGTAAATTTTATTGTTACTATTTTAAATATAAGTTTTAGATTTAAAAATATAGATAAATTTCCTTTGTTTGTGTGATCAATTATAATTACTGCAATTTTATTACTTCTATCTTTACCTGTCTTAGCAGGTGCAATTACTATATTATTAACAGATCGAAATTTAAATACTAGATTTTTTGACCCCTCAGGGGGCGGTGATCCAATTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.865, -85.415, 540 meters, caterpillar collection date: 13/i/2011, wasp eclosion date: 05/ii/2011. Depository:
Host data. Episimus ortygia (Tortricidae) feeding on Vismia baccifera (Hypericaceae).
Caterpillar and holotype voucher codes. 11-SRNP-180, DHJPAR0043010.
Paratypes
None.
Ascogaster guillermopereirai , sp. nov.
Diagnostics
BOLD:ACJ4048. Consensus barcode. AATTTTATATTTTATATTTGGAATTTGATCAGGAATAATTGGATTATCTTTAAGTTTATTAATTCGTATAGAATTAAGTTCAGTTTCTTCTTATTTAGGTAATGATCAAATTTATAATAGAATTGTAACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGCTTCGGAAATTGATTAGTTCCTTTAATATTAGGGGCTCCTGATATATCTTTCCCTCGATTGAATAATTTGAGATTTTGATTATTAATTCCTTCACTTTTTTTTTTATTAATTGGTGGTATTTTGAATTCAGGAGTTGGAACTGGATGAACAGTTTATCCACCGTTGTCATTAGGTATTTATCATAGAGGTATTTCTGTAGATTTAAGTATTTTTTCTTTACATATAGCTGGAATATCTTCAATTTTAGGGGCCTTAAATTTTATTATTACTATTTATTGTATATGGTTAGGTTCAAAAAATATGGATAAGTTATCTTTATTTGTTTGATCAGTAATAATTACTGCATTTTTATTAATTACATCTTTACCAGTTCTAGCAGGCGCAATTACTATATTATTAACTGATCGTAATTTAAATACTAGATTTTTTGATCCTTCTGGTGGTGGTGATCCTGTTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal, Rio Blanco Abajo, 10.90037 -85.37254, 500 meters, caterpillar collection date: 27/ix/2012, wasp eclosion date: 15/x/2012.
Host data. Mictopsichia Janzen330 (Tortricidae) feeding on Marcgravia nepenthoides (Marcgraviaceae).
Caterpillar and holotype voucher codes. 12-SRNP-5277, DHJPAR0051332.
Paratype
Host = Mictopsichia Janzen330: DHJPAR0051327.
Ascogaster gustavoecheverrii , sp. nov.
Diagnostics
BOLD:AAL0574. Consensus barcode. TATTTTATATTTTATATTTGGAATTTGATCTGGTATAGTTGGTTTATCCTTAAGTTTATTAATTTGTATAGAATTAAGATTAGTTTCTTCTTATTTAGGTAATGATCAAATTTATAATAGAGTAGTTACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGCGGTTTCGGAAATTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATGTCTTTTCCTCGTTTAAATAATTTAAGTTTTCGACTTTTGGTTCCTTCTCTATTTTATTCAATATTAAGAGGTATTTTAAATACTGGGAATGGTACAGGATGAACAGTATATCCACCATTATCATTAGGAATATATCATAGAGGTATTTCTGTAGATCTTAGAATTTTTTCTTTACATATAGCTGGTATATCATCAATTTTAGGAGCTTTAAATTTTATTATTACTATTTATTGTATATGAATTGGTTTTAAAAATATAAATAAATTATCATTATWTGTTTGATCTGTATTAATTACTACTTTTTTATTAATTACATCTTTACCTGTATTAGCAGAAGCTATTACTATATTATTAACTGATCGTAATTTAAATACTAGATTTTTTGATCCATCTGGTGGAGGAGATCCTGTATTATATCAACATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93, -85.372, 527 meters, caterpillar collection date: 18/xi/2013, wasp eclosion date: 12/xii/2013. Depository:
Host data. Megalota crassana (Tortricidae) feeding on Croton schiedeanus (Euphorbiaceae).
Caterpillar and holotype voucher codes. 13-SRNP-6613, DHJPAR0054560.
Paratypes
Hosts = Megalota ochreoapex, Megalota spinulosa, Megalota simpliciana, Megalota delphinosema, Megalota crassana. DHJPAR0042852, DHJPAR0042856, DHJPAR0045414, DHJPAR0046941, DHJPAR0049003, DHJPAR0053678, DHJPAR0054566, DHJPAR0062572. Depository:
Ascogaster katyvandusenae , sp. nov.
Diagnostics
BOLD:AAK1026. Consensus barcode. AATTTTGTATTTTTTATTTGGCATTTGATCCGGTGTATTTGGGTTGTCTTTAAGATTAATTATTCGAATAGAATTAAGTTCAGTCACTTCTTATTTAGGTAATGATCAGATTTATAATAGTATTGTCACCCTTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGTTTTGGAAATTGATTAGTTCCTTTAATGTTAGGGGGTCCAGATATATCATTCCCTCGTATAAATAATTTAAGATTTTGATTATTGGTTCCTTCAATTATTTTACTAATTAATAGAAGTTTTATTAATGTTGGGGTAGGGACAGGATGAACTGTTTATCCACCTTTATCTCTTATTATTGGACATAGAGGAGCTTCTGTAGATATAAGAATTTTTTCTTTACATTTAGCTGGTATATCTTCAATTATAGGGGCTGTAAATTTTATTGTTACTATTTTAAACATAAGATTTAGGTTTAAAAATATGGATAAATTTCCTTTATTTGTTTGATCAATTATAATTACTGCAATTTTATTATTATTATCTTTACCTGTTTTAGCAGGTGCAATTACTATATTATTAACAGATCGAAATTTAAATACAAGTTTTTTTGACCCTTCAGGGGGGGGAGACCCAATTTTATATCAGCATTTATTT.
Holotype ♂
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.865, -85.415, 540 meters, caterpillar collection date: 18/viii/2010, wasp eclosion date: 31/viii/2010. Depository:
Host data. Megalota spinulosa (Tortricidae) feeding on Croton schiedeanus (Euphorbiaceae).
Caterpillar and holotype voucher codes. 10-SRNP-4345, DHJPAR0055824.
Paratype
Host = Amorbia decerptana: DHJPAR0035314. Depository:
Ascogaster luisdiegogomezi , sp. nov.
Diagnostics
BOLD:ACC1221. Consensus barcode. AATTTTGTATTTTTTATTTGGTATTTGATCAGGTATAATTGGTTTATCTTTAAGTTTATTAATTCGTATAGAATTAAGATCAGTTTCTTCTTATTTAGGAAATGATCAAATTTATAATAGGGTAGTAACAATGCATGCTTTTATCATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGTTTTGGGAATTGATTAGTTCCTTTAATATTAGGATCTCCTGATATATCTTTTCCTCGATTAAATAATTTAAGTTTTTGATTATTAATTCCTTCTTTATTTTTTTTATTAATTAGAGGTATTTTAAATATTGGAGTTGGTACAGGATGAACTGTTTATCCACCATTATCATTAGGTATATTTCATAGAGGAATTTCAGTTGATTTAAGAATTTTCTCTTTACATATAGCAGGCATATCTTCTATTTTAGGAGCTTTAAATTTTATTATTACTATTTTTTGTATATGATTTGGATTTAAATATATAGATAAATTATCTTTATTTATTTGATCAGTAGTAATTACTGCATTTTTATTAATTACATCTTTACCCGTTTTAGCAGGTGCTATTACAATATTATTAACAGATCGAAATTTAAATACTAGTTTTTTTGACCCTTCTGGTGGAGGTGACCCAATTTTATATCAACATTTGTTT.
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93, -85.372, 527 meters, caterpillar collection date: 09/vi/2012, wasp eclosion date: 04/vii/2012. Depository:
Host data. Stenoma Janzen414 (Depressariidae) feeding on Ocotea atirrensis (Lauraceae).
Caterpillar and holotype voucher codes. 12-SRNP-2366, DHJPAR0049931.
Paratypes
None.
Chelonus Panzer, 1806
Chelonus is worldwide in distribution with ca. 1000 described species and many more undescribed. Hosts include a wide range of lepidopterous families. Here we follow
Chelonus alejandrozaldivari , sp. nov.
Diagnostics
BOLD:ACO0288. Consensus barcode. TACATTATATTTTATTTTTGGTATTTGATCTGGAATATTTGGTTTATCTTTAAGTTTACTTATTCGAATAGAATTAAGGATAGGAGGTAGATTATTATTAAATGATCAATTATATAATAGTTTAGTAACATTRCATGCTTTTGTAATAATTTTTTTTATAGTTATACCTGTTATAATTGGAGGYTTTGGTAATTGATTAATTCCTTTAATATTAGGATTACCTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGGTTATTAATTCCTTCATTAACTTTATTAATTTTAAGGGGATTTATTAATATAGGGGTAGGTACAGGTTGAACTGTTTATCCTCCTTTATCTTCTTTAATTGGTCATGGAGGAATTTCTGTTGATTTATCTATTTTTTCTTTACATTTAGCAGGAGCTTCTTCTATTATAGGCGCAATTAATTTTATTGTTACTATTTTAAATACTTGAATATTAAGTAAATATTTAGATAAGTTTCCTTTATTTTCATGATCTGTTTTTATTACTGCTATTTTATTATTATTATCATTRCCAGTATTAGCYGGTGCAATTACTATATTATTAAGAGATCGTAATTTAAATACTAGATTTTTTGATCCTTCTGGTGGAGGRGAYCCTGTTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 05-18/xii/2014, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher code. BIOUG44692-B02.
Paratypes
BIOUG43804-H09, BIOUG44551-E11. Depository:
Chelonus gustavogutierrezi , sp. nov.
Diagnostics
BOLD:AAA6603. Consensus barcode. AATATTATATTTTATATTTGGAATTTGATGTGGRATAATAGGATTGTCATTAAGAGTAWTAATTCGTATAGAATTAAGAAGAGTAATAAGATTATTTTATAATGATCAATTATATAATAGAATTGTTACTATGCATGCTTTTATTATAATTTTTTTTATAGTTATACCATTAATAATTGGTGGATTTGGRAATTGATTAATTCCACTAATATTAGGATTATCAGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGATTGTTACTACCTTCAATTATTTTATTAATTATAGGTGGATTTGTTAATATAGGAGCAGGTACAGGATGAACAGTTTATCCACCTTTATCATTATTAATAGGTCATAGAGGAATCTCAGTAGATTTATCTATTTTTTCTTTACATTTAGCTGGTATATCTTCAATTATAGGTTCAATTAATTTTATTGTTACTATTATTAATACATGATTAAATATAAAATATATGGATAAATATCCTTTATTTGTTTGATCAGTATTTATTACTACTATTTTATTATTATTATCTTTGCCAGWTTTGGCTGGAGCAATTACTATATTATTAAGTGATCKAAATTTAAATACTAGTTTTTTTGATCCTTCAGGTGGTGRTGATCCAGTATTATATCAACAYTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Flecha, 10.947, -85.315, 491 meters, caterpillar collection date: 15/ix/2009, wasp eclosion date: 27/ix/2009. Depository:
Host data. Omiodes cuniculalis (Crambidae) feeding on Gliricidia sepium (introduced) (Fabaceae), and a variety of other woody species.
Caterpillar and holotype voucher codes. 09-SRNP-80021: DHJPAR0037990.
Paratypes
Host = Omiodes cuniculalis, Pyralidae 09-SRNP-71481: DHJPAR0037185, DHJPAR0037164, DHJPAR0029095, DHJPAR0029096, DHJPAR0029097, DHJPAR0029099, DHJPAR0029100, DHJPAR0029084, DHJPAR0029083, DHJPAR0029081, DHJPAR0029080, DHJPAR0029092, DHJPAR0029091, DHJPAR0029090, DHJPAR0029089, DHJPAR0029101, DHJPAR0037985. Depository:
Chelonus gustavoinduni , sp. nov.
Diagnostics
BOLD:ADA0356. Consensus barcode. GTATTATATTTTATTTTTGGTATTTGATCTGGAGTTTTAGGATTATCATTAAGATTAATTTTACGAATAGAATTAAGAGTGTTAGGAAGGTTATTAAAAAATGATCAATTATATAATAGGGTAGTTACTTTACATGCTTTTGTAATAATTTTTTTTATGGTTATACCAGTAATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATATTAGGATTACCAGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGGTTATTAATTCCTTCATTAATAATGTTATTATTGAGAAGATTTGTAAATATAGGTGTAGGTACAGGATGAACGGTTTATCCACCATTATCTTCATTAATAGGACATGGTGGTATTTCAGTAGATTTATCAATTTTTTCTTTACATTTAGCAGGTGCATCTTCTATTATGGGGGCAATTAATTTTATTGTAACGGTAATAAATACTAATTTTAAGATTGGGTTTATAGATAAATTTCCATTATTTGTTTGATCAGTTTTAATTACGGCTATTTTATTATTATTATCATTGCCAGTATTAGCCGGTGCTATTACTATATTATTA.
Holotype ♂
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, 22‒29/vi/2015, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28148-H10.
Paratypes
None.
Chelonus harryramirezi , sp. nov.
Diagnostics
BOLD:AAE2698. Consensus barcode. AATATTATATTTTATTTTTGGAATTTGATGKGGAATGTTAGGTTTATCATTAAGAGTTATAATTCGTATAGAATTAAGAAGGACATTAAGTTTATTCTATAATGATCAATTATATAATAGAATTGTTACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCATTAATAATTGGAGGGTTTGGAAATTGATTAATTCCTTTAATATTAGGTTTATCAGATATAATTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCAATTATTTTATTAATTATAGGGGGTTTTGTAAATACAGGGGCAGGTACAGGATGAACAGTTTATCCTCCTTTATCATTATTGATAGGTCATAGAGGGGTATCAGTAGATTTATCAATCTTTTCTTTACATTTAGCTGGAGCATCATCTATTATGGGTTCTATTAATTTTATTGTAACTATTATTAATACTTGGTTGAATATAAAATATATAGATAAATATCCTTTATTTGTTTGATCTGTTTTTATTACCACAATTTTGTTATTATTATCTTTACCAGTTTTAGCTGGGGCAATTACTATATTATTAAGAGATCGAAATTTAAATACTAGATTTTTTGATCCTTCAGGGGGTGGAGAYCCAGTATTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Quebrada Bambu, 10.93, -85.252, 109 meters, caterpillar collection date: 14/ii/2013, wasp eclosion date: 01/iii/2013. Depository:
Host data. Omiodes humeralis (Crambidae) feeding on Inga sapindoides (Fabaceae).
Caterpillar and holotype voucher codes. 13-SRNP-75542, DHJPAR0051927.
Paratypes
Hosts = Omiodes humeralis. DHJPAR0023700, DHJPAR0020624, DHJPAR0020623, DHJPAR0020802, DHJPAR0038012, DHJPAR0038013, DHJPAR0038014, DHJPAR0020804, DHJPAR0046938, DHJPAR0051326, DHJPAR0051328, DHJPAR0051330, DHJPAR0055229, DHJPAR0054564, DHJPAR0054938. Depository:
Chelonus hartmanguidoi , sp. nov.
Diagnostics
BOLD:AAD1994. Consensus barcode. AATATTATATTTTATTTTTGGAATTTGGTGTGGAATAATAGGATTATCCTTAAGAGTTATGATTCGTATAGAGTTAAGAAGAACAATAAGATTATTTTGTAATGATCAATTATATAATAGAATTGTTACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCATTAATAATTGGGGGGTTTGGAAATTGATTAATYCCTTTAATATTAGGTTTATCAGATATAATTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATCCCTTCAATTATTTTATTAATTATAGGAGGATTTGTAAATATAGGTGCAGGTACAGGATGAACAATTTATCCGCCTTTATCATTATTAATAGGTCATAGTGGGATTTCAGTAGATTTATCAATTTTTTCTTTACATTTAGCTGGGGCATCATCTATTATAGGTTCAATTAATTTTATTGTGACTATTATTAATACTTGATTAAATATAAAGTATATGGATAAATATCCTTTATTTGTTTGATCTGTTTTTATTACTACAATTTTATTATTATTATCTTTRCCTGTTTTAGCTGGGGCAATTACTATACTATTAAGAGATCGAAATTTAAATACTAGATTTTTTGATCCTTCAGGGGGTGGGGACCCAGTATTATACCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Palomo, 10.962, -85.28, 96 meters, caterpillar collection date: 19/i/2011, wasp eclosion date: 10/ii/2011. Depository:
Host data. Omiodes Janzen05 (Crambidae) feeding on Entada gigas (Fabaceae).
Caterpillar and holotype voucher codes. 11-SRNP-67022, DHJPAR0042543.
Paratypes
Hosts = Omiodes Janzen03, spiloBioLep01 BioLep577, Piletosoma thialis, Omiodes Janzen05, Phostria cyrisalis. DHJPAR0029154, DHJPAR0029166, DHJPAR0035222, DHJPAR0035307, DHJPAR0035308, DHJPAR0035312, DHJPAR0035313, DHJPAR0036398, DHJPAR0046801, DHJPAR0046947, DHJPAR0052896, DHJPAR0053677, DHJPAR0053686, DHJPAR0054439, DHJPAR0054570, DHJPAR0054571, DHJPAR0056957, DHJPAR0056958, DHJPAR0056963, DHJPAR0056968, DHJPAR0056969. Depository:
Chelonus hazelcambroneroae , sp. nov.
Diagnostics
BOLD:AAE9128. Consensus barcode. TATATTATATTTTATTTTTGGAGTTTGAAGTGGGATAATAGGTTTATCATTAAGAGTTATAATTCGTATAGAATTAAGGAGTGTTATAAGATTATTTAGTAATGATCAGTTATATAATAGGATTGTTACAATACATGCCTTTGTAATAATTTTTTTTATAGTTATACCTTTAATAATTGGAGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGATTATCTGATATAATTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCACTTATTTTATTAATTATAGGTGGATTTGTTAATATAGGTGCTGGGACAGGATGAACAGTTTATCCTCCTTTATCATTATTAATAGGTCATAGAGGAATTTCAGTAGATTTATCTATTTTTTCTTTACATTTAGCTGGAATATCATCAATTATAGGTTCAATTAATTTTATTGTTACTATTATAAATACTTGATTACATGTAAAATATATAGATAAATATCCATTATTTGTTTGATCTGTATTTATTACAACTATTTTATTATTGTTATCATTGCCAGTTTTAGCAGGAGCAATTACTATATTATTAAGTGATCGAAATTTAAATACAAGATTTTTTGATCCATCAGGTGGAGGTGATCCAGTATTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93, -85.372, 527 meters, caterpillar collection date: 27/vii/2012, wasp eclosion date: 25/viii/2012. Depository:
Host data. Crambidae: Prenesta scyllalisDHJ02 (Crambidae) feeding on Allomarkgrafia plumeriiflora (Apocynaceae).
Caterpillar and holotype voucher codes. 12-SRNP-3201, DHJPAR0049930.
Paratypes
Host = Prenesta scyllalisDHJ02: DHJPAR0030990, DHJPAR0030989, DHJPAR0030988, DHJPAR0022199. Depository:
Chelonus iangauldi , sp. nov.
Diagnostics
BOLD:AAD4709. Consensus barcode. AATATTATATTTTATTTTTGGAGTTTGAAGAGGRATAATAGGGTTATCTTTAAGAGTAATAATTCGTATAGAATTAAGAAGAGTAATAAGATTATTTTATAATGATCAATTATATAATAGAATTGTAACGATACATGCTTTTATTATAATTTTTTTTATAGTTATACCATTAATAGTAGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGATTATCTGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCAATTATTTTATTAATTATAGGTAGATTTGTTAATATAGGAGCTGGAACAGGGTGAACAGTATATCCTCCATTATCATTATTAGTAAGACATAGAGGAATTTCTGTAGATTTATCTATTTTTTCTTTACATTTAGCTGGAATATCATCAATTATAGGTTCAATTAATTTTATTGTTACTATTTTAAATACTTGAATATATAAAAAATATATAGATAAATATCCATTATTTGTGTGATCTATTTTTATTACAACAATTTTATTATTATTATCATTACCAGTTTTGGCTGGTGCAATTACTATATTATTAAGTGATCGAAATTTAAATACAAGATTTTTTGATCCATCAGGAGGAGGAGATCCAGTATTATACCAACATTTATTT.
Holotype♀
Alajuela, Sector San Cristobal, Quebrada Garcia, 10.861, -85.426, 495 meters, caterpillar collection date: 20/vii/2009, wasp eclosion date: 11/x/2009. Depository:
Host data. Prenesta scyllalisDHJ01 (Crambidae) feeding on Forsteronia spicata (Apocynaceae).
Caterpillar and holotype voucher codes. 09-SRNP-4322, DHJPAR0037165.
Paratypes
Host = Prenesta scyllalisDHJ01: DHJPAR0037160, DHJPAR0037183, DHJPAR0036726, DHJPAR0037168, DHJPAR0037175, DHJPAR0037169, DHJPAR0040017. Depository:
Etymology
Chelonus isidrochaconi , sp. nov.
Diagnostics
BOLD:AAC8260. Consensus barcode. AATATTATATTTTGTATTTGGAATTTGAAGAGGTATAATTGGGYTATCATTAAGAGTTATAATTCGTATAGAATTAAGTAGGGTAATAAGATTATTTTCTAAYGATCAATTGTATAATAGAATTGTGACTATACATGCTTTTATTATAATTTTTTTTATAGTAATACCTTTRATAATTGGRGGTTTTGGAAATTGGYTAATTCCTTTAATATTAGGATTATCTGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGGTTATTAATTCCTTCAATTATTTTATTAATTATAGGCGGATTTGTAAATATAGGAGCTGGGACAGGATGAACAGTGTATCCYCCTTTATCTTTATTAATAGGTCATAGAGGGGTGTCAGTTGATTTATCTATTTTTTCTTTACATTTAGCTGGGGCTTCATCAATTATAGGKTCAATTAATTTTATTGTTACTATTATAAATACTTGGTTACATATTAAGTATATAGATAAATAYCCATTATTTGTTTGATCTGTATTTATTACAACTATTCTTTTATTATTATCATTACCAGTTTTAGCTGGAGCAATTACTATATTATTAAGTGATCGAAATTTAAATACAAGATTTTTTGAYCCTTCAGGAGGGGGRGAYCCTGTATTGTATCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Estación San Gerardo, 10.88, -85.389, 575 meters, caterpillar collection date: 26/v/2012, wasp eclosion date: 16/vi/2012. Depository:
Host data. Parastenia retractalis (Crambidae) feeding on Trichostigma octandrum (Phytolaccaceae).
Caterpillar and holotype voucher codes. 12-SRNP-2158, DHJPAR0049008.
Paratypes
Host = Parastenia retractalis: DHJPAR0037159, DHJPAR0037174, DHJPAR0037181, DHJPAR0037166, DHJPAR0037172, DHJPAR0037173, DHJPAR0037162, DHJPAR0037176, DHJPAR0037179, DHJPAR0048983, DHJPAR0048984, DHJPAR0048986, DHJPAR0048987, DHJPAR0048988. Depository:
Etymology
Chelonus isidrochaconi is named to honor Sr. Isdro Chacon of BioAlfa for his decades of dedicated support and action for the taxonomy and biodiversity of Costa Rica in general and specifically INBio, Costa Rica’s Instituto Nacional de Biodiversidad and the Museo Nacional de Costa Rica.
Chelonus janecheverriae , sp. nov.
Diagnostics
BOLD:AAL5555. Consensus barcode. GTAATAGGCTTATCTTTAAGTGTAATAATTCGTATAGAATTAAGAAGTGTAATAAGATTATTTTATAATGATCAATTATATAATAGAATTGTAACTATACATGCTTTTATTATAATTTTTTTTATAGTTATGCCTTTAATAATTGGGGGGTTTGGAAATTGATTAATTCCTTTAATGTTAGGTTTATCTGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCAATTATTTTATTAATTATAGGAGGATTTGTTAATATAGGGGCTGGCACAGGATGAACAGTTTATCCGCCATTATCATTATTAATAGGTCATAGTGGTGTTTCAGTAGATTTATCTATTTTTTCTTTACATTTGGCAGGAGCCTCATCTATTATAGGTTCAATTAATTTTATTGTGACTATTATAAATACTTGGATGTATTATAAATACATAGATAAATATCCATTATTTGTTTGATCAGTATTTATTACAACTATTTTATTATTATTATCATTACCAGTTTTAGCTGGTGCAATTACTATATTATTAAGAGACCGAAATTTGAACACAAGATTTTTTGATCCATCAGGGGGGGGGGG.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, San Lucas, 10.918, -85.303, 320 meters, caterpillar collection date: 02/ii/2010, wasp eclosion date: 20/ii/2010. Depository:
Host data. crambidJanzen01 Janzen27 (Crambidae) feeding on Calathea lutea (Marantaceae).
Caterpillar and holotype voucher codes. 10-SRNP-40463, DHJPAR0039179.
Paratypes
None.
Etymology
Chelonus janecheverriae is named to honor Sra. Jane Echeverri for her lifetime support of Sr. Gustavo Echeverri in their high-quality management of Costa Rican farms and their financial support for decades of ACG’s Programa de Educación Biológica for the school children in the schools surrounding ACG.
Chelonus jeffmilleri , sp. nov.
Diagnostics
BOLD:ACF0845. Consensus barcode. AATATTATATTTTTTATTTGGAATTTGAAGTGGGATAATAGGTTTATCTTTAAGTGTAATAATTCGTATAGAATTAAGAAGTGTAATAAGATTATTTTATAATGATCAATTATATAATAGAATTGTAACTATACATGCTTTTATTATAATTTTTTTTATAGTTATGCCTTTAATAATTGGGGGATTTGGAAATTGATTAATCCCTTTAATATTAGGATTATCTGATATAATTTTTCCTCGAATAAATAATATAAGATTTCGATTATTAATTCCTTCAATTATTTTATTAATTATAGGAGGATTCGTTAATATAGGGGCTGGTACGGGATGAACAGTTTATCCTCCATTATCATTATTAATAGGTCATAGTGGAGTTTCAGTAGATTTATCTATTTTTTCTTTACATTTGGCAGGGGCTTCATCTATTATAGGTTCAATTAATTTTATTGTGACTATTATAAATACTTGGTTACATTATAAATATATAGATAAATATCCATTATTTGTTTGATCAGTATTTATTACAACTATTTTATTATTATTATCATTACCAGTTTTAGCTGGTGCAATTACTATATTATTAAGAGATCGAAATTTAAATACAAGATTTTTTGATCCATCAGGGGGAGGAGACCCAGTATTATATCAGCATTTATTT.
Holotype ♂
Guanacaste, Sector Del Oro, Quebrada Trigal, 11.027, -85.495, 290 meters, caterpillar collection date: 04/viii/2009, wasp eclosion date: 18/viii/2009. Depository:
Host data. Phostria oajacalis (Crambidae) feeding on Merremia umbellata (Convolvulaceae).
Caterpillar and holotype voucher codes. 09-SRNP-22201, DHJPAR0040375.
Paratypes
Host = Eulepte Solis15: DHJPAR0042859, DHJPAR0042862. Depository:
Chelonus jennyphillipsae , sp. nov.
Diagnostics
BOLD:ACL6996. Consensus barcode. AATGTTATATTTTTTATTTGGAATTTGGAGTGGGATAATAGGTTTATCTTTGAGTGTAATAATTCGTATAGAATTAAGAAGTGTAATAAGATTATTTTATAATGATCAATTATATAATAGAATTGTAACTATACATGCTTTTATTATAATTTTTTTTATAGTTATGCCTTTAATAATTGGGGGATTTGGGAATTGGTTAATCCCTTTAATATTAGGATTATCTGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGGTTACTAATTCCTTCAATTATTTTATTAATTATAGGAGGATTTGTTAATATGGGGGCTGGTACGGGATGAACAGTTTATCCTCCATTATCATTATTAATAGGTCATAGTGGAGTTTCAGTAGATTTATCTATTTTTTCTTTACATTTGGCAGGGGCTTCATCTATTATAGGTTCAATTAATTTTATTGTGACTATTATAAATACTTGATTGCATTTTAAATATATAGATAAATATCCATTATTTGTTTGATCAGTATTTATTACAACTATTTTATTATTATTATCATTACCAGTTTTAGCTGGTGCAATTACTATATTATTAAGAGATCGAAATTTAAATACAAGATTTTTTGATCCATCAGGGGGAGGGGATCCAGTATTATACCAGCATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Malaguenya, 10.956, -85.284, 221 meters, caterpillar collection date: 14/x/2013, wasp eclosion date: 30/x/2013. Depository:
Host data. Eulepte Janzen12 (Crambidae) feeding on Vitex cooperi (Lamiaceae).
Caterpillar and holotype voucher codes. 13-SRNP-79751, DHJPAR0053682.
Paratypes
None.
Etymology
Chelonus jennyphillipsae is named to honor Dr. Jenny Phillips of BioAlfa for her decades of enthusiastic taxonomic and administrative support of INBio, parataxonomists, and all matters biodiversity of Costa Rica coupled with intensely developing BioAlfa of Costa Rica.
Chelonus jeremydewaardi , sp. nov.
Diagnostics
BOLD:AAB1653. Consensus barcode. AATATTATATTTTTTATTTGGGATTTGAAGTGGAATAATAGGTYTATCTTTAAGTGTAATAATTCGTATAGAATTAAGAAGTGTAATRAGATTATTTTATAATGATCAATTATATAATAGAATTGTAACTATACATGCTTTTATTATAATTTTTTTTATAGTTATGCCTTTAATAATTGGGGGATTTGGAAATTGATTAATCCCTTTAATATTAGGATTATYTGATATAATTTTTYCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCAATTATTTTATTAATTATAGGAGGATTTGTTAATATAGGGGCTGGWACAGGGTGAACAGTTTATCCYCCATTATCATTATTAATAGGTCATAGTGGAGTTTCAGTAGATTTATCTATTTTTTCTTTACATTTGGCAGGGGYTTCATCTATTATAGGTTCAATTAATTTTATTGTGACTATTATAAATACTTGRYTACATTATAAATATATAGATAAATATCCGTTATTTGTTTGGTCAGTATTTATTACAACTATTTTATTATTATTATCATTACCAGTTTTAGCTGGTGCAATTACTATATTATTAAGAGATCGAAAYTTAAATACAAGATTTTTTGATCCATCAGGGGGAGGAGAYCCAGTATTATATCAGCATTTATTT.
Holotype ♂
Guanacaste, Sector Pitilla, Bullas, 10.987, -85.385, 440 meters, caterpillar collection date: 29/v/2014, wasp eclosion date: 15/vi/2014. Depository:
Host data. Syllepis marialis (Crambidae) feeding on Allophylus psilospermus (Sapindaceae).
Caterpillar and holotype voucher codes. 14-SRNP-70940, DHJPAR0055376.
Paratypes
Hosts = Crambidae: Asturodes fimbriauralis, Asturodes junkoshimurae, Ceratocilia sixolalis, Desmia benealisDHJ02, Desmia Janzen03, Desmia ploralisDHJ10, Desmia Solis100, Eulepte Solis15, Herpetogramma salbialis, Herpetogramma Solis10, Leucochromodes melusinalisDHJ02, Microthyris prolongalisDHJ02, Omiodes fulvicauda, Patania Solis03, Patania Solis04, Phostria cyrisalis, Psara obscuralisDHJ01, Salbia cassidalis, spiloBioLep01 BioLep311, spiloJanzen01 Janzen14DHJ02, Syllepis marialis. DHJPAR0029069, DHJPAR0029070, DHJPAR0029117, DHJPAR0035309, DHJPAR0035311, DHJPAR0035315, DHJPAR0036400, DHJPAR0037182, DHJPAR0037862, DHJPAR0037979, DHJPAR0037981, DHJPAR0037982, DHJPAR0037989, DHJPAR0039121, DHJPAR0039502, DHJPAR0040364, DHJPAR0040368, DHJPAR0042084, DHJPAR0042093, DHJPAR0042096, DHJPAR0042857, DHJPAR0042860, DHJPAR0045402, DHJPAR0045403, DHJPAR0045408, DHJPAR0046798, DHJPAR0046935, DHJPAR0048091, DHJPAR0048092, DHJPAR0048093, DHJPAR0048094, DHJPAR0048097, DHJPAR0048104, DHJPAR0048169, DHJPAR0048973, DHJPAR0050072, DHJPAR0051337, DHJPAR0052887, DHJPAR0052891, DHJPAR0053674, DHJPAR0053681, DHJPAR0055370, DHJPAR0056949, DHJPAR0058149, DHJPAR0062180, DHJPAR0062181, DHJPAR0062182, DHJPAR0062183, DHJPAR0062184, DHJPAR0062185, DHJPAR0062186, DHJPAR0062187, DHJPAR0062188, DHJPAR0062189, DHJPAR0062190, DHJPAR0062191. Depository:
Other material
A specimen registered in BOLD (NHM749343) from Belize shares the same BIN and is likely conspecific. It was not examined and is not part of the type series.
Etymology
Chelonus jeremydewaardi is named to honor Dr. Jeremy deWaard for his many years laboring in execution and administration of the field and laboratory actions performed by the Centre for Biodiversity Genomics to rapidly and accurately DNA barcode tens of thousands of Costa Rican insects.
Chelonus jessiehillae , sp. nov.
Diagnostics
BOLD:AAW4588. Consensus barcode. TATATTATATTTTATATTTGGAATTTGATGTGGGATATTAGGGTTATCTTTAAGATTAATGATTCGTATGGAATTAAGAAGAGTAATAAGTTTATTTTTTAATGATCAATTATATAATAGAATTGTTACTATACATGCTTTTGTAATAATTTTTTTTATAGTAATACCAGTAATAATTGGGGGGTTTGGTAATTGATTAGTTCCTTTAATATTAGGGTTATCTGATATAATTTTTCCACGAATGAATAACATAAGTTTTTGGTTATTAATTCCTTCAATTATTTTATTAATTATAGGAGGATTTGTAAATATAGGGGCAGGTACTGGGTGGACAGTTTATCCACCATTATCTTTATTAATAGGTCATAGAGGGGTATCTGTTGATTTATCAATTTTTTCTTTACATTTAGCAGGTATATCATCAATTATAGGTTCAATTAATTTTATTGTTACTATTGTTAATACATGATTATTAATTAATTATATAGATAAATATCCTTTATTTGTATGGTCAGTGTTTATTACTACAATTTTATTATTATTATCTTTACCGGTTTTAGCTGGTGCAATTACTATATTATTAAGAGATCGTAATTTAAATACTAGATTTTTTGATCCTTCAGGAGGAGGGGATCCAGTTTTATACCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Sendero Albergue Crater, 10.849, -85.328, 980 meters, caterpillar collection date: 10/viii/2010, wasp eclosion date: 31/viii/2010. Depository:
Host data. gelJanzen01 Janzen349 (Gelechiidae) feeding on Mikania banisteriae (Asteraceae).
Caterpillar and holotype voucher codes. 10-SRNP-4459, DHJPAR0042088.
Paratypes
None.
Chelonus jesusugaldei , sp. nov.
Diagnostics
BOLD:ABY3312. Consensus barcode. TATAATATATTTTGTATTTGGAATTTGATGTGGGATATTAGGTTTATCTTTGAGATTAATAATTCGTATAGAATTAAGAAGAGTAATAAGTTTATTTTATAATGATCAATTATATAATAGAATTGTTACTATACATGCTTTTGTAATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGTTTATCAGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGATTATTACTTCCTTCAATTATTTTATTAATTATAGGAGGATTTGTAAATATAGGAGCAGGTACTGGTTGAACAGTTTATCCACCATTATCTTTATTAATAGGTCATAGTGGAATTTCTGTTGATTTATCAATTTTTTCTTTACATTTAGCAGGTATATCTTCTATTATAGGTTCAATTAATTTTATTGTTACTATTATTAATACATGAATATTAATAAATTATATAGATAAATATCCTTTATTTGTATGATCAGTATTTATTACTACAATTTTATTATTATTATCTTTACCAGTTTTGGCTGGTGCTATTACAATGTTATTAAGAGATCGAAATTTAAATACAAGATTTTTTGATCCATCAGGAGGAGGTGATCCAGTGTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector El Hacha, Estación Los Almendros, 11.032, -85.528, 290 meters, caterpillar collection date: 01/iv/2011, wasp eclosion date: 08/v/2011. Depository:
Host data. Herpetogramma phaeopteralis (Crambidae) feeding on Scleria latifolia (Cyperaceae).
Caterpillar and holotype voucher codes. 10-SRNP-4459, DHJPAR0048100.
Paratypes
None.
Etymology
Chelonus jesusugaldei is named to honor Sr. Jesus Ugalde for his many years of dedicated administrative support to the national biodiversity inventory under way by INBio, the National Biodiversity Institute of Costa Rica, the collection from which now resides in the Museo Nacional de Costa Rica.
Chelonus jimlewisi , sp. nov.
Diagnostics
BOLD:AAF2610. Consensus barcode. TATATTATATTTTTTATTTGGAATTTGATCAGGCATATTAGGATTATCTTTAAGTTTAATAATTCGTATAGAACTAAGTAGGGTTATAAGATTATTTTATAATGATCAGTTATATAATAGGATTGTAACTATACATGCATTTGTAATAATTTTTTTTATAGTTATACCAGTAATAATTGGTGGATTTGGRAATTGATTAATTCCTTTAATATTAGGCTTATCTGATATAATTTTCCCTCGAATAAATAACATAAGATTTTGATTATTATTACCTTCAATTTTTTTATTAATTATAGGAGGATTTGTTAATATGGGGGCAGGAACTGGTTGAACRGTTTATCCACCATTATCTCTAATAATGGGACATAGGGGAATTTCTGTAGATTTATCAATTTTTTCTTTACATTTAGCAGGCATATCTTCAATTATAGGATCAATTAATTTTATTGTAACTATTATTAATACTTGATTGATTTATAATTATATGGATAAATATCCTTTATTTGTATGATCAGTATTTATTACTACAATTTTATTATTATTATCTTTACCAGTATTAGCTGGGGCTATTACTATATTATTAAGAGATCGAAATTTAAATACTAGGTTTTTTGATCCTTCAGGGGGAGGAGACCCAGTTTTATATCAACATTTRTTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Palomo, 10.962, -85.28, 96 meters, caterpillar collection date: 16/x/2012, wasp eclosion date: 01/xi/2012. Depository:
Host data. Neoleucinodes Janzen02 (Crambidae) feeding on Heliconia irrasa (Heliconiaceae).
Caterpillar and holotype voucher codes. 12-SRNP-68630, DHJPAR0051336.
Other material
Several specimens that were not examined but that are in BOLD (e.g., DHJPAR0054551) and in the Janzen and Hallwachs database are recorded from Neoleucinodes alegralis (Crambidae). Depository:
Chelonus jimmilleri , sp. nov.
Diagnostics
BOLD:ADM8275. Consensus barcode. TATATTATATTTTTTATTTGGAATTTGATCTGGTATATTAGGATTATCTTTAAGATTAATAATTCGTATAGAATTAAGAAGAGTAATAAGATTATTTTATAATGATCAATTATATAATAGTATTGTTACTATACATGCATTTGTAATAATTTTTTTTATGGTTATACCAGTAATAATTGGGGGATTTGGGAATTGGTTAGTTCCTTTAATATTAGGATTATCTGATATAATTTTTCCTCGAATAAATAATTTAAGATTTTGGTTGTTAGTGCCTTCAATTATTTTATTAATTATAGGTGGATTTGTAAATATGGGTGCAGGAACTGGTTGAACAGTTTATCCTCCATTATCTTTATTAATAGGTCATAGAGGAATTTCTGTTGATTTATCAATTTTTTCTTTACATTTGGCTGGTATATCTTCAATTATAGGTTCAATTAATTTTATTGTAACTATTATTAATACTTGATTAATAATTAATTATATAGATAAATATCCTTTATTTGTATGATCAGTATTTATTACTACAATTTTATTATTATTATCTTTACCAGTGTTAGCTGGTGCTATTACAATATTATTAAGAGATCGTAATTTAAATACTAGATTTTTTGATCCTTCAGGAGGGGGAGATCCAGTTTTATATCAGCATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Palomo, 10.9619, -85.2804, 96 meters, caterpillar collection date: 02/xi/2017, wasp eclosion date: 07/xii/2017. Depository:
Host data. Antaeotricha Janzen78 (Depressariidae) feeding on Hylaeanthe hoffmannii (Marantaceae).
Caterpillar and holotype voucher codes. 17-SRNP-46525, DHJPAR0062576.
Paratypes
Host = Antaeotricha Janzen78: DHJPAR0062577, DHJPAR0062578. Depository:
Etymology
Chelonus jimwhitfieldi , sp. nov.
Diagnostics
BOLD:ACC1273. Consensus barcode. TATATTATATTTTATATTTGGAATATGATGTGGGATTTTAGGTTTATCTTTAAGATTGATGATTCGGATAGAGTTAAGAAGTGTAATGAGTTTATTTTCTAATGATCAATTATATAATAGGATTGTCACTATACATGCTTTTGTAATAATTTTTTTTATGGTTATACCAATTATAATTGGCGGATTTGGAAATTGGTTAATTCCATTAATATTAGGATTATCAGATATGATTTTTCCTCGGATAAATAATATAAGATTTTGATTATTGGTTCCTTCAATTATATTATTAATTATAAGAGGATTTGTGAATATGGGAGCAGGGACTGGGTGGACTGTGTATCCTCCTTTGTCGTTGTTGATAGGTCATAGAGGTATTTCAGTAGATTTATCTATTTTTTCTTTGCATTTGGCAGGAGTTTCTTCAATTATAGGCTCAATTAATTTTATTGTAACTATTGTTAGTACTTGAATAAGTTTAAAATATATAGATAAATATTCGTTATTTGTTTGATCTGTGTTTATTACTACTATTTTATTATTATTATCTTTACCAGTGTTAGCTGGGGCAATTACTATATTATTAAGTGATCGAAATTTAAATACAAGTTTTTTTGATCCTTCTGGTGGGGGAGATCCTGTATTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Medrano, 11.016, -85.381, 380 meters, caterpillar collection date: 22/viii/2012, wasp eclosion date: 13/ix/2012. Depository:
Host data. Prenesta scyllalis (Crambidae) feeding on Forsteronia spicata (Apocynaceae).
Caterpillar and holotype voucher codes. 12-SRNP-72051, DHJPAR0050070.
Paratype
Host = Prenesta scyllalisDHJ01: DHJPAR0050073. Depository:
Etymology
Chelonus jimwhitfieldi is named to honor Dr. Jim Whitfield of the University of Illinois for his decades of mentorship to DHJ about Braconidae taxonomy and stimulus for their collection and rearing from ACG, while carrying out many braconid taxonomic projects for ACG specimens.
Chelonus johanvalerioi , sp. nov.
Diagnostics
BOLD:AAF2611. Consensus barcode. TATATTATATTTTATATTTGGAATTTGGKSCGGCATATTAGGYTTATCATTAAGATTAATAATTCGTATAGAATTAAGAAGTGTAATAAGATTATTTTATAATGATCAATTGTATAATAGAATTGTAACAATACATGCTTTTGTTATAATTTTTTTTATAGTTATGCCAGTGATAATTGGAGGATTTGGTAATTGGTTGGTTCCTTTAATATTAGGGTTATCAGATATAATTTTTCCTCGTATAAATAATATAAGATTTTGGTTACTAATTCCTTCAATTATTTTATTAATTATGGGTGGTTTTGTAAATATAGGGGCAGGTACAGGTTGGACAGTTTATCCACCTTTATCTCTATTAAGAAGACATAGAGGAATTTCGGTGGATTTATCTATTTTTTCTTTGCATTTAGCAGGGGCTTCTTCAATTATAGGGTCTATTAATTTTATTATTACTATTATTAATACATGGATAGTGTTAAGTAACATAGATAAATATTCTTTATTTGTTTGATCTGTTTTTATTACAACAATTTTATTGTTATTATCTTTGCCTGTTTTAGCAGGGGCTATYACTATATTATTAAGTGACCGAAATTTAAATACTAGTTTTTTTGATCCTTCTGGAGGGGGTGAYCCAGTGTTGTACCAACATTTATTT.
Holotype ♂
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.865, -85.415, 540 meters, caterpillar collection date: 19/viii/2010, wasp eclosion date: 13/ix/2010. Depository:
Host data. Ceratocilia Janzen02 (Crambidae) feeding on Cosmibuena grandiflora (Rubiaceae).
Caterpillar and holotype voucher codes. 10-SRNP-4589, DHJPAR0042090.
Paratypes
Hosts = Ceratocilia Janzen02, Phostria latiapicalis, Pilocrocis purpurascens (all Crambidae). DHJPAR0029142, DHJPAR0036388, DHJPAR0036728, DHJPAR0039119, DHJPAR0042087, DHJPAR0042092, DHJPAR0048106, DHJPAR0052096, DHJPAR0055381, DHJPAR0055397, DHJPAR0055398, DHJPAR0055951, DHJPAR0056967, DHJPAR0057408, DHJPAR0057409. Depository:
Chelonus johnburnsi , sp. nov.
Diagnostics
BOLD:ACJ4065. Consensus barcode. AATATTATATTTTATTTTTGGTATATGGGCAGGTATAATAGGTTTATCATTAAGAATAATGATTCGAATGGAATTAAGAAGTGTTATGAGATTATTTTTTAGGGATCAATTATATAATAGATTAGTAACTATACATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGGTTGGTGCCATTGATATTGGGTTTATCAGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCATCTTTATTTTTATTAATTATAGGTGGATTTGTAAATACAGGGGCTGGTACAGGATGAACTGTTTATCCTCCATTATCTTTATTAATAGGTCATGGAGGAGTTTCAGTTGATTTATCAATTTTTTCTTTACATTTAGCTGGTGCATCATCTATTATAGGATCTATAAATTTTATTGTTACTATTTTAAATACATGAATAAAATTAAAGTATATAGATAAATTTTCATTATTTATTTGGTCAGTATTAATTACTACTATTTTATTACTTTTGTCATTGCCTGTATTAGCTGGGGCAATTACAATATTGTTAAGAGATCGTAATTTAAATACTAGATTTTTTGATCCTTCAGGAGGGGGGGACCCTGTATTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Palomo, 10.962, -85.28, 96 meters, caterpillar collection date: 02/i/2013, wasp eclosion date: 05/ii/2013. Depository:
Host data. Spilomela pantheralis (Crambidae) feeding on Celtis iguanaea (Ulmaceae).
Caterpillar and holotype voucher codes. 13-SRNP-67036, DHJPAR0051324.
Paratypes
None.
Etymology
Chelonus johnburnsi is named to honor Dr. John Burns of the Smithsonian Institution for his decades of taxonomic projects and mentorship of DHJ and WH about the Hesperiidae of ACG, as well as happily accepting their DNA barcoding as part of their taxonomic discovery.
Chelonus johnnoyesi , sp. nov.
Diagnostics
BOLD:ACF7386. Consensus barcode. AATATTATATTTTATTTTTGGTATTTGATGTGGAGTTTTAGGTTTATCTTTAAGTTTATTAATTCGTATGGAATTAAGAAGGGTAATAAGTTTATTTATAAATGATCAATTGTATAATAGAGTTGTTACTATGCATGCTTTTGTAATAATTTTTTTTATGGTTATACCAGTTATAGTTGGTGGGTTTGGAAATTGATTAGTACCTTTAATATTAGGGCTATCTGATATAATTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTATTTCCTTCAATTATTTTATTAGTTATAGGTGGATTTATTAATATGGGGGTTGGAACAGGGTGAACTGTTTATCCCCCATTATCTTTAATGATGGGTCATAGGGGGATTTCCGTTGATTTATCAATTTTTTCTTTACATTTGGCTGGGATATCGTCAATTATAGGTTCTATTAATTTTATTGTTACAACAATTAATACATGATTAAAGGTTACTTATATAGATAAATATTCATTATTTATTTGATCTATTTTTATTACTACAATTTTATTGTTATTATCTTTACCTGTCTTAGCTGGGGCAATTACTATATTATTAAGTGATCGAAATTTAAATACTAGATTTTTTGATCCTTCAGGAGGAGGAGA--------.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 11-18/vi/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG05181-G09.
Paratypes
None.
Chelonus jorgebaltodanoi , sp. nov.
Diagnostics
BOLD:ACK4767. Consensus barcode. AATTCTTTATTTCATTTTTGGGATTTGATCTGGTGTAATAGGGTTATCTTTAAGTTTATTAATTCGTATGGAAYTAAGGAGAGTAATAAGATTAYTTATAAATGATCAATTATATAATAGTATTGTTACTATACATGCTTTTATTATAATTTTTTTTATGGTTATACCTGTAATAGTTGGGGGATTCGGAAATTGATTAATTCCTTTAATATTAGGTTTATCTGATATAATTTTTCCTCGTATAAATAATTTAAGATTTTGATTACTGATTCCATCAATTATTTTATTAYTATTAGGAGGYTTTACTAATACAGGGGCTGGAACAGGATGAACTGTGTATCCTCCATTATCTTTAATAATAGGACATAGAGGAATTTCAGTTGATTTGTCAATYTTTTCTTTGCATTTAGCTGGGGCTTCTTCTATTATAGGATCTATTAATTTTATTGTAACAACAATTAATACATGATTAAAAATAACTAAYATAGATAAATATTCTTTATTTGTATGATCTATTTTTATCACTACAATTTTACTTTTATTATCTTTACCAGTTTTAGCYGGGGCAATTACTATATTATTAAGAGATCGAAATTTAAATACTAGGTTTTTTGATCCTTCTGGTGGTGGGGACCCTGTATTATACCAGCATTTATTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-2, 10.7634, -85.335, 824 meters, 10‒17/iv/2014, Malaise trap PL12-2A. Depository:
Host data. None.
Holotype voucher code. BIOUG30966-F11.
Paratypes
Both Malaise-trapped. BIOUG08913-A09, BIOUG28821-H05. Depository:
Etymology
Chelonus jorgebaltodanoi is named to honor Sr. Jorge Baltodano (RIP) for his many years as a neighboring collaborator with ACG and for accepting the challenge of being the first President of the Comité Local for ACG; part of his ranch is now a permanent part of ACG thanks to his foresight and spirit for conservation.
Chelonus jorgehernandezi , sp. nov.
Diagnostics
BOLD:ADB2931. Consensus barcode. ATTCTTTATTTCATTTTTGGGATTTGATCTGGTRTAATAGGGTTATCTTTAAGTTTACTAATTCGTATGGAACTAAGAAGAGTAATAAGGTTACTTATAAATGATCAATTATATAATAGTATCGTTACTATGCATGCTTTTATTATAATTTTTTTTATGGTTATGCCTGTAATAGTTGGTGGATTTGGTAATTGATTAATTCCTTTAATATTAGGTTTATCTGATATAATTTTCCCCCGTATAAATAATTTAAGATTTTGATTATTGATTCCGTCAATTATTTTATTATTATTAGGAGGTTTTATTAATAYAGGGGCTGGAACAGGATGAACTGTGTATCCTCCATTATCTTTAATAATAGGACATAGAGGAATTTCAGTTGATTTGTCAATTTTTTCTTTGCATTTAGCTGGAGYTTCTTCTATTATAGGATCTGTTAATTTTATTGTAACAACAATTAATACATGATTAAAAATAACTAATATAGATAAATATTCTTTATTTGTATGATCTATTTTTATTACCACAATTTTACTTTTATTATCTTTACCAGTTTTAGCTGGGGCAATCACTATATTATTAAGAGACCGAAATTTAAATACTAGGTTTTTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 05‒12/xii/2013, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher codes: BIOUG29282-F08.
Paratype
Malaise-trapped. BIOUG29803-C11. Depository:
Chelonus josealfredohernandezi , sp. nov.
Diagnostics
BOLD:ADC6444. Consensus barcode. AATTCTTTATTTTATTTTTGGAATTTGATCTGGTATAATAGGGTTATCTTTAAGTTTATTAATTCGTATGGAATTA---AGAAGAGTAACAAGGTTATTTATAAATGATCAATTATATAATAGTATTGTTACTATACATGCTTTTGTAATAATTTTTTTTATAGTTATACCTGTAATAGTAGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGTTTGTCCGATATAATTTTTCCTCGTATGAATAATTTAAGATTTTGATTATTGATTCCATCAATTATTTTATTAATAATAGGAGGTTTTATTAATATAGGGGCTGGAACGGGATGAACTGTTTATCCCCCATTATCATTAATAATAGGACATAGGGGAATTTCAGTTGATTTATCAATTTTTTCTTTACATTTAGCTGGGGCTTCTTCTATTATAGGGTCAATTAATTTTATTGTTACAGTGATTAATACATGATTAAAAATAACTAATATAGATAAATATTCTTTATTTGTTTGATCTATTTTTATCACTACAATTTTACTTTTATTATCTTTACCAGTTTTAGCTGGAGCTATTACAATATTATTAAGAGACCGAAATTTAAATACTAGATTTTTTGATCCTTCGGGTGGGGGAGATCC-------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 15‒22/v/2014, Malaise trap PL12-9A. Depository:
Host data. None.
Holotype voucher code. BIOUG29216-H07.
Paratypes
None.
Chelonus josefernandeztrianai , sp. nov.
Diagnostics
BOLD:ACJ1335. Consensus barcode. AATTTTGTATTTTATTTTTGGAATATGATCAGGGATAATAGGATTATCATTAAGGTTATTAATTCGAATAGAATTAAGTATTGTGGGTAGATTATTTATAAATGATCAGTTATATAATAGAATTGTAACAATACATGCATTTATTATAATTTTTTTTATAGTCATACCAATTATAATTGGTGGTTTTGGGAATTGATTAATTCCTTTAATATTAGGATTACCAGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCAATTATTTTATTAATTATAGGGGGATTTGTAAATATAGGAGTTGGAACTGGTTGAACTGTATATCCACCTTTATCTTTATTAATAAGACATAGAGGTATTTCAGTAGATTTATCAATTTTTTCTTTGCATTTAGCTGGGATATCTTCAATTATAGGGTCTATTAATTTTATTGTTACAATTTTAAATACATGAATAAATAAAAAAATGATAGATAAATTTCCTTTATTTGTTTGATCAGTATTTATTACAACAATTTTATTATTATTATCTTTACCAGTTTTAGCTGGAGCAATTACAATGTTGTTAAGTGATCGAAATTTAAATACAAGATTTTTTGATCCTTCTGGTGGAGGAGATCCAGTATTATATCAACATTTATTT------.
Holotype ♂
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 02‒09/iv/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG07458-A09.
Paratypes
None.
Chelonus josehernandezcortesi , sp. nov.
Diagnostics
BOLD:ACR6579. Consensus barcode. ATATTATATTTTATTTTTGGTATATGATCTGGAATATTAGGTTTATCATTAAGTTTATTAATTCGAATAGAATTAAGAATTACGGGTAGATTATTTATAAATGATCAATTATATAATAGAATTGTAACTATACATGCTTTTATTATAATTTTTTTTATGGTTATACCAATTATAATTGGTGGTTTTGGGAATTGACTAATTCCACTAATATTAGGTTTACCTGATATGATTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATACCTTCGTTATTATTGTTATTATTAGGAGGATTTGTAAATATAGGAGTAGGAACAGGTTGAACAGTTTATCCACCATTATCTTTATTAATAGGTCATAGTGGAATTTCAGTTGATTTATCAATTTTTTCTTTACATTTAGCTGGGGCATCTTCAATTATAGGATCAATTAATTTTATTGTAACAATTATTAATACATGAATAAATAATAAATTTATAGATAAATTTCCATTATTTGTTTGATCAGTATTTATTACTACAATTTTGTTATTATTATCTTTACCAGTATTAGCTGGTGCAATTACAATATTATTAAGTGATCGAAATTTAAAT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 13‒20/ii/2014, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher code. BIOUG29681-C10.
Paratype
Malaise-trapped. BIOUG17842-H09. Depository:
Chelonus josemanuelperezi , sp. nov.
Diagnostics
BOLD:ACZ8757. Consensus barcode. AATATTATATTTTTTATTTGGTTTATGAAGAGGAATTTTAGGTTTGTCATTAAGTTTGTTAATTCGTATAGAATTAAGAATAGTTGGTAGATTATTTATAAATGATCAGCTATATAATAGAATTGTAACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTAATAATTGGAGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGGTTATCAGATATAATTTTTCCACGAATAAATAATATAAGATTTTGATTGTTAATTCCTTCTATTTTGTTATTAATTATAGGAGGATATGTGGGAACAGGGGTAGGTACTGGTTGAACTGTTTATCCACCTTTATCTTTATTAATTGGCCATAGTGGAATTTCTGTAGATTTATCTATTTTTTCTTTACATTTAGCTGGAGTATCATCAATTATGGGATCTGTAAATTTTATTGTTACTATTATAAATACTTGAATAGAAAAAATAAACATGGATAAATATTCTTTATTTATTTGATCTGTTTTTATTACTACTATTTTATTATTATTATCTTTACCTGTGTTAGCAGGTGCTATTACTATATTATTAAGTGATCGTAATTTGAATACAAGATTTTTTGATCCTTCAGGTGGGGGGGATCCAGTATTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.88, -85.389, 575 meters, 29/xii/2014‒05/i/2015, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28039-F05.
Paratypes
All Malaise-trapped. BIOUG27759-B05, BIOUG28037-E12, BIOUG28039-G01. Depository:
Chelonus josephinerodriguezae , sp. nov.
Diagnostics
BOLD:ADG0772. Consensus barcode. ATATTATATTTCATTTTTGGTTTATGAAGCGGAATTTTAGGTTTATCATTAAGTTTATTAATTCGAATAGAATTAAGAATAGTAGGAAGTTTATTTTTAAATGATCAATTATATAATAGAATTGTAACAATACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGGAATTGATTAGTTCCATTAATATTAGGATTATCTGATATAATTTTTCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCAATTTTATTATTAATTTTAGGAAGATATGTAGGTACTGGAGCAGGAACAGGATGAACTGTATATCCACCATTATCTTTATTAATAGGTCATAGAGGAATATCTGTAGATTTATCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATTATAGGTTCAATTAATTTTATTGTGACAATTATAAATACTTGAATATTAAAAAAAAATATAGATAAATATTCATTATTTATTTGATCTGTTTTTATTACAACTATTTTATTGTTATTATCTTTACCTGTATTAGCTGGTGCAATTACAATATTATTAAGTGATCGTAAT----------------------------.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, 25/xii/2014‒01/i/2015, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG31680-G05.
Paratypes
None.
Chelonus juanmatai , sp. nov.
Diagnostics
BOLD:ABY3313. Consensus barcode. AATATTATATTTTTTATTTGGTTTATGAAGAGGAATTTTAGGTTTGTCATTAAGTTTGTTAATTCGTATAGAATTAAGAATAGTTGGTAGATTATTTATAAATGATCAGCTATATAATAGAATTGTAACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTAATAATTGGAGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGGTTATCAGATATAATTTTTCCACGAATAAATAATATAAGATTTTGATTGTTAATTCCTTCTATTTTGTTATTAATTATAGGAGGATATGTGGGAACAGGGGTAGGTACTGGTTGAACTGTTTATCCACCTTTATCTTTATTAATTGGCCATAGTGGAATTTCTGTAGATTTATCTATTTTTTCTTTACATTTAGCTGGAGTATCATCAATTATGGGATCTGTAAATTTTATTGTTACTATTATAAATACTTGAATAGAAAAAATAAACATGGATAAATATTCTTTATTTATTTGATCTGTTTTTATTACTACTATTTTATTATTATTATCTTTACCTGTGTTAGCAGGTGCTATTACTATATTATTAAGTGATCGTAATTTGAATACAAGATTTTTTGATCCTTCAGGTGGGGGGGATCCAGTATTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal, Finca San Gabriel, 10.878, -85.393, 645 meters, caterpillar collection date: 13/viii/2011, wasp eclosion date: 29/viii/2011. Depository:
Host data. immidjanzen01 Janzen23 (Immidae) feeding on Pentagonia donnell-smithii (Rubiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-3192, DHJPAR0048103.
Paratypes
None.
Chelonus junkoshimurae , sp. nov.
Diagnostics
BOLD:AAF2633. Consensus barcode. AATATTATATTTTATTTTTGGTATATGATGTGGTATACTTGGATTATCATTAAGTATATTAATTCGAATAGAATTAAGTATAGTTTCAAGTTTATTAATAAATGATCAATTATATAATAGAATTGTTACAATACATGCATTTGTAATAATTTTTTTTATGGTTATACCARTTATAATTGGTGGATTTGGGAATTGGTTAATTCCATTAATATTAGGATTATCAGATATAATATTTCCACGAATAAATAATATAAGATTTTGATTGTTAGTACCGTCAATTATATTATTAATTTTAAGAGGTTTTATTAATACTGGTGTTGGAACTGGATGAACAGTTTATCCCCCATTATCTTTATTAATAGGTCATGGGGGAATTTCTGTAGATTTATCAATTTTTTCTTTACATTTAGCAGGGGCTTCATCAATTATAGGTTCAATTAATTTTATTGTTACAGTAATAAATTCTTGAATAAAATTAAGTTATATAGATAAATTTGCTTTATTTGTTTGATCAGTTTTTATTACAACAATTTTATTATTATTATCTTTACCGGTATTAGCAGGAGCTATTACTATATTATTAAGGGATCGAAATTTAAATACAAGATTTTTTGATCCTTCAGGAGGAGGAGATCCTGTATTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Orosi, Maderos, 11.005, -85.475, 510 meters, caterpillar collection date: 19/vi/2010, wasp eclosion date: 07/vii/2010. Depository:
Host data. Diaphania hyalinataDHJ02 (Crambidae) feeding on Gurania makoyana (Cucurbitaceae).
Caterpillar and holotype voucher codes. 10-SRNP-21385, DHJPAR0040366.
Paratypes
Hosts = Diaphania hyalinataDHJ01, Diaphania hyalinataDHJ02. BIOUG17755-H07, DHJPAR0021626, DHJPAR0029175, DHJPAR0029176, DHJPAR0039504, DHJPAR0040367, DHJPAR0040373, DHJPAR0040374. Depository:
Chelonus kateperezae , sp. nov.
Diagnostics
BOLD:ACR8387. Consensus barcode. TATTTTATGTTTGGTATATGATCAGGTATATTAGGTTTATCTTTAAGATTAATTATTCGAATAGAATTAGGAACTTTAAAAACTTTATTGTTTAATGATCAATTATATAATAGATTAGTTACTTTACATGCTTTTGTAATAATTTTTTTTATGGTTATGCCAGTAATAATTGGTGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGTTTATCAGATATATTATTTCCACGTATAAATAATATAAGTTTTTGATTATTAGTGCCATCAATGATTTTATTATTAATAAGTGGTTTTGTAAATGTTGGAGTTGGTACAGGTTGAACGGTTTATCCTCCTTTATCTTCTTTAATTGGTCATAGGGGTGTATCGGTTGATATATCGATTTTTTCTTTACATTTAGCAGGGATATCATCAATTATGGGAGCAATTAATTTTATTGTAACTGTTATAAATACTTGATTAAAATATAATTTTATAGATAAATTTCCTTTATTTGTTTGATCAGTGTATATTACAGCTATTTTATTATTATTGTCTTTACCAGTTTTAGCAGGGGCTATTACTATATTGTTAAGGGAT---------------------------------------------------.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 04‒11/vi/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG17967-C10.
Paratypes
None.
Etymology
Chelonus kateperezae is named to honor Ms. Kate Perez for her many years laboring in execution and administration of the field and laboratory actions performed by the Centre for Biodiversity Genomics to rapidly and accurately DNA barcode tens of thousands of Costa Rican insects from Malaise traps installed by ACG and BioAlfa.
Chelonus luciariosae , sp. nov.
Diagnostics
BOLD:ACM2853. Consensus barcode. TACTTTATATTTAATTTTTGGTATATGATCAGGTATATTTGGTTTATCTTTAAGTTTATTAATTCGTTTAGAATTAAGTGGATTAATAAGATTTTTTAATAATGATCAATTATATAATAGAATAATTACTTTACATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATAGGTGGTTTTGGAAATTGATTAATTCCAATAATATTGGGTTTAATAGATATAGCTTTTCCTCGATTAAATAATTTAAGGTTTTGATTATTAATTCCTTCAATTATATTATTAGTTATAAGTAGATTTGTAAATAGTGGTTCTGGTACAGGTTGAACAGTTTATCCTCCATTATCTTCATTAATAGGTCATGGGGGGGTATCAGTAGATATAACTATTTTTTCTTTACATTTAGCAGGTGTGTCTTCAATTTTAAGATCAATTAATTTTATTGTAACTATTTTAAATAGTTGAATAAATGTAAAGTATATAGATAAATTTCCTTTATTTATTTGATCTGTTTTTCTTACAGCAATTTTATTATTATTATCTTTACCTGTATTAGCAGGGGCAATTACTATATTATTAAGAGATCGAAATATAAATACTAGATTTTTTGATTCTTCAGGAGGAGGTGATCCTATTTTGTATCAGCATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal, Jardin Estrada, 10.865, -85.397, 722 meters, caterpillar collection date: 30/xii/2013, wasp eclosion date: 24/i/2014. Depository:
Host data. chryBioLep01 BioLep174 (Pyralidae) feeding on Mansoa hymenaea (Fabaceae).
Caterpillar and holotype voucher codes. 13-SRNP-7596, DHJPAR0054557.
Paratypes
Host = chryBioLep01: BioLep174, DHJPAR0054549, DHJPAR0054550, DHJPAR0054552, DHJPAR0054555, DHJPAR0054559, DHJPAR0054561, DHJPAR0054563, DHJPAR0054567, DHJPAR0054782. Depository:
Chelonus luzmariaromeroae , sp. nov.
Diagnostics
BOLD:AAM1444. Consensus barcode. GACATTATATTTTTTATTTGGTATATGATCAGGGTTTTTTGGTTTATCATTAAGAATATTAATTCGTATAGAATTAAGTTCTTTAGGTAGATTATTAATAAATGATCAATTGTATAATAGAATTGTAACTATGCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGTAATTGGATAGTTCCTTTAATGTTAGGTATATGTGATATAATATTTCCTCGAATAAATAATATAAGATTTTGGATATTAATACCTTCTTTAATAATATTAATTTTAGGAAGGTTAGTAAATATGGGTGTTGGGACAGGATGAACAGTTTATCCTCCTTTATCTTTATTAATAGGACATGGGGGGATYTCTTTAGATTTATCAATTTTTTCTTTACATTTAGCAGGGATTTCTTCAATTTTAGGTTCTATTAATTTTATTGTAACTATTTTAAATACTTGRATAGAAAAAAAATTTTTGGATATATTTCCTTTATTTATTTGATCAGTATTTATTACAACAATTTTATTATTATTATCATTACCAGTATTAGCTGGCGCAATTACTATACTTTTAAGTGATCGTAATTTAAATACAAGATTTTTTGATCCTGCTGGTGGAGGGGATCCTGTATTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal, Tajo Angeles, 10.865, -85.415, 540 meters, caterpillar collection date: 12/viii/2011, wasp eclosion date: 28/viii/2011. Depository:
Host data. Dichomeris Janzen703 (Gelechiidae) feeding on Koanophyllon hylonoma (Asteraceae).
Caterpillar and holotype voucher codes. 11-SRNP-3178, DHJPAR0048167.
Paratypes
Hosts = Dichomeris Janzen703, Dichrorampha Janzen322. DHJPAR0037974, DHJPAR0037975. Depository:
Etymology
Chelonus luzmariaromeroae is named to honor Sra. Luz Maria Romero for her decades of conducting and developing the ACG Biological Education Program, followed by more than a decade of mentoring the parataxonomists as to how to better manage and polish their field databases and their Species Pages on the ACG web site.
Chelonus manuelpereirai , sp. nov.
Diagnostics
BOLD:ACP7116. Consensus barcode. AACATTATATTTTTTATATGGAATATGATCTGGATTTTTTGGTCTATCACTAAGTATATTAATTCGAATAGAATTAAGATCTTTAGGAAGATTTTTATTAAATGATCAATTATATAATAGAATTGTTACAATACATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAGTAGGTGGTTTTGGAAATTGAATAGTTCCTATAATATTAGGTTTACCAGATATAATTTTTCCACGAATAAATAATTTAAGATTTTGATTATTAATTCCATCATTAATTTTATTAATTTTAAGTAGTTTAGTTAATTCAGGAGTTGGTACAGGTTGAACCGTTTATCCACCTTTATCATTATTAATAGGTCATAGAGGAATTTCAGTTGATTTATCAATTTTTTCTTTACATTTAGCTGGAGCATCTTCAATTTTAGGATCAATTAATTTTATTGTAACTACATTAAATACTTGAATAAAAAAAAATATTATAGATACTTATCCATTATTTATTTGATCAATTTTTATTACTACAATTTTATTATTATTATCTTTACCTGTTTTAGCTGGTGCAATTACAATATTATTAAGAGATCGAAATTTAAATACAAGATTTTTTGACCCAGCAGGTGGGGGAGATCCTGTACTTTATCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Sendero Juntas, 10.907, -85.288, 400 meters, caterpillar collection date: 30/iv/2014, wasp eclosion date: 12/v/2014. Depository:
Host data. gelJanzen01 Janzen501 (Gelechiidae) feeding on Gouania polygama (Rhamnaceae).
Caterpillar and holotype voucher codes. 14-SRNP-42146, DHJPAR0055499.
Paratypes
None.
Chelonus manuelzumbadoi , sp. nov.
Diagnostics
BOLD:ACW6597. Consensus barcode. GTTTTATATTTTATTTTTGGAATATGGAGGGGGGTTTTGGGTTTGTCTTTAAGATTAATTTTACGAATAGAATTAAGTGTTTTAGGTAGATTAATTAAAAATGATCAATTATATAATAGTGTAGTAACTATACATGCTTTTGTAATAATTTTTTTTATGGTTATACCAGTTATAATTGGTGGTTTTGGATTAGTTCCATTAATGTTGGGTTTGCCAGATATAGCATTTCCTCGAATGAATAATATAAGATTTTGATTATTAATTCCTTCATTATTATTATTATTATTAGGGGGGTTTATTAATAGAGGAGTGGGAACAGGGTGAACTGTATATCCTCCATTATCGTCATTAATTGGTCACGGTGGAATTTCTGTAGATTTATCAATTTTTTCTTTACATTTAGCAGGGATATCTTCAATTATAGGAGCTATTAATTTTATTGTAACAGTAATAAATATAAGTTTTGATTTAAAATGTATAGATAAATATTCATTATTTGTATGGTCAGTTTTGATTACAGCGGTTTTATTGTTGTTATCATTACCTGTTTTGGCTGGTGCAATTACTATATTATTAAGTGAT.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.3889, 575 meters, 23‒30/xii/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG23153-E04.
Paratypes
None.
Chelonus marianopereirai , sp. nov.
Diagnostics
BOLD:ADC0083. Consensus barcode. TTTATTTTTGGAATTTGAGCAGGAGTTTTAGGTTTATCTTTAAGATTAATTTTACGAATAGAATTAAGTGTATTAGGAAGCTTATTAAAAAATGATCAATTATATAATAGTGTGGTTACTATACATGCTTTTGTAATAATTTTTTTTATGGTGATACCAATTATGATTGGGGGATTTGGTAATTGATTAATTCCATTAATATTAGGATTACCTGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATATTATTGTTATTAAGAAGATTTGTAAATACAGGTGTAGGAACTGGATGAACAGTTTATCCACCATTATCATCATTAATAGGTCATGGAGGAATTTCCGTGGATTTATCAATTTTTTCTTTACATTTAGCAGGTATATCTTCAATTATAGGAGCAATTAATTTTATTGTTACAGTTTTAAATACAAATTTTGATATAAATTATATAGACAAATTTCCATTATTTGTATGATCTGTATTAATTACAGCAATTTTATTATTATTGTCATTACCTGTTTTAGCTGGTGCAATTACTATA------------------------------------------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 06‒13/xi/2014, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher codes: BIOUG30255-D09.
Paratypes
None.
Chelonus maribellealvarezae , sp. nov.
Diagnostics
BOLD:AAM1066. Consensus barcode. AGTATTATATTTTATTTTTGGKATATGATCGGGGGTTTTGGGTTTATCTYTAAGGTTAATAATTCGTATAGAATTRAGTATATTAGGTAGTATATTTATAAATGATCAATTATATAATAGAATTGTTACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTACCATTAATATTAGGTTTACCAGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGATTAYTAATTCCTTCTTTATTATTATTAATTATAAGAAGTTTYGTAAATACAGGTGTAGGAACTGGATGAACAGTTTATCCTCCATTATCTTTATTAATAGGTCATAGGGGTATTTCAGTAGATTTATCAATTTTTTCTTTACATTTAGCAGGTATRTCTTCAATTATRGGGTCAATTAATTTTATTGTTACTGTYTTAAATACATGAATAAATAATAAATTAATAGATAARTTTTCTTTATTTGTTTGATCRATTTTTATTACAACAATTTTATTATTGTTATCTTTACCTGTTTTAGCGGGRGCTATTACTATATTAYTAAGGGATCGAAATTTAAATACTAGATTTTTTGATCCTTCTGGAGGGGGGGATCCTATTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Mundo Nuevo, Vado Miramonte, 10.772, -85.434, 305 meters, caterpillar collection date: 18/xii/2009, wasp eclosion date: 31/xii/2009. Depository:
Host data. Antaeotricha Janzen221 (Depressariidae) feeding on Desmodium procumbens (Fabaceae).
Caterpillar and holotype voucher codes. 09-SRNP-58092, DHJPAR0038214.
Paratype
Host = elachJanzen01 Janzen38: DHJPAR0051929.
Chelonus markmetzi , sp. nov.
Diagnostics
BOLD:AAA4444. Consensus barcode. TATATTATATTTTATKTTTGGTATATGGGCTGGATTTTTGGGATTATCATTAAGATTATTAATTCGGATAGAATTGAGAGTGCTAGGYAGAGTATTTACTAATGAYCAATTATATAATAGAATTGTAACTATACATGCTTTTGTAATAATTTTTTTTATGGTTATACCAATTATGATTGGTGGATTTGGRAATTGGTTGGTACCTTTGATATTAGGATTACCGGATATAATTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCATCTTTAATATTATTAATATTAGGGGGATTTGTCAATATAGGGGTAGGTACTGGTTGAACAGTTTACCCCCCATTATCTTTATTAATAGGTCATAGAGGAGTTTCTGTTGATTTATCAATTTTTTCTTTACATATAGCGGGAGTYTCATCTATTATAGGATCRGTAAATTTTATTGTAACTATTTTAAATACTTGAATAAAAAATGATTTTATAGATAAATTTCCTTTATTTATTTGATCTGTATTTATTACTACTATTTTATTATTATTATCGTTACCWGTATTAGCAGGGGCTATTACTATATTATTAAGTGAYCGTAATTTAAATACAAGATTTTTTGATCCATCAGGTGGGGGGGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Mundo Nuevo, Estación La Perla, 10.767, -85.433, 325 meters, caterpillar collection date: 18/v/2011, wasp eclosion date: 31/v/2011. Depository:
Host data. Herpetogramma phaeopteralis (Crambidae) feeding on Axonopus fissifolius (Poaceae).
Caterpillar and holotype voucher codes. 11-SRNP-5549, DHJPAR0045350.
Paratypes
Hosts = Herpetogramma phaeopteralis. DHJPAR0045335, DHJPAR0045355, DHJPAR0055996, DHJPAR0055997, DHJPAR0056052, BIOUG17653-H06, BIOUG18660-H04. Depository:
Other material
Specimens from Mexico (BIOUG19755-A08, BIOUG20526-D01) share the same BIN on BOLD and may be conspecific. They were not examined.
Chelonus markshawi , sp. nov.
Diagnostics
BOLD:ACL6642. Consensus barcode. GATACTTTATTTTATTTTTGGTATTTGATCAGGAATATTAGGTTTATCTTTGAGAATAATTATTCGTTTAGAGTTAATTTATGTTGGGAGATTTTTAACAAATGATCAGTTATATAATAATATTGTAACTATACATGCATTTATTATAATTTTTTTTATAGTAATGCCAGTAATAATTGGGGGGTTTGGAAATTGATTGGTTCCTTTAATATTAGGATTATCAGATATATCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCAATTTTAATAATGTTAATAAGGGGGTTTGTAAATATGGGTGCAGGAACTGGATGAACAGTGTATCCTCCTTTATCTTTAAGTATGGGTCATAGTGGTATTTCTGTAGATATAGTAATTTTTTCTTTACATTTAGCTGGGATATCTTCTATTATGGGTGCTATTAATTTTATTACTACTGTAATGAATACTTGAATAAATGTAAAATTAATAGATAAATTTCCTCTATTTATTTGATCTGTTTATATTACAGCAATTTTATTATTGTTATCATTGCCTGTGTTGGCAGGAGCAATTACTATATTATTAAGAGATCGTAATATAAATACAAGATTTTTTGATTCATCTGGTGGTGGAGATCCTGTATTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Quebrada Escondida, 10.899, -85.275, 420 meters, caterpillar collection date: 01/vii/2013, wasp eclosion date: 04/ix/2013. Depository:
Host data. Stenoma Janzen142 (Depressariidae) feeding on Calyptrogyne trichostachys (Arecaceae).
Caterpillar and holotype voucher codes. 13-SRNP-42923, DHJPAR0053672.
Paratypes
None.
Chelonus martajimenezae , sp. nov.
Diagnostics
BOLD:ADA0878. Consensus barcode. GTATTATATTTTTTATTTGGATTATGATCAGGTGTAATAGGTTTATCATTAAGAATAATTATTCGTTTTGAGTTAATATATCCTGGAGCATTTATAGGAAATGATCAATTATATAATAGAGTTGTTACTATACATGCATTTGTAATGATTTTTTTTATAGTTATACCGGTAATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGTTTACCGGATATAGCTTTTCCACGAATAAATAATATAAGGTTTTGATTATTAATACCTTCAATATTATTYTTAATTATAGGTAGATTATTAAATAGAGGCACTGGTACTGGATGAACCGTTTATCCACCTTTATCTTTAATTATAGGTCATAGTGGTATTTCAGTTGATTTTACAATTTTTTCTTTACATTTGGCTGGAATATCATCAATTATAGGTGCTTTAAATTTTATTGTTACTATTATTAATACTTGAATAAGAAAAATTTATATAGATAAATTTTCTTTRTTTGTATGATCAGTATTTATTACAGCAATTTTGTTATTAATATCTTTACCTGTTTTAGCTGGGGCAATTACTATATTATTAAGTGATCGTAATATTAAT.
Holotype ♂
Alajuela, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, 25/viii‒01/ix/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG27764-E10.
Paratypes
None.
Chelonus mayrabonillae , sp. nov.
Diagnostics
BOLD:ADB0889. Consensus barcode. GTATTATATTTTTTATTTGGATTATGATCAGGTGTAATAGGTTTATCATTAAGAATAATTATTCGTTTTGAGTTAATATATCCTGGAGCATTTATAGGAAATGATCAATTATATAATAGAGTTGTTACTATACATGCATTTGTAATGATTTTTTTTATAGTTATACCGGTAATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGTTTACCGGATATAGCTTTTCCACGAATAAATAATATAAGGTTTTGATTATTAATACCTTCAATATTATTYTTAATTATAGGTAGATTATTAAATAGAGGCACTGGTACTGGATGAACCGTTTATCCACCTTTATCTTTAATTATAGGTCATAGTGGTATTTCAGTTGATTTTACAATTTTTTCTTTACATTTGGCTGGAATATCATCAATTATAGGTGCTTTAAATTTTATTGTTACTATTATTAATACTTGAATAAGAAAAATTTATATAGATAAATTTTCTTTRTTTGTATGATCAGTATTTATTACAGCAATTTTGTTATTAATATCTTTACCTGTTTTAGCTGGGGCAATTACTATATTATTAAGTGATCGTAATATTAAT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 03‒10/vii/2014, Malaise trap PL12-9A. Depository:
Host data. None.
Holotype voucher code. BIOUG29390-E08.
Paratypes
All Malaise-trapped. BIOUG28728-G11, BIOUG28876-C11, BIOUG29391-H05, BIOUG29395-G08, BIOUG29397-E02, BIOUG29501-H09, BIOUG29509-E11. Depository:
Chelonus meganmiltonae , sp. nov.
Diagnostics
BOLD:ADA8853. Consensus barcode. ATATTATATTTTATTTTTGGAATTTGGTCTGGTATTTTGGGTTTATCATTAAGTATAATAATTCGAATAGAATTAAATTGTACTGGAAGTTTGTTTATTAATGATCAATTATATAATAGATTAGTAACTATACATGCGTTTGTAATAATTTTTTTTATGGTTATACCAGTAATAATTGGTGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGATTATCTGATATAGCTTTTCCTCGAATAAATAATATGAGTTTTTGATTATTAATTCCTTCAATTTTTATATTAATTATTAGTAGATTTGTAAATATAGGTGTTGGTACTGGTTGGACTGTATATCCACCTTTATCTTCATTATTAGGTCATAGTGGTGTATCTGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGAATATCATCAATTATAGGTGCTATTAATTTTATTGTTACTATTTTAAATACATGAATAATAAATTTATTAATAGATAAATTTCCATTATTTGTTTGATCAGTATTTATTACTGCTTTATTATTATTATTATCATTACCTGTATTAGCTGGTGCAATTACTATATTATTAAGAGATCGTAAT---------------------------------------.
Holotype ♂
Guanacaste, Pailas Dos, PL12-1, 10.7642, -85.335, 828 meters, 20‒27/ii/2014, Malaise trap PL12-1A. Depository:
Host data. None.
Holotype voucher code. BIOUG28769-G03.
Paratypes
None.
Etymology
Chelonus meganmiltonae is named to honor Sra. Megan Milton for her many years laboring in execution and administration of the field and laboratory actions performed by the Centre for BioDiversity Genomics to rapidly and accurately DNA barcode tens of thousands of Costa Rican insects found in ACG.
Chelonus melaniamunozae , sp. nov.
Diagnostics
BOLD:AAM1107. Consensus barcode. AATTTTATACTTTATTTTTGGAATATGATCAGGTATATTAGGTTTATCTTTAAGTATAATAATTCGTTTAGAATTAGCTTTTACTGGAAGTTTATTTATTAATGATCAATTATATAATTTATTAGTAACTTTACATGCTTTTGTAATAATTTTTTTTATAGTAATGCCATTAATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGTTTACCTGATATAGCTTTTCCTCGAATGAATAATATAAGATTTTGACTATTAATTCCTTCAATTTTTTTATTAATTATAAGAAATTTTGTTAATATAGGAGTAGGTACTGGATGAACAGTATATCCTCCATTATCTTCTTTAATAGGTCATAGAGGTATTTCTGTTGATTTGGCTATTTTTTCTTTACATTTGGCAGGAATATCTTCTATTATAGGAGCTATTAATTTTATTGTAACTGTAATTAATWCTTGAATAAAAAATTTATATATAGATAAATTATCTTTATTTGTTTGATCTGTTTTTATTACTGCAATTTTATTATTATTATCATTACCTGTATTAGCAGGAGCTATTACTATATTATTAAGAGATCGTAATATAAATACAAGATTTTTTGATTCATCTGGGGGAGGAGATCCTATTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Pitilla, Sendero Mismo, 10.988, -85.42, 680 meters, caterpillar collection date: 10/iii/2010, wasp eclosion date: 25/iii/2010. Depository:
Host data. gelJanzen01 Janzen758 (Gelechiidae) feeding on Coccoloba porphyrostachys (Polygonaceae).
Caterpillar and holotype voucher codes. 10-SRNP-30674, DHJPAR0039043.
Paratypes
Hosts = gelJanzen01 Janzen758. DHJPAR0039052, DHJPAR0039064, DHJPAR0039065. Depository:
Chelonus michaelstroudi , sp. nov.
Diagnostics
BOLD:ACR3626. Consensus barcode. TATATTATATTTTATTTTTGGAATATGATCTGGAATATTTGGTTTATCATTGAGTATAATTATTCGTTTAGAATTAATTTATCCTGGAAGT---TTATTAATGAATGATCAATTGTATAATAGG------GTAATTACTATACATGCTTTTGTAATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGTTTTGGAAATTGATTAATTCCTTTAATGTTGGGTTTACCGGATATAGCATTTCCTCGTATAAATAATATAAGATTTTGATTATTAATCCCTTCATTAATTATATTAATTATAGGAGGTTTTATTAATATAGGAGCAGGTACTGGTTGAACTATTTATCCTCCATTATCATCATTAATAGGACATAGTGGAATATCTGTAGATTTATCTATTTTTTCTTTGCATTTAGCTGGAATATCTTCAATTATAGGTGCTATTAATTTTATTGTTACAGTATTAAATACTTGAATAGATATT-AAATTTATAGATAAATATCCATTATTTGTATGATCAGTATTTATTACAGCTTTTTTATTATTATTATCATTACCTGTATTAGCAGGAGCAATTACTATATTATTAAGGGATCGAAATATAAATACAAGATTTTTTGATTCTTCTGGAGGGGGAGATCCAGTTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Pitilla, Medrano, 11.016, -85.381, 380 meters, caterpillar collection date: 11/vi/2014, wasp eclosion date: 26/vi/2014. Depository:
Host data. gelJanzen01 Janzen485 (Gelechiidae) feeding on Rinorea deflexiflora (Violaceae).
Caterpillar and holotype voucher codes. 14-SRNP-71020, DHJPAR0056003.
Paratypes
None.
Chelonus michellevanderbankae , sp. nov.
Diagnostics
BOLD:AAT8838. Consensus barcode. TGTTTTATATTTTATTTTTGGAATATGATCAGGTATATTAGGATTATCTTTAAGATTATTAATTCGTATGGAATTGAGAATAACTGGAAGATTATTAATAAATGATCAATTATATAATAGTATTGTTACATTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGTAATTGATTAATTCCTTTAATATTAGGGTYACCTGATATAGCTTTTCCWCGAATAAATAATATAAGATTTTGGTTATTAATTCCGTCAATTTTTTTATTGATTTTAGGTGGATTTGTTAATATAGGAGCTGGGACTGGTTGAACAGTTTATCCTCCATTATCTTTATTAATTGGTCATAGTGGAGCTTCAGTGGATATATCAATTTTTTCATTACATTTGGCTGGAATATCTTCAATTATAGGAGCAATTAATTTTATTGTAACAGTAATTAATACATGAATAGATATAAAATTAATAGATAAATTTTCTTTATTTGTTTGATCAGTATTTATTACTGCAATTTTATTATTGTTGTCTTTGCCTGTATTAGCTGGTGCTATTACTATATTATTAAGAGATCGTAATTTAAATACAAGATTTTTTGATCCATCTGGTGGGGGGGATCCTGTTTTATATCAGCATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Mismo, 10.988, -85.42, 680 meters, caterpillar collection date: 16/ii/2010, wasp eclosion date: 08/iii/2010. Depository:
Host data. Gregarious parasitoid of Anacrusis nephrodes (Tortricidae) feeding on Zygia palmana (Fabaceae).
Caterpillar and holotype voucher codes. 10-SRNP-30487, DHJPAR0038977.
Paratypes
Host = Anacrusis nephrodes: DHJPAR0029134, DHJPAR0029135, DHJPAR0039447, DHJPAR0040018, DHJPAR0040021, DHJPAR0040406, DHJPAR0047069, DHJPAR0049118, DHJPAR0050964, DHJPAR0055415. Depository:
Chelonus mingfangi , sp. nov.
Diagnostics
BOLD:ADA5459. Consensus barcode. ATTTTATATTTTTTATTTGGTATATGGTCAGGAATATTAGGATTATCTTTAAGAATATTAATTCGTATAGAATTAAGGTTTTCTGGAAGGTTATTTTTAAATGATCAATTATATAATAGGATTGTTACTATACATGCTTTTGTTATAATTTTTTTTATAGTGATACCAATTATAATTGGGGGATTTGGAAATTGATTAATTCCTTTAATATTAGGATTACCTGATATAGCTTTTCCTCGAATAAATAATATAAGATATTGATTATTAATTCCTTCTTTATTAATATTAATAATAAGTGGATTTATTAATATAGGAGTAGGTACTGGATGAACTATTTATCCTCCTTTATCATTATTAATAGGTCATGGAGGTATTTCTGTGGATATATCAATTTTTTCTTTACATTTAGCTGGGATATCTTCTATTATAGGAGCTATTAATTTTATTATTACTATTATTAATACTTGAATAGATATTAAATTTATAGATAAGTTTCCTTTATTTGTTATGTCAGTTTTTATTACTGCTATTTTATTATTATTATCATTACCAGTATTAGCAGGGGCTATTACTATATTATTAAGAGATCGTAAT------------------------------------.
Holotype ♂
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, 24‒31/iii/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28344-E03.
Paratypes
None.
Chelonus minorcarmonai , sp. nov.
Diagnostics
BOLD:ADB0320. Consensus barcode. ATTTTATATTTTTTATTTGGTATATGATCTGGTATATTAGGATTATCATTAAGTATATTAATTCGTATAGAATTAAGATTTTCTGGTAGGTTATTTATAAATGATCAATTATATAATAGAATTATTACTTTACATGCTTTTGTTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAGTTCCTTTAATATTAGGTTTACCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTATTTATATTATTAATAAGAGGATTTATTAATATAGGTGTAGGTACAGGTTGAACAGTTTATCCACCTTTATCATTATTAATTGGTCATGGTGGAATTTCTGTTGATATATCAATTTTTTCTTTACATTTAGCTGGTATATCATCAATTATAGGTGCAATTAATTTTATTGTTACTATTATTAATACATGATTAAATATTAATTATATAGATAAATTTTCTTTATTTGTAATATCAGTTTTTATTACTGCAATTTTATTATTATTATCTTTACCAGTTTTAGCAGGTGCTATTACTATATTATTAAGGGATCGTAATATAAATACAAGATTTTTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 05‒12/xii/2013, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher code. BIOUG29282-H09.
Paratypes
All Malaise-trapped. BIOUG28697-B07, BIOUG29112-C06, BIOUG29112-F08, BIOUG29415-F01. Depository:
Chelonus monikaspringerae , sp. nov.
Diagnostics
BOLD:ACU3782. Consensus barcode. ATATTATATTTTATATTTGGAATATGATCTGGTATATATGGTTTGTCTTTAAGTTTATTAATTCGTATGGAATTAAGGATAGCAGGTAGGTTATTTGTGAATGATCAATTATACAATAGAGTTGTTACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGTGGTTTTGGAAATTGATTAATTCCATTAATATTAGGTTTACCTGATATAGCATTTCCTCGTATAAATAATATAAGATTTTGGTTATTGATTCCATCTTTAATTATGTTAACATTAAGAGGATTTGTTAATATAGGGGCAGGTACTGGATGAACTGTTTATCCTCCTTTATCTTTATTAATTGGTCACGGGGGTATTTCTGTTGATATATCAATTTTTTCTTTACATTTAGCAGGGATATCTTCTATTATAGGGGCTATTAATTTTATTGTTACTATTATTAATACATGAATAAATAATAAATTTATAGATAAATTTTCTTTATTTGTTTGATCAGTTTTTATTACTGCTATTTTATTATTATTATCTTTACCTGTATTAGCAGGTGCTATTACTATATTATTAAGTGATCGTAAT------------------------------------------.
Holotype ♀
Alajuela, Sector San Cristobal, Estación San Gerardo, 10.88, -85.389, 575 meters, 19‒26/viii/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG19587-A04.
Paratypes
None.
Chelonus moniquegilbertae , sp. nov.
Diagnostics
BOLD:AAJ0370. Consensus barcode. TATATTATATTTTATTTTTGGAATATGGTCAGGAATATTAGGATTGTCGTTGAGTTTATTGATTCGTATAGAATTAAGGTTAACAGGTAGGTTATTTTTAAATGATCAACTTTATAATAGAGTAGTAACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGAAATTGATTGGTTCCATTAATGTTAGGTTTACCTGATATAGCATTTCCTCGAATAAATAATATAAGATTTTGATTGTTAATTCCTGCTATTATTTTGATGATTTTAAGAGGGTTTGTTAATATAGGAGCTGGTACTGGTTGAACAGTTTATCCTCCTTTATCTTCTTTGATAGGACATGGTGGAATTTCTGTTGATATATCAATTTTTTCTTTACATTTAGCTGGAGCTTCATCAATTATAGGTGCTATTAATTTTATTGTTACAGTAATTAATACTTGAATAAATATTAAATTTATAGATAAATTTTCTTTATTTGTTTGATCAGTATTTATTACTGCTATTTTATTATTATTATCTTTACCTGTATTAGCGGGTGCTATTACTATATTATTAAGAGATCGTAATATAAATACAAGATTTTTTGATCCTTCTGGTGGTGGAGATCCAGTTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Pitilla, Estación Quica, 10.997, -85.397, 470 meters, caterpillar collection date: 09/viii/2009, wasp eclosion date: 31/viii/2009. Depository:
Host data. Gregarious parasitoid of Antaeotricha spurca (Depressariidae) feeding on Cassia grandis (introduced) (Fabaceae).
Caterpillar and holotype voucher codes. 09-SRNP-71792, DHJPAR0040022.
Paratypes
Hosts = Omiodes humeralis (Crambidae) elachJanzen01 Janzen235 (Depressariidae), elachJanzen01 Janzen38 (Depressariidae). DHJPAR0020801, DHJPAR0020805, DHJPAR0042517, DHJPAR0051323. Depository:
Chelonus motohasegawai , sp. nov.
Diagnostics
BOLD:AAW3555. Consensus barcode. TATTTTATATTTTATATTTGGTATATGAGCAGGTATATTAGGTTTATCATTAAGTTTATTAATTCGTATAGAATTAAGATTAAGTGGTAGTTTATTTATAAATGATCAATTATATAATAGTGTAGTTACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGGTTTGGAAATTGATTAGTTCCTTTAATATTAGGATTACCTGATATAGCATTTCCTCGAATAAATAATATAAGATTTTGATTATTAGTTCCTTCTTTATTTTTAATATTATTAAGTAGAATAGTTAATATAGGTGTAGGAACTGGATGAACAGTTTATCCTCCTTTATCTTCATTAGTTGGTCATGGAGGAATTGCAGTAGATATATCAATTTTTTCTTTACATATAGCTGGGGCATCTTCTATTATAGGAGCAATTAATTTTATTGTTACTATTTTAAATTCTTGAATTGAAACTAAATTTATCGATAAATTTCCTTTATTTGTATGATCAGTATTTATTACTGCAATTTTATTATTGTTATCTTTACCTGTATTAGCTGGTGCAATTACAATGTTATTAAGAGATCGTAATTTAAATACAAGATTTTTTGATCCTTCAGGGGGAGGAGACCCAGTTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Cacao, Estación Cacao, 10.927, -85.468, 1150 meters, caterpillar collection date: 07/i/2011, wasp eclosion date: 04/ii/2011. Depository:
Host data. Gregarious parasitoid of Anacrusis ellensatterleeae (Tortricidae) feeding on Rondeletia costaricensis (Rubiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-35011, DHJPAR0042920.
Paratype
Host = Ardeutica Brown002 (Tortricidae): DHJPAR0030992. Depository:
Etymology
Chelonus motohasegawai is named for Dr. Motohiro Hasegawa of JICA, Japan in recognition of his many years of facilitating and causing the integration of Costa Rican SINAC activities with ICE activities through his outstanding example for the Pailas II Geothermal Development Project beginning in 2013.
Chelonus nataliaivanovae , sp. nov.
Diagnostics
BOLD:AAF2632. Consensus barcode. TATATTATATTTTATTTTTGGAATATGGTCAGGTATATTAGGGTTATCTTTAAGATTAATAATTCGAATAGAATTGAGATTAAGGGGAAGATTATTTATAAATGATCAATTATATAATAGAATTGTTACTTTACATGCTTTTGTTATAATTTTTTTTATAGTAATACCAATTATAATTGGGGGATTTGGAAATTGRTTAGTTCCTTTAATATTAGGGYTRCCAGATATAGCTTTTCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCACTTTTTTTAATAATTTTAAGGAGGTTTATTAATATAGGTGTAGGGACAGGTTGAACAGTTTATCCTCCATTATCATCTTTAATTGGTCATGGTGGTATTTCAGTTGATATATCAATTTTTTCTTTACATTTAGCAGGYATATCTTCAATTATAGGAGCAATTAATTTTATTGTAACTATTTTAAATACATGAATTAATAATAAATTTATGGATAAATTCCCTTTATTTGTTTGATCTGTATTTATTACTGCAATTTTATTATTGTTATCTTTACCTGTATTAGCTGGGGCTATTACTATATTATTAAGTGATCGTAATTTAAATACTAGATTTTTTGATCCTTCTGGAGGGGGGGATCCTGTATTGTATCAGCATTTWTTT.
Holotype ♂
Guanacaste, Sector Pitilla, Bullas, 10.987, -85.385, 440 meters, caterpillar collection date: 28/iv/2011, wasp eclosion date: 13/v/2011. Depository:
Host data. Gregarious parasitoid of Antaeotricha Janzen140DHJ03 (Depressariidae) feeding on Zygia longifolia (Fabaceae).
Caterpillar and holotype voucher codes. 11-SRNP-65195, DHJPAR0042960.
Paratypes
Hosts = Omiodes humeralis (Crambidae), Spilomela pantheralis (Crambidae), Antaeotricha BioLep38 (Depressariidae), Antaeotricha ianthinaEPR01 (Depressariidae), Antaeotricha Janzen140DHJ03 (Depressariidae), Antaeotricha Janzen401 (Depressariidae), Antaeotricha spurca (Depressariidae). Amorbia productana (Tortricidae), Amorbia revolutana (Tortricidae), Anacrusis nephrodes (Tortricidae), Anacrusis turrialbae (Tortricidae), Megalota vulgaris (Tortricidae), Strepsicrates Brown37 (Tortricidae). DHJPAR0029133, DHJPAR0034268, DHJPAR0035316, DHJPAR0035415, DHJPAR0035559, DHJPAR0039111, DHJPAR0040019, DHJPAR0040020, DHJPAR0045406, DHJPAR0049124, DHJPAR0049786, DHJPAR0052897, DHJPAR0053679, DHJPAR0055461, DHJPAR0056713. Depository:
Etymology
Chelonus nataliaivanovae is named to honor Dr. Natalia Ivanova for her many years laboring in execution and administration of the laboratory actions performed by the Centre for Biodiversity Genomics to rapidly and accurately DNA barcode tens of thousands of Costa Rican insects.
Chelonus nelsonzamorai , sp. nov.
Diagnostics
BOLD:ADB4135. Consensus barcode. ATTTTATATTTTATTTTTGGTATATGATCTGGAATATTAGGATTATCATTAAGATTATTAATTCGTTTAGAATTAGGTATATTAGGTAGTTTATTAATAAATGATCAATTATATAATAGAATAGTTACTTTACATGCATTTGTAATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGTTTTGGTAATTGATTAATTCCTTTAATGTTAGGTTTACCAGATATAGCATTTCCTCGAATGAATAATATAAGATTTTGATTATTAATTCCATCTATATTTATATTGTTAATAGGGGGATTTGTTAATGCTGGTGCTGGAACTGGTTGAACAGTTTATCCACCTTTATCTTCAATTATAGGACATAGGGGGGTATCTGTTGATATATCAATTTTTTCATTACATTTGGCTGGGATATCATCTATTATAGGTGCAATTAATTTTATTATTACTTCAATAAATACTTGAATATTAAATAAATATATTGATAAATTTCCTTTATTTGTGTGATCGGTTTTAATTACAGCTATTTTATTATTATTATCATTACCTGTATTAGCAGGGGCTATTACTATGCTTTTAAGTGATCGTAATTTAAATACA------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 19‒26/vi/2014, Malaise trap PL12-9A. Depository:
Host data. None.
Holotype voucher code. BIOUG29298-C10.
Paratypes
None.
Chelonus normwoodleyi , sp. nov.
Diagnostics
BOLD:AAW3564. Consensus barcode. GATTTTGTATTTTATATTTGGGATATGAGCTGGAATATTAGGTTTATCATTAAGAATATTGATCCGAATAGAGTTGAGTGTAATTGGAAGATTAATATTAAATGATCAATTGTATAATAGAATTGTAACTTTGCATGCTTTTGTAATAATTTTTTTTATGGTGATACCAATTATAATTGGTGGTTTTGGAAATTGATTAGTTCCATTAATAATTGGTTTACCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTATTGATTGTTGATTCCTTCTTTATTTTTATTATTATCAAGGGGATTTGTAAATACAGGAGTAGGTACTGGTTGGACAGTTTATCCTCCTTTATCTTCACTAACGGGTCATGGGGGTGTTTCTGTTGATTTGTCTATTTTTTCATTACATTTAGCTGGTTCTTCTTCTATTATAGGTGCTATTAATTTTATTGTTACAAGRTTGAATACTTGATTGAGTATTTATTATATGGATAAGATTTCTTTATTTGTTTGATCTGTATTAATTACTGCTATTTTATTATTGTTATCCTTACCGGTRTTAGCGGGTGCTATTACTATACTTTTAAGAGACCGAAATTTAAATACTAGATTTTTTGATCCTTCTGGTGGGGGTGACCCAGTATTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93, -85.372, 527 meters, caterpillar collection date: 22/ii/2013, wasp eclosion date: 28/iii/2013. Depository:
Host data. Stenoma BioLep82 (Depressariidae) feeding on Luehea seemannii (Malvaceae).
Caterpillar and holotype voucher codes. 13-SRNP-880, DHJPAR0051920.
Paratypes
Host = Stenoma BioLep82: DHJPAR0036402, DHJPAR0048089, DHJPAR0048090, DHJPAR0048196. Depository:
Chelonus osvaldoespinozai , sp. nov.
Diagnostics
BOLD:ABU8005. Consensus barcode. TGTATTATATTTTATTTTTGGTATATGATCTGGGATATTAGGTTTATCACTAAGTATATTAATTCGAATAGAATTAAGTTTGGTAGGTAGATTATTAATAAATGATCAGTTATATAATAGAATTGTTACTTTACATGCCTTTGTTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGTAATTGATTAATTCCTTTAATATTAGGTTTACCTGATATAGCATTTCCACGTATAAATAATATAAGATATTGACTATTAATTCCTTCTTTATTTATATTATTATTAAGTGGATTTGTTAATATAGGGGTAGGTACTGGATGAACAGTTTATCCTCCTTTATCTTTATTGATTGGTCATGGGGGAATTTCTGTAGATTTATCTATTTTTTCTYTACATTTAGCTGGAATATCTTCTATTATAGGGGCYATTAATTTTATTACTACTAGATTAAATACTTGAATTAATAATAAGTATATGGATAAATTYCCTTTATTTGTTTGGTCAGTATTAATTACTGCTGTTTTATTATTATTGTCYTTACCTGTATTGGCAGGTGCTATTACTATATTATTAAGGGATCGAAATTTAAATACCAGATTTTTTGATCCATCTGGTGGGGGGGATCCAGTTTTATATCAGCATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Sendero Venado, 10.897, -85.27, 420 meters, caterpillar collection date: 24/iv/2011, wasp eclosion date: 21/v/2011. Depository:
Host data. Stenoma Janzen230 (Depressariidae) feeding on Xylopia frutescens (Annonaceae).
Caterpillar and holotype voucher codes. 11-SRNP-41837, DHJPAR0042858.
Paratype
Host = Stenoma Janzen230: DHJPAR0052184. Depository:
Chelonus pamelacastilloae , sp. nov.
Diagnostics
BOLD:AAK1568. Consensus barcode. GATATTATATTTTATTTTTGGTATATGATCGGGAATTTTAGGTTTATCTTTGAGAATATTAATTCGTATAGAGTTAAGATTAAGAGGAAGTTTGTTAATAAATGATCAGTTATATAATAGAATTGTTACTTTACATGCTTTTATTATAATTTTTTTTATGGTTATACCAATTATAATTGGGGGTTTTGGAAATTGATTAGTTCCTTTAATGTTAGGAGTTCCAGATATAGCATTTCCACGAATAAATAATATAAGATATTGGTTATTAATTCCGTCTTTATTTATATTAATTATAAGAAGATTTGTTAATGTAGGTGTAGGTACGGGTTGAACAGTTTATCCACCATTATCTTTATTAATTGGTCATGGTGGAGTTTCTGTAGATATATCAATTTTTTCTTTACATTTAGCTGGGATATCTTCAATTATAGGTGCTATTAATTTTATTGTTACTGGGGCAAATAGATGAATAAAGAATAAATTTATAGATAAATATCCATTATTTGTATGGTCAGTATTGATTACTGCGTTATTATTACTATTATCTTTGCCAGTATTGGCGGGAGCTATTACTATATTATTAAGAGATCGTAATATAAATACTAGGTTTTTTGATCCTTCAGGTGGTGGGGATCCAGTATTATATCAACATTTGTTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Sendero Venado, 10.897, -85.27, 420 meters, caterpillar collection date: 29/vii/2011, wasp eclosion date: 26/viii/2011. Depository:
Host data. Antaeotricha BioLep91 (Depressariidae) feeding on Tachigali costaricensis (Fabaceae).
Caterpillar and holotype voucher codes. 11-SRNP-43517, DHJPAR0045401.
Paratypes
None.
Chelonus paulgoldsteini , sp. nov.
Diagnostics
BOLD:AAE9140. Consensus barcode. GGTGTTATATTTTATTTTTGGGATATGGGCCGGAGTATTGGGCTTATCATTAAGTATATTAATTCGAATGGAATTAAGGGTGAGAGGTAGGTTATTTATAAATGATCAGTTATATAATAGAATTGTGACTTTACATGCTTTTGTTATAATTTTTTTTATAGTTATGCCAATTATAATTGGGGGGTTTGGGAATTGGYTGATTCCTTTAATATTAGGGGTTCCGGATATGGCTTTTCCTCGAATGAATAATATAAGTTATTGGTTATTAATTCCTTCTTTATTTATATTGGTAGTTAGGAGGTTTATTAATATGGGGGTAGGTACGGGATGAACCGTTTATCCTCCTTTATCATTATTAATTGGTCATGGGGGYGTGTCAGTGGATATATCAATTTTTTCTTTACATTTAGCTGGTGCGTCTTCAATTATAGGGGCGATTAATTTTATTGTGACAGGGGTTAATACGTGGATAAGTAATAAATTAATAGATAAGTTCCCTTTATTTGTGTGATCAGTAATAATTACTGCGGTATTATTGCTATTGTCTTTACCAGTTTTAGCTGGGGCAATTACAATGTTGTTAAGGGATCGTAATTTAAATACAAGATTTTTCGATCCTYMRGGGGGGGGGGRTCCTGTATTATATCAGCATTTATTT.
Holotype ♀
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.865, -85.415, 540 meters, caterpillar collection date: 23/ii/2011, wasp eclosion date: 23/iii/2011. Depository:
Host data. elachBioLep01 BioLep754 (Depressariidae) feeding on Tapirira brenesii (Anacardiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-830, DHJPAR0042851.
Paratypes
Hosts = Stenoma BioLep86 (Depressariidae), elachBioLep01 BioLep754 (Depressariidae). DHJPAR0035526, DHJPAR0035527, DHJPAR0035528, DHJPAR0039120, DHJPAR0042855, DHJPAR0046940, DHJPAR0048102, DHJPAR0048980, DHJPAR0051921, DHJPAR0051923, DHJPAR0051928, DHJPAR0056954, DHJPAR0056976. Depository:
Etymology
Chelonus paulgoldsteini is named for Dr. Paul Goldstein of the Smithsonian Institution and USDA in recognition of his intense care and curation and taxonomic processing of the thousands of species of DNA barcoded ACG moths being deposited in the collections of the National Museum of Natural History.
Chelonus paulhansoni , sp. nov.
Diagnostics
BOLD:ACB1254. Consensus barcode. TGTATTATATTTTATTTTTGGTATATGATGTGGGGTTTTGGGTTTGTCTATAAGGGTATTAATTCGTATAGAATTAAGAATATCTGGAAGATTATTGTTAAATGATCAATTATATAATAGAATTGTGACTTTACATGCTTTTATTATAATTTTTTTTATGGTTATACCAATTATGATTGGGGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGGTTACCTGATATAGCATTTCCTCGAATAAATAATATAAGATATTGATTATTAATTCCATCTTTATTTTTGTTATTAATAAGTGGATTTATTAATGTGGGGGTAGGTACTGGGTGAACAGTTTATCCTCCATTATCTTTATTAATTGGACATGGAGGAATTTCAGTAGATATATCAATTTTTTCATTACATTTAGCTGGGGTTTCATCAATTATAGGTGCAATTAATTTTATTACTACAATTATAAATATATGATTAAAAATAAAATTTATAGATAAATTTCCTTTATTTGTATGATCAGTTTTGATTACTGCATTTTTATTATTATTATCTTTACCAGTTTTGGCAGGTGCTATTACTATATTATTAAGTGATCGTAATATAAATACAAGATTTTTTGATCCTTCAGGTGGGGGGGATCCAATTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Quebrada Bambu, 10.93, -85.252, 109 meters, caterpillar collection date: 27/x/2012, wasp eclosion date: 14/xi/2012. Depository:
Host data. Antaeotricha Janzen204 (Depressariidae) feeding on Miconia xalapensis (Melastomataceae).
Caterpillar and holotype voucher codes. 12-SRNP-77452, DHJPAR0051343.
Paratypes
Hosts = Antaeotricha Janzen204. DHJPAR0048978, DHJPAR0049326, DHJPAR0061491. Depository:
Chelonus paulheberti , sp. nov.
Diagnostics
BOLD:AAN2554. Consensus barcode. TATATTATATTTTATTTTTGGTATATGATGTGGTATATTGGGGTTGTCTTTAAGTATATTGATTCGTATAGAGTTAAGAATATCAGGTAGACTTTTATTAAATGATCAATTATATAATAGAATTGTAACTYTACATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTAATAATTGGGGGATTTGGTAATTGATTAATTCCTTTAATATTAGGATTACCTGATATGGCTTTTCCTCGAATAAACAATATAAGTTAYTGATTATTAATTCCTTCATTATTTATACTAATAATAAGAGGATTTATTAATACAGGAGTTGGAACCGGTTGAACTGTTTATCCTCCATTATCATTATTAATTGGACATGGGGGGATTTCTGTAGATATATCAATTTTTTCTTTACATTTAGCAGGGGCTTCTTCAATTATGGGGGCAATTAATTTTATTACTACTATTATAAATATGTGATTAGTAAAAAGTTTTATGGATAAATATCCTTTATTTGTATGATCAGTTTTAATTACTGCAGTTTTGTTATTATTATCTCTTCCTGTGTTAGCAGGGGCTATTACTATATTATTAAGAGATCGTAATATAAATACTAGATTTTTTGATCCTTCAGGAGGGGGGGATCCTATTTTATATCAACATTTGTTT.
Holotype ♀
Alajuela, Sector San Cristobal, Quebrada Garcia, 10.861, -85.426, 495 meters, caterpillar collection date: 27/ii/2011, wasp eclosion date: 22/iii/2011. Depository:
Host data. elachBioLep01 BioLep754 (Depressariidae) feeding on Tapirira brenesii (Anacardiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-869, DHJPAR0042850.
Paratypes
Hosts = Antaeotricha Janzen233 (Depressariidae), elachBioLep01 BioLep754 (Depressariidae), elachBioLep01 BioLep55 (Depressariidae), Stenoma BioLep86 (Depressariidae). DHJPAR0039503, DHJPAR0042854, DHJPAR0042861, DHJPAR0042863, DHJPAR0048105, DHJPAR0048972. Depository:
Etymology
Chelonus paulheberti is named for Dr. Paul Hebert founder and major motor of the Centre for Biodiversity Genomics at the University of Guelph, Canada, and his invention of the DNA barcoding concept for identification of all species anywhere anytime by anyone.
Chelonus petronariosae , sp. nov.
Diagnostics
BOLD:ACB1255. Consensus barcode. TATATTATATTTTATTTTTGGTATATGATGTGGAGTTTTAGGTTTATCTTTAAGTATATTAATTCGAATAGAATTAAGGGTATCTGGTAGATTATTATTAAATGATCAATTATATAATAGTATTGTAACCCTACATGCCTTTATTATAATTTTTTTTATAGTTATACCAGTAATAATTGGTGGATTTGGAAATTGATTAGTACCTTTAATATTAGGTTTACCTGATATAGCATTTCCTCGAATAAATAATATAAGATATTGATTATTAATTCCTTCATTATTTATATTATTAATGAGGGGATTTATTAATATGGGGGTAGGTACTGGTTGAACAGTTTATCCCCCTTTATCATTATTGATTGGTCATGGAGGTATTTCTGTAGATATATCAATTTTTTCATTACATTTAGCAGGGGCTTCTTCAATTATAGGAGCTATTAATTTTATTACTACTATTATAAATATATGAATAAATAGGAGATTTATAGATAAATATCCATTATTTGTATGATCAGTATTAATTACAGCGTTTTTATTATTATTATCTTTACCGGTATTAGCAGGAGCTATTACTATATTATTAAGGGATCGTAATATAAATACAAGATTTTTTGATCCTTCAGGTGGAGGTGACCCAATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Brasilia, Moga, 11.012, -85.349, 320 meters, caterpillar collection date: 13/iii/2012, wasp eclosion date: 04/iv/2012. Depository:
Host data. Antaeotricha Janzen146 (Depressariidae) feeding on Lonchocarpus guatemalensis (Fabaceae).
Caterpillar and holotype voucher codes. 12-SRNP-65223, DHJPAR0048976.
Paratypes
None.
Chelonus ramyamanjunathae , sp. nov.
Diagnostics
BOLD:ABA9309. Consensus barcode. AACATTATATTTTATTTTTGGTATATGATGTGGAGTTTTAGGTTTATCTTTAAGTATACTAATTCGAATAGAATTAAGTATATCTGGGAGATTATTATTAAATGATCAGTTATATAATAGTATTGTAACTTTACATGCTTTTATTATAATTTTTTTTATGGTTATGCCGGTAATAATTGGGGGATTTGGTAATTGATTAGTACCTTTAATATTAGGTTTACCTGATATAGCATTTCCTCGGATGAATAATATAAGATATTGGTTATTAATTCCTTCATTATTTATATTATTAATAAGTGGTTTTATTAATATAGGGGTGGGGACAGGTTGAACGGTTTATCCTCCTTTATCTTTGTTAATTGGTCATGGGGGTATTTCAGTAGATATATCAATTTTTTCATTACATTTAGCTGGGGCTTCTTCAATTATAGGTGCTATTAATTTTATTACTACAATTTTAAATATATGAATAAAAAGAAAATTTATGGATAAATTTCCTTTATTTGTTTGATCTGTATTAATTACTGCATTTTTACTTTTGTTATCATTACCTGTTTTGGCAGGGGCTATTACTATATTATTAAGTGATCGAAATATAAATACTAGATTTTTTGATCCTTCAGGGGGAGGTGATCCAATTTTATACCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Malaguenya, 10.956, -85.284, 221 meters, caterpillar collection date: 07/vii/2011, wasp eclosion date: 25/vii/2011. Depository:
Host data. Antaeotricha Janzen321 (Depressariidae) feeding on Combretum fruticosum (Combretaceae).
Caterpillar and holotype voucher codes. 11-SRNP-67386, DHJPAR0045410.
Paratypes
Hosts = Antaeotricha Janzen126, Antaeotricha Janzen146, Antaeotricha Janzen321. DHJPAR0040016, DHJPAR0045411, DHJPAR0045412, DHJPAR0045413, DHJPAR0048971, DHJPAR0048975, DHJPAR0055366, DHJPAR0055369. Depository:
Chelonus randallgarciai , sp. nov.
Diagnostics
BOLD:ADR1783. Consensus barcode. AACATTATATTTTATTTTTGGGATATGATGTGGAGTTTTAGGTTTATCTTTAAGTATACTAATTCGAATGGAATTAAGTGTATCTGGGAGGTTATTATTAAATGATCAGTTATATAATAGTATTGTAACTTTACATGCTTTTATTATAATTTTTTTTATGGTTATGCCGGTAATAATTGGGGGATTTGGTAATTGATTAGTACCTTTAATATTAGGTTTACCTGATATAGCATTTCCTCGTATGAATAATATAAGATATTGGTTATTAATTCCTTCATTATTTATATTATTAATAAGGGGTTTTATTAATATAGGGGTGGGGACAGGTTGAACGGTTTATCCCCCTTTATCTTTATTAATTGGTCATGGGGGTATTTCAGTAGATATATCAATTTTTTCATTACATTTAGCTGGGATTTCTTCAATTATAGGTGCTATTAATTTTATTACTACAATTTTAAATATATGAATAAAAAGAAAATTTATGGATAAATTTCCTTTATTTGTTTGGTCTGTATTAGTTACTGCATTTTTACTTTTATTATCATTACCTGTTTTGGCAGGGGCTATTACTATATTATTAAGTGACCGAAATATAAATACTAGATTTTTTGATCCTTCAGGGGGAGGTGACCCAATTTTATATCAGCATTTATTT.
Holotype ♀
Guanacaste, Sector San Cristobal, Sendero Huerta, 10.9305, -85.3722, 527 meters, caterpillar collection date: 25/v/2018, wasp eclosion date: 14/vi/2018. Depository:
Host data. Antaeotricha Janzen146 (Depressariidae) feeding on Lonchocarpus guatemalensis.
Caterpillar and holotype voucher codes. 18-SRNP-926, DHJPAR0062693.
Paratypes
None.
Chelonus rebeccakittelae , sp. nov.
Diagnostics
BOLD:ACB1121. Consensus barcode. GTGGAGTTTTGGGATTATCTTTAAGGATATTAATTCGTATGGAGTTAAGAATATCTGGAAGGTTATTGTTGAATGATCAGTTATATAATAGAATCGTTACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATGCCTGTAATAATTGGTGGGTTTGGTAATTGATTAATTCCTTTAATATTAGGGTTGCCTGATATAGCTTTTCCACGTATAAATAATATAAGTTATTGATTATTAATTCCTTCATTATTTATATTATTAATAAGGGGATTTATTAATATAGGAGTAGGAACTGGTTGAACGGTTTATCCTCCATTATCATTATTAATTGGACATGGAGGTATTTCAGTAGATATATCAATTTTTTCTTTACATTTGGCTGGGGCTTCTTCTATTATAGGGGCTATTAATTTTATTACTACAATTATAAATATATGAGTAATTAAAAGATTTATAGATAAATATCCTTTATTTGTATGATCAGTATTTATTACTGCATTTTTATTATTATTATCATTACCAGTATTGGCTGGGGCTATTACTATATTATTAAGAGATCGTAATATAAATACAAGATTTTTTGATCCTTCAGGGGGGGGGGA-------------------------.
Holotype ♀
Guanacaste, Sector El Hacha, Estación Los Almendros, 11.032, -85.528, 290 meters, caterpillar collection date: 20/ii/2012, wasp eclosion date: 10/v/2012. Depository:
Host data. Antaeotricha Janzen224 (Depressariidae) feeding on Microdesmia arborea (Chrysobalanaceae).
Caterpillar and holotype voucher codes. 12-SRNP-20443, DHJPAR0048982.
Paratypes
None.
Chelonus robertoespinozai , sp. nov.
Diagnostics
BOLD:AAD4678. Consensus barcode. GATATTATATTTTATTTTTGGCATATGGTGTGGAGTATTAGGATTATCTTTAAGTATATTAATTCGAATAGAATTAAGGATATCTGGTAGTTTATTAATAAATGATCAATTATATAATAGTATTGTAACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTTATGATTGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGATTACCTGATATAGCTTTYCCTCGAATGAATAATATAAGTTATTGATTATTAATTCCTTCATTATTTTTATTATTAATAAGAGGATTTATTAATATAGGGGTTGGTACTGGATGAACAGTTTATCCTCCATTATCATTATTAATTGGGCATGGGGGAATTTCAGTAGATATATCAATTTTTTCTTTACATTTAGCTGGGGCATCTTCAATTATAGGTGCTATTAATTTTATTACTACAATTATAAATATATGGGTGGTTAGGAGGTTTATAGATAAATATCCTTTATTTGTGTGGTCGGTGTTAATTACTGCCTTTTTATTATTATTATCTTTACCTGTATTAGCAGGGGCTATTACTATATTATTAAGTGATCGTAATATGAATACAAGATTTTTTGATCCTTCAGGAGGGGGGGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Brasilia, Moga, 11.012, -85.349, 320 meters, caterpillar collection date: 24/x/2011, wasp eclosion date: 18/xi/2011. Depository:
Host data. elachJanzen01 Janzen211 (Depressariidae) feeding on Miconia argentea (Melastomataceae).
Caterpillar and holotype voucher codes. 11-SRNP-66100, DHJPAR0046939.
Paratypes
Host = Ategumia lotanalis (Crambidae): DHJPAR0029174, DHJPAR0029177. Depository:
Chelonus robertofernandezi , sp. nov.
Diagnostics
BOLD:ACJ5332. Consensus barcode. AATATTATATTTTATTTTTGGTATATGGTGTGGAGTTTTAGGGTTATCTTTGAGAATATTAATTCGGATAGAATTAAGTATATCTGGAAGTTTATTATTAAATGATCAATTATATAATAGTATCGTAACTTTACATGCTTTTATTATAATTTTTTTTATGGTTATACCTGTTATAATTGGTGGATTTGGTAATTGATTAATTCCTTTAATGTTAGGATTACCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTATTGATTATTGATTCCTTCATTATTTATACTATTAATAAGGGGATTTGTTAATATAGGAGTTGGTACTGGTTGAACAGTTTATCCTCCTTTATCCTTATTAATTGGGCATGGAGGTATTTCAGTAGATATATCAATTTTTTCTTTACATTTAGCTGGTGCTTCATCAATTATAGGTGCTATTAATTTTATTACTACAGTTATAAATATATGGGTGGTTAGGAGATTTATAGATAAATATCCTTTATTTGTATGATCAGTATTAATTACTGCATTTTTATTATTATTATCATTACCTGTATTAGCGGGGGCTATTACTATGTTATTGAGTGATCGTAATATAAATACAAGATTTTTTGATCCCTCAGGGGGGGGAGACCCAATTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Vado Rio Francia, 10.901, -85.289, 400 meters, caterpillar collection date: 26/vii/2013, wasp eclosion date: 13/viii/2013. Depository:
Host data. Antaeotricha Janzen110 (Depressariidae) feeding on Ardisia compressa (Primulaceae).
Caterpillar and holotype voucher codes. 13-SRNP-42816, DHJPAR0052883.
Paratypes
None.
Etymology
Chelonus robertofernandezi is named to honor Sr. Roberto Fernandez of GDFCF and BioAlfa for his years of environmental planning for ICE followed by his current facilitation of the interaction between BioAlfa and ICE and other sectors of Costa Rican society while administrating the field aspects of the SINAC participation in BioAlfa Malaise-trapping of national parks.
Chelonus rocioecheverriae , sp. nov.
Diagnostics
BOLD:ACB1122. Consensus barcode. AATATTATATTTTATTTTTGGAATATGGTGTGGAGTTTTAGGGTTATCTTTAAGTATTTTAATTCGAATAGAATTAAGTATAACTGGAAGATTATTTATAAATGATCAATTATATAATAGAATTGTAACTTTGCATGCTTTTATTATAATTTTTTTTATAGTTATACCCGTTATGATTGGTGGATTTGGTAATTGATTAATTCCTTTGATATTAGGGTTACCTGATATGGCATTTCCACGAATGAATAATATAAGTTATTGATTATTAATTCCTTCATTATTTATATTATTAATAAGAGGATTTATTAATATAGGAGTAGGTACAGGATGAACAGTTTATCCTCCTTTATCTTTATTAATTGGGCATGGTGGTATTTCCGTAGATATATCAATTTTTTCTTTACATTTAGCTGGGGCTTCTTCAATTATAGGTGCTATTAATTTTATTACTACGATTATAAATATATGAATAAATAAAAAGTTTATAGATAAATATCCTTTATTTGTGTGATCAGTATTAATTACTGCATTTTTATTATTATTATCTTTACCTGTATTAGCAGGAGCTATTACTATATTATTAAGAGATCGTAATATAAATACGAGATTTTTTGATCCTTCAGGAGGGGGTGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Brasilia, Moga, 11.012, -85.349, 320 meters, caterpillar collection date: 13/iii/2012, wasp eclosion date: 01/iv/2012. Depository:
Host data. Antaeotricha Janzen146 (Depressariidae).
Caterpillar and holotype voucher codes. 12-SRNP-65224, DHJPAR0048970.
Paratypes
None.
Chelonus rodrigogamezi , sp. nov.
Diagnostics
BOLD:AAJ0362. Consensus barcode. AATATTATATTTTATTTTTGGGATATGATGTGGGGTTTTAGGRTTRTCTYTAAGTATACTAATTCGAATGGAATTAAGAATAACTGGGAGGTTATTTATAAATGATCAATTATATAATAGAATTGTAACTTTACATGCTTTTATTATAATTTTTTTTATGGTTATGCCTGTYATGATTGGYGGATTTGGTAATTGATTAATTCCYYTAATGTTGGGTTTACCTGATATAGCATTYCCTCGAATGAATAATATAAGTTATTGATTATTAATYCCYTCATTATTTATATTATTAATAAGGGGATTTATTAATATAGGTGTGGGYACTGGATGAACAGTTTATCCYCCTTTATCTTTATTAATTGGTCATGGGGGTATTTCAGTAGATATRTCAATTTTTTCTTTACATTTAGCTGGGGCTTCTTCAATTATAGGTGCTATTAATTTTATTACTACYATTATAAATATATGAATAAATAAGAAATTTATAGATAAATATCCTTTATTTGTATGATCTGTATTAATTACAGCATTTTTATTATTATTATCTTTRCCTGTATTGGCAGGGGCTATTACTATATTATTAAGTGATCGTAATATGAATACAAGRTTTTTTGATCCYTCAGGAGGAGGAGAYCCAATTTTATAYCAACAYTTWWTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Sendero Juntas, 10.907, -85.288, 400 meters, caterpillar collection date: 03/vi/2010, wasp eclosion date: 19/vi/2010. Depository:
Host data. Antaeotricha Janzen88 (Depressariidae).
Caterpillar and holotype voucher codes. 10-SRNP-41977, DHJPAR0040365.
Paratypes
Hosts = Antaeotricha Janzen88. DHJPAR0029168, DHJPAR0040359, DHJPAR0040360, DHJPAR0040361, DHJPAR0040362, DHJPAR0040363, DHJPAR0040370, DHJPAR0042097, DHJPAR0046937, DHJPAR0048098, DHJPAR0048099, DHJPAR0050074, DHJPAR0050121, DHJPAR0051329, DHJPAR0051335, DHJPAR0051341, DHJPAR0052880, DHJPAR0052884, DHJPAR0052885, DHJPAR0052888, DHJPAR0052892, DHJPAR0052893, DHJPAR0052894, DHJPAR0052895, DHJPAR0053675, DHJPAR0053680, DHJPAR0054554, DHJPAR0054558, DHJPAR0054562, DHJPAR0054565, DHJPAR0055230, DHJPAR0055358, DHJPAR0055359, DHJPAR0055361, DHJPAR0055362, DHJPAR0055374, DHJPAR0055375, DHJPAR0055378, DHJPAR0055383, DHJPAR0055385, DHJPAR0055394, DHJPAR0055401, DHJPAR0055993, DHJPAR0055994, DHJPAR0055999, DHJPAR0056001, DHJPAR0056349, DHJPAR0056352, DHJPAR0056951, DHJPAR0056953, DHJPAR0056956, DHJPAR0056959, DHJPAR0056971, DHJPAR0062574, DHJPAR0062575.
Etymology
Chelonus rodrigogamezi is named to honor Dr. Rodrigo Gámez for his founding role for ACG both logistically and philosophically, and then the same for INBio and its enormous impact on Costa Rica and the international world of conservation through national biodevelopment.
Chelonus ronaldzunigai , sp. nov.
Diagnostics
BOLD:AAK1016. Consensus barcode. AATACTATATTTTATTTTTGGAATATGATGTGGAGTTTTAGGATTATCATTAAGTATATTAATTCGAATAGAATTAAGAATAACTGGAAGATTATTTATAAATGATCAGTTATATAATAGTATTGTGACTTTACATGCTTTTATTATAATTTTTTTTATGGTTATACCTGTTATAATTGGTGGATTTGGTAATTGATTAATTCCTTTAATATTAGGATTACCTGATATAGCATTTCCTCGAATGAATAATATAAGTTATTGATTATTAATTCCTTCATTATTTATATTATTGATAAGGGGTTTTATTAATATAGGAGTTGGTACTGGATGAACAGTTTATCCCCCATTATCATTATTAATTGGGCATGGAGGTATTTCAGTGGATATATCAATTTTTTCTTTACATTTAGCGGGGGCTTCCTCAATTATAGGTGCTATTAATTTTATTACTACGATTATAAATATGTGGATAATTAAAAGATTTATAGATAAATATCCTTTATTTGTATGATCAGTATTAATTACTGCATTTTTATTATTATTATCTTTACCTGTATTGGCAGGGGCTATTACTATATTATTAAGTGATCGTAATATAAATACAAGATTTTTTGATCCTTCTGG----------------------------------.
Holotype ♀
Alajuela, Sector San Cristobal, Puente Palma, 10.916, -85.379, 460 meters, caterpillar collection date: 16/ix/2009, wasp eclosion date: 07/x/2009. Depository:
Host data. Antaeotricha Janzen134 (Depressariidae) feeding on Calophyllum brasiliense (Calophyllaceae).
Caterpillar and holotype voucher codes. 09-SRNP-4814, DHJPAR0037167.
Paratypes
None.
Etymology
Chelonus ronaldzunigai is named to honor Sr. Ronald Zuñiga for his decades of taxonomic curation and support of the Hymenoptera portion of the INBio and Museo Nacional insect collection, and now the same role for BioAlfa for Costa Rica in general and ACG specifically.
Chelonus rosibelelizondoae , sp. nov.
Diagnostics
BOLD:AAM1446. Consensus barcode. AATATTATATTTTATTTTTGGAATATGGTGTGGAGTTTTAGGATTATCATTAAGTATATTAATTCGAATAGAATTAAGAATAACTGGAAGATTATTTATAAATGATCAGTTATATAATAGGATTGTGACTTTACATGCTTTTATTATAATTTTTTTTATGGTTATACCTGTAATAATTGGTGGATTTGGTAATTGATTAATTCCTTTAATGTTAGGATTACATGATATAGCATTTCCTCGAATGAATAATATAAGTTATTGATTATTAATTCCTTCATTATTTATATTATTGATAAGGGGATTTATTAATATAGGGGTTGGTACTGGATGAACAGTTTATCCTCCATTGTCATTATTAATTGGGCATGGAGGTATTTCAGTGGATATATCAATTTTTTCTTTACATTTGGCGGGGGCTTCTTCAATTATAGGTGCCATTAATTTTATTACTACGATTATAAATATGTGGATAATTAAAAGATTTATAGATAAATATCCTTTATTTGTATGATCAGTATTAATTACTGCATTTTTATTATTATTATCTTTACCTGTATTGGCGGGGGCTATTACTATATTATTAAGTGATCGTAATATAAATACAAGATTTTTTGACCCTTCTGGTGGGGGGGATCCAATTTTGTATCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Puente Palma, 10.916, -85.379, 460 meters, caterpillar collection date: 17/x/2012, wasp eclosion date: 21/xi/2012. Depository:
Host data. Antaeotricha renselariana (Depressariidae) feeding on Pterocarpus rohrii (Fabaceae).
Caterpillar and holotype voucher codes. 12-SRNP-4584, DHJPAR0051334.
Paratype
Host = Antaeotricha Janzen146 (Depressariidae): DHJPAR0038004. Depository:
Chelonus rostermoragai , sp. nov.
Diagnostics
BOLD:ACJ3551. Consensus barcode. AATATTATATTTTATTTTTGGAATATGGTGTGGAGTTTTGGGATTATCATTAAGTATATTAATTCGAATAGAATTAAGAATAACYGGAAGATTATTTATAAATGATCAGTTGTATAATAGGATTGTRACTTTACATGCTTTTATCATAATTTTTTTTATGGTTATACCTGTTATAATTGGTGGATTTGGTAATTGATTAATTCCATTAATGTTAGGATTACCTGATATAGCATTTCCTCGAATGAATAATATAAGTTATTGATTATTAATTCCTTCATTATTTATATTATTGATAAGAGGATTTATYAATATAGGAGTTGGTACTGGATGAACAGTTTATCCTCCATTATCATTATTAATTGGACATGGGGGTATTTCGGTAGATATATCAATTTTTTCTTTGCATTTAGCGGGAGCTTCTTCAATTATAGGTGCCATTAATTTTATTACTACTATTATAAATATGTGGATAATTAAAAGATTTATAGATAAATATCCTTTATTTGTATGATCAGTATTAATTACTGCATTTTTAYTATTATTATCTTTACCGGTATTAGCAGGAGCTATTACTATATTATTAAGTGATCGTAATATAAATACAAGATTTTTTGATCCTTCTGGGGGGGGAGATCCAATTTTGTATCAACATCTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93, -85.372, 527 meters, caterpillar collection date: 08/ii/2013, wasp eclosion date: 01/iii/2013. Depository:
Host data. Antaeotricha stigmatiasDHJ01 (Depressariidae) feeding on Tetrapterys tinifolia (Malpighiaceae).
Caterpillar and holotype voucher codes. 13-SRNP-616, DHJPAR0051922.
Paratypes
Host = Antaeotricha stigmatiasDHJ01: DHJPAR0051925, DHJPAR0051926, DHJPAR0053673, DHJPAR0053685, DHJPAR0056000. Depository:
Chelonus ruthfrancoae , sp. nov.
Diagnostics
BOLD:AAW4589. Consensus barcode. AATATTATATTTTATTTTTGGRATATGGTGTGGRGTTTTAGGRTTATCTTTAAGTGTATTAATTCGAATAGAATTAAGAATAACTGGAAGGTTATTTATAAATGATCAGTTATATAATAGGATTGTAACTTTACATGCTTTTATTATAATTTTTTTTATGGTTATACCTGTAATGATTGGKGGGTTTGGTAATTGATTAATTCCCTTAATGTTAGGATTACCTGACATAGCATTTCCTCGTATGAATAATATAAGTTATTGATTAYTAATTCCTTCATTATTTATATTATTAATAAGGGGATTTATTAATATAGGAGTTGGGACTGGATGAACAGTTTATCCTCCATTATCATTATTAATTGGYCATGGRGGTATTTCAGTAGATATATCAATTTTTTCATTACATTTAGCYGGGGCTTCTTCAATTATAGGTGCTATTAATTTTATTACTACTATTATGAATATATGAATAATTAAGAGATTTATGGATAAATATCCTTTATTTGTATGATCAGTGTTAATTACTGCATTTTTGTTATTATTATCTTTACCTGTTTTGGCWGGAGCTATTACTATATTATTAAGTGATCGTAATATAAATACAAGATTTTTTGATCCCTCAGGAGGGGGGGATCCAATTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93, -85.372, 527 meters, caterpillar collection date: 27/i/2013, wasp eclosion date: 27/i/2013. Depository:
Host data. Antaeotricha similisEPR02 (Depressariidae) feeding on Inga oerstediana (Fabaceae).
Caterpillar and holotype voucher codes. 12-SRNP-5796, DHJPAR0051094.
Paratypes
Hosts = Antaeotricha BioLep46, Antaeotricha Janzen13, Antaeotricha Janzen290, Antaeotricha Janzen31, Antaeotricha similisEPR02, Antaeotricha similisEPR03, Cerconota Janzen82 (Depressariidae), Stenoma Janzen09 (Depressariidae). DHJPAR0042089, DHJPAR0042091, DHJPAR0050136, DHJPAR0051344, DHJPAR0055363, DHJPAR0055399, DHJPAR0056353, DHJPAR0056354, DHJPAR0056950, DHJPAR0056962.DHJPAR0056965. Depository:
Chelonus scottmilleri , sp. nov.
Diagnostics
BOLD:ABY5286. Consensus barcode. AATATTATATTTTATTTTTGGAATATGGTGTGGAGTTTTAGGATTATCTTTAAGTATATTAATTCGAATAGAATTAAGAATAACTGGGAGGCTATTTATAAATGATCAGTTATATAATAGAATTGTGACCTTACATGCTTTTATTATAATTTTTTTTATGGTTATACCTGTAATGATTGGTGGATTTGGTAATTGATTAATTCCTTTAATGTTAGGATTACCTGATATAGCATTTCCTCGTATGAATAATATAAGTTATTGATTATTAATTCCTTCATTATTTATATTATTGATAAGGGGATTTATTAATATAGGAGTTGGAACTGGATGAACAGTTTATCCTCCATTATCATTATTAATTGGTCATGGGGGTATTTCTGTAGATATATCAATTTTTTCTTTACATTTAGCTGGTGCTTCTTCAATTATAGGTGCTATTAATTTTATTACTACAATTATAAATATATGAATAATTAAAAGGTTTATAGATAAATATCCTTTATTTGTATGATCAGTATTAATTACTGCATTTTTATTATTATTATCTTTACCTGTGTTGGCTGGGGCTATTACTATATTATTAAGTGATCGTAATATAAATACAAGATTTTTTGATCCCTCAGGAGGAGGGGATCCAATTTTATATCAGCATTTATTT.
Holotype ♂
Guanacaste, Sector Pitilla, Estación Quica, 10.997, -85.397, 470 meters, caterpillar collection date: 22/vi/2010, wasp eclosion date: 09/vii/2010. Depository:
Host data. Antaeotricha radicalisEPR03 (Depressariidae) feeding on Miconia trinervia (Melastomataceae).
Caterpillar and holotype voucher codes. 10-SRNP-71898, DHJPAR0040369.
Paratypes
Hosts = Antaeotricha Janzen04, Antaeotricha Janzen204, Antaeotricha marmorea, Antaeotricha radicalis, Antaeotricha radicalisEPR02, Antaeotricha radicalisEPR03. DHJPAR0035310, DHJPAR0039122, DHJPAR0045285, DHJPAR0048095, DHJPAR0048096, DHJPAR0051325, DHJPAR0051339, DHJPAR0051345, DHJPAR0053670, DHJPAR0055384, DHJPAR0055388, DHJPAR0055389, DHJPAR0061492. Depository:
Etymology
Chelonus scottmilleri is named to honor Dr. Scott Miller of the Smithsonian Institution for his decades of peripheral and direct support of the establishment of ACG and the permanent deposit of the ACG inventory voucher specimens, both DNA barcoded and not, as well as facilitating their collateral information integration with that of older specimens in the same Smithsonian collections.
Chelonus scottshawi , sp. nov.
Diagnostics
BOLD:ABX5499. Consensus barcode. AATATTATATTTTATTTTTGGAATATGATGTGGAGTTTTAGGRTTATCTTTAAGTATRTTAATTCGAATAGAATTAAGAATAACTGGAAGATTATTTATAAATGATCAGTTATATAATAGAATTGTRACTTTRCAYGCTTTTATTATAATTTTTTTTATGGTTATACCTGTAATGATTGGTGGRTTTGGTAATTGATTRATTCCTTTAATGYTAGGATTACCTGATATAGCATTYCCTCGTATGAATAATATAAGTTAYTGATTATTAATYCCTTCATTATTTATATTATTGATAAGGGGRTTTATTAATATAGGAGTTGGAACTGGATGAACAGTTTATCCTCCATTGTCATTATTAATTGGTCATGGRGGTATTTCTGTAGATATATCAATTTTTTCTTTACATTTAGCTGGTGCTTCTTCAATTATAGGTGCTATTAATTTTATTACTACAATTATAAATATATGAATAATTAAGAGATTTATAGATAAATAYCCTTTATTTGTATGATCAGTATTAATTACTGCAYTTTTGTTATTATTATCTTTACCTGTGTTGGCTGGGGCTATTACCATATTATTAAGTGAYCGTAATATAAATACAAGATTTTTTGAYCCCTCAGGAGGAGGGGAYCCAATTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Quebrada Guarumo, 10.904, -85.284, 400 meters, caterpillar collection date: 3/iii/2011, wasp eclosion date: 30/iii/2011. Depository:
Host data. Antaeotricha Janzen146 (Depressariidae) feeding on Lonchocarpus oliganthus (Fabaceae).
Caterpillar and holotype voucher codes. 11-SRNP-41083, DHJPAR0042853.
Paratypes
Hosts = Antaeotricha Janzen146, Antaeotricha Janzen24, Antaeotricha Janzen245, Antaeotricha Janzen40, Antaeotricha Janzen49, Antaeotricha renselariana. DHJPAR0036381, DHJPAR0036382, DHJPAR0036383, DHJPAR0036384, DHJPAR0036386, DHJPAR0036387, DHJPAR0036389, DHJPAR0036393, DHJPAR0036395, DHJPAR0036403, DHJPAR0037988, DHJPAR0037992, DHJPAR0037995, DHJPAR0039505, DHJPAR0048979, DHJPAR0048981, DHJPAR0048992, DHJPAR0049007, DHJPAR0049018, DHJPAR0051924, DHJPAR0052182, DHJPAR0052183, DHJPAR0052185, DHJPAR0052186, DHJPAR0052187, DHJPAR0052280, DHJPAR0054573, DHJPAR0055360, DHJPAR0055365, DHJPAR0055372, DHJPAR0055377, DHJPAR0055379, DHJPAR0055380, DHJPAR0055386, DHJPAR0055387, DHJPAR0055391, DHJPAR0055392, DHJPAR0055393, DHJPAR0055395, DHJPAR0055396, DHJPAR0055991, DHJPAR0056955, DHJPAR0062573, DHJPAR0062697. Depository:
Chelonus sergioriosi , sp. nov.
Diagnostics
BOLD:ACA6835. Consensus barcode. AATATTATATTTTATTTTTGGTATATGATCTGGTATATTTGGTTTATCATTAAGTTTATTAATTCGTATAGAATTGAGAATTTCTGGAACTTTATTTGGTAATGATCAATTATATAATAGTATTGTAACTTTRCATGCATTTATTATAATTTTTTTTATAGTTATRCCAGTTATAATTGGTGGTTTTGGAAATTGATTAGTTCCATTAATATTAGGATTACCAGATATAGCTTTTCCTCGTATAAATAATATAAGGTTTTGATTATTAATACCTTCATTATTTATATTAATTATAAGAGGATTTGTAAATATAGGAGTAGGAACTGGTTGAACTGTTTATCCTCCRTTATCTTTATTAATAGGACATGGTGGAGTTTCGGTAGATATATCAATTTTTTCTTTACATTTAGCTGGTATATCTTCAATTATAGGAGCTATTAATTTTATTGTTACTATTATGAATATATGAATAAGAATAGTTTTTTTTGATAAATATCCTTTATTTGTATGATCAGTATTAATTACTGCGGTATTATTATTATTATCATTACCAGTTTTAGCTGGTGCTATTACTATATTATTAAGAGATCGAAACTTAAATACAAGATTTTTTGATCCTTCAGGKGGWGGTGAYCCAGTATTATATCAACATTTATTT. Distribution restricted to Central America.
Holotype ♂
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 25/ii‒11/iii/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG10752-A09.
Paratypes
None.
Etymology
Chelonus sergioriosi is named to honor Sr. Sergio Rios of GDFCF and ACG for his many years as a dedicated inventory parataxonomist for ACG.
Note
A specimen from Collier County, Florida, United States (BOLD code BIOUG02575-C01) shares the same BIN, but the color of the hind leg is dramatically different making it unlikely to be conspecific. The hind femur and tibia of the Florida specimen are pale yellow, whereas the tibia of C. sergioriosi is brown. More specimens from the two localities will need to be barcoded and examined to confirm the consistency of this difference.
Chelonus sigifredomarini , sp. nov.
Diagnostics
BOLD:AAM1445. Consensus barcode. AATTTTATATTTTATTTTTGGTATTTGAAGAGGTATATTTGGTTTATCTTTAAGAATAATAATTCGTTTAGAATTATCATTTACTGGTAATTTAATAATAAATGATCAATTATATAATAGTATTGTTACTATACATGCTTTTATTATGATTTTTTTTTTAGTTATACCAGTTATAATTGGTGGTTTTGGTAATTGATTAATTCCATTAATATTAGGTTTATGTGATATATGTTTTCCTCGAATGAATAATATAAGATTTTGATTATTAATACCTTCWTTATTTATATTAATTATAAGAGGATTTGTAAATACAGGTGTAGGTACAGGTTGGACTTTGTATCCTCCATTATCYTTATTAATAGGTCATAGGGGAATTTCAGTAGATATATCTATTTTTTCTTTACATTTAGCTGGAATATCTTCAATTATAGGAGCTATTAATTTTATTGTAACAATTGTTAAYACTTGGATAATAAAAGTTTATATAGATAAAATTTCATTATTTGTTTGGTCAGTTTTAATTACTGCTGTATTATTATTATTATCTTTACCTGTTTTAGCAGGAGGTATTACTATATTATTAAGTGATCGGAATTTTAATACAACATTTTTTGATTCTTCAGGAGGTGGTGATCCTATTTTGTATCAACATTTATTT.
Host data. Euacidalia certissa (Geometridae) feeding on Leucaena multicapitula (Fabaceae).
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Estación Llanura, 10.933, -85.253, 135 meters, caterpillar collection date: 28/xi/2009, wasp eclosion date: 19/xii/2009. Depository:
Host data. None.
Caterpillar and holotype voucher codes. 09-SRNP-76655, DHJPAR0037978.
Partypes
None.
Etymology
Chelonus sigifredomarini is named to honor Sr. Sigifredo Marin for many decades of intense conservation administration of ACG itself followed by the same intelligent care and development for GDFCF field projects and the members of the ACG Parataxonomist Program.
Chelonus stevearonsoni , sp. nov.
Diagnostics
BOLD:AAW3558. Consensus barcode. TGTTTTATATTTTATTATAGGTATATGATCTGGTGTATTTGGATTGTCATTAAGTATACTTATTCGTTTAGAATTATCTTTTCCTGGAAGTTTAATAATAAATGATCAATTATATAATAGTTTAATTACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCAGTTATAATTGGTGGTTTTGGTAATTGATTAGTTCCATTAATATTAGGTTTATGTGATATATGTTTTCCTCGAATAAATAATATAAGATTTTGATTATTGGTTCCTTCTTTAATTTTATTAATTTCAAGTTCTTTTATTAATACTGGGGTTGGTACTGGATGAACTGTTTATCCTCCATTATCTTTATTAATAGGTCATAGAGGTATTTCAGTAGATTTATCTATTTTTTCTCTTCATTTGGCTGGTATATCATCAATTATAGGAGCAATTAATTTTATTGTAACTGTAATTAATACTTGAATATTAAAAAATTATATAGATAAATTATCTTTATTTGTATGATCTGTAATAATTACAGCTGTATTATTATTATTATCTTTACCAGTATTAGCTGGTGCTATTACTATATTATTAAGAGATCGAAATATAAATACAACATTTTTTGATTCTTCAGGTGGYGGAGATCCTGTATTGTATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Gazo, 10.98, -85.386, 485 meters, caterpillar collection date: 18/iii/2014, wasp eclosion date: 06/iv/2014. Depository:
Host data. Leuciris fimbriaria (Geometridae) feeding on Mimosa watsonii (Fabaceae).
Caterpillar and holotype voucher codes. 14-SRNP-70585, DHJPAR0055364.
Paratypes
Host = Leuciris fimbriaria: DHJPAR0016436, DHJPAR0029120, DHJPAR0042849. Depository:
Chelonus stevestroudi , sp. nov.
Diagnostics
BOLD:AAW3557. Consensus barcode. TATTTTATATTTTATTTTTGGCATATGATGTGGTATATTCGGATTATCTTTAAGTATATTAATTCGTATAGAATTAGGTACTTTAAAAAGATTATTTATAAATGATCAGTTATATAAYGGRTTAGTTACTATACATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGTAATTGATTAGTTCCATTAATATTAGGATTACCTGATATATCTTTTCCTCGTATAAATAATATAAGTTATTGATTATTGATTCCTTCATTATTTATATTAATTATAAGAAGTTTTGTAAATATGGGGGTTGGTACAGGTTGAACTGTRTATCCACCATTATCTTTATTAATGGGACATAGGGGAGTTTCTGTAGATATAGCTATTTTTTCTTTACATTTAGCGGGRGCTTCATCTATTATAGGTGCAATTAATTTTATTATTACAAGGTTAAATACTTATATAAAATTAAGTTATATAGAAAAATTTCCTTTATTTGTTTGATCAGTATTAATTACTGCAGTATTATTATTATTATCTTTACCAGTATTAGCGGGGGCTATTACAATATTGTTAAGAGATCGAAATATAAATACTAGATTTTTTGATCCAGCAGGAGGAGGGGATCCATTATTGTATCAACATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93, -85.372, 527 meters, caterpillar collection date: 03/viii/2014, wasp eclosion date: 23/viii/2014. Depository:
Host data. Antaeotricha Janzen13 (Depressariidae) feeding on Inga oerstediana (Fabaceae).
Caterpillar and holotype voucher codes. 14-SRNP-3582, DHJPAR0056351.
Paratypes
Host = Antaeotricha BioLep46, Antaeotricha Janzen290, Antaeotricha similisEPR01: DHJPAR0020803, DHJPAR0056002, DHJPAR0056004. Depository:
Etymology
Chelonus stevestroudi is named to honor Mr. Steve Stroud of Escazu, Costa Rica for his decades of support, both financial and psychological, for all things ACG and the subsequent spin-out of BioAlfa, and its exploration of private ecotourism at Hacienda Baru.
Chelonus sujeevanratnasinghami , sp. nov.
Diagnostics
BOLD:ABW9828. Consensus barcode. TGTATTATATTTTATTTTTGGGATATGGTCTGGTATATTAGGTTTATCTTTAAGTATATTAATTCGTATAGAATTAGTATATCCTGGTAGATTATTAATAAATGATCAATTATATAATAGATTAGTTACTATACATGCATTTATTATAATTTTTTTTATAGTGATACCTATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATGTTAGGATTACCTGATATAATTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATAATATTAATTTTAAGAATATTAGTTAATATGGGGGTAGGGACAGGATGAACAGTTTATCCTCCTTTATCTTCATTAATAGGTCATAGGGGAGTAAGTGTAGACATAGCTATTTTTTCTTTGCATTTAGCTGGGATATCATCAATTATAGGAGCTATTAATTTTATAGTAACTGTGTTAAATAGATGGATACATTATAGTTATATAGATGTAATACCTTTATTTGTTTGATCAGTTTTTATTACGGCAATTTTATTATTATTATCATTACCTGTATTAGCCGGTGCTATTACTATATTATTAAGAGATCGTAATATAAATACTACATTTTTTGATCCTACGGGAGGGGGAGACCCAATTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Brasilia, Gallinazo, 11.018, -85.372, 360 meters, caterpillar collection date: 09/i/2012, wasp eclosion date: 24/i/2012. Depository:
Host data. Rhobonda gaurisanaDHJ04 (Choreutidae) feeding on Ficus donnell-smithii (Moraceae).
Caterpillar and holotype voucher codes. 12-SRNP-65012, DHJPAR0046942.
Paratypes
Host = Rhobonda gaurisanaDHJ04: DHJPAR0046943, DHJPAR0046944, DHJPAR0046945, DHJPAR0046946, DHJPAR0047222, DHJPAR0047223, DHJPAR0047229, DHJPAR0047230. Depository:
Etymology
Chelonus sujeevanratnasinghami is named to honor Dr. Sujeevan Ratnasingham of the Centre for Biodiversity Genomics at the University of Guelph for designing and executing the intellectual and logistic guts of BOLD at the CBG, without which CBG would have no purpose.
Chelonus sureshnaiki , sp. nov.
Diagnostics
BOLD:AAW3556. Consensus barcode. TATATTATATTTTATTTTTGGTATATGAAGAGGTATACTAGGTTTATCTTTAAGTTTATTAATTCGTATAGAATTAAGATTTAGGGGAAGTTTATTTCAAAATGATCAATTATATAATAGAATTGTAACTATACATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAGTAGGGGGTTTTGGAAATTGATTGATTCCTTTAATATTAGGTATACCTGATATAATATTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTGTTAATATTAATTTTTGGGGGATTTATTAATATAGGTGTAGGTACTGGATGAACATTATATCCTCCATTATCATTATTAATGAGACATGGAGGAATGTCAGTAGATATTTCAATTTTTTCGTTACATTTAGCTGGTATATCTTCAATTATAGGATCAATTAATTTTATTGTAACTATTATAATAGTATGAATTGAAAAAATATATATAGATAAATTTTCTTTATTTTTATGATCTATTTTAATTACTACAATTTTATTGTTATTATCTTTACCTGTATTGGCTGGTGCTATTACAATATTATTAAGAGATCGTAATATAAATACTAGATTTTTTGATCCCGCTGGGGGCGGGGATCCAGTATTGTATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Medrano, 11.016, -85.381, 380 meters, caterpillar collection date: 05/ix/2014, wasp eclosion date: 27/ix/2014. Depository:
Host data. Microsca hedialis (Thyrididae) feeding on Alchornea latifolia (Euphorbiaceae).
Caterpillar and holotype voucher codes. 14-SRNP-71641, DHJPAR0056350.
Paratype
Host = Rhodoneura terminalis (Thyrididae): DHJPAR0029162. Depository:
Chelonus torbjornekremi , sp. nov.
Diagnostics
BOLD:ACZ0402. Consensus barcode. AATATTATATTTTTTATTTGGAATTTGAAGTGGAGTTATAGGTTTGTCTTTAAGTATAATAATTCGTATAGAATTGAGAAGTGTAATAAGATTATTTTATAATGATCAGTTATATAATAGAATTGTTACTATACATGCTTTTATTATAATTTTTTTTATAGTAATACCTTTAATAATTGGGGGATTTGGAAATTGATTAATTCCATTGATGTTAGGATTGTCTGATATAATTTTTCCTCGAATAAATAATATAAGATTTTGGTTATTAATTCCTTCAATTATTTTATTAATTATAGGGGGATTTGTAAATATAGGGTCTGGAACAGGGTGAACAATTTATCCTCCATTATCACTATTAATAGGCCATAGGGGAGTTTCAGTAGATTTATCTATTTTTTCTTTACACTTAGCAGGTATATCATCTATTATAGGTTCAATTAATTTTATTGTTACTATTATAAATACTTGATTACACATTAAATATATAGATAAATATCCTTTATTTGTTTGATCTGTATTTATTACAGTTATTTTATTATTAATATCATTACCAGTTTTAGCTGGGGCAATTACTATATTATTAAGTGATCGAAATTTAAATACAAGATTTTTTGATCCATCAGGAGGGGGAGATCCGGTATTGTATCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Cafecito, 10.94404, -85.31738, 455 meters, caterpillar collection date: 10/x/2014, wasp eclosion date: 26/x/2014. Depository:
Host data. pyrausJanzen01 Janzen26 (Crambidae) feeding on Baccharis trinervis (Asteraceae).
Caterpillar and holotype voucher codes. 14-SRNP-81300, DHJPAR0056964.
Paratypes
Hosts = pyrausJanzen01 Janzen26, Aponia Janzen469 (Crambidae). DHJPAR0045404, DHJPAR0060594, DHJPAR0061493. Depository:
Etymology
Chelonus torbjornekremi is named to honor Dr. Torbjorn Ekrem of the NTNU University of Museum in Norway for his eager and high quality administrative and science participation in the iBOL international meetings in South Africa and in Trondheim, Norway, and especially in support of BioAlfa and the ACG-ICE project to biomonitor the Pailas II geothermal site.
Chelonus yeimycedenoae , sp. nov.
Diagnostics
BOLD:ADW6358. Consensus barcode. TATTATATTTCATATTTGGTATATGATCTGGTATATTTGGATTATCATTAAGTTTATTAATTCGAATAGAATTAAGAGTATCGGGTAGTTTATTTATAAATGATCAATTATATAATAGAATTGTTACTTTACATGCATTTATTATAATTTTTTTTATAGTTATGCCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTGGGTTTACCTGATATATTATTTCCACGAATAAATAATATAAGTTTTTGATTATTAATACCTTCATTAATTATATTAATATTGAGTAGATTTGTAAATATAGGAGTAGGTACAGGTTGAACTATTTATCCTCCATTATCTTTATTAATTGGTCATGGTGGAATTTCAGTGGATATATCAATTTTTTCTTTACATTTAGCTGGAATATCTTCAATTATAGGTGCTATTAATTTTATAGTTACAATTATTAATACATGAATAAGTAATAATTATATAGATAAATTTTCTTTATTTGTTTGATCAGTTTTTATTACTGCAATTTTATTATTATTATCTTTACCTGTTTTAGCTGGTGCTATTACTATATTATTAAGAGATCGAAATATAAATACAAGATTTTTTGATCCTTCAGGTGGGGGTGATCCAATTTTATATCAGCATTTAT------------------------------------------.
Holotype ♂
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 05‒18/xii/2014, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher code. BIOUG44134-G02.
Paratypes
None.
Leptodrepana Shaw, 1983
Leptodrepana species are found throughout the New World. Dadelahi and Shaw (2018) described the first 24 Costa Rican species of the genus. Only one of these, L. alexisae, was captured in our Malaise traps and none was reared. Its identification was based on morphological inspection rather than its barcode. Hosts are unknown for the genus, and we do not treat any reared species here. This strongly suggests that the hosts are leaf-miners since these were not included in the Janzen and Hallwachs rearing program.
Leptodrepana alexisae
Diagnostics
BOLD:ABV9918. Consensus barcode. GGTATTTTATATTTTATTTTTGGTATATGATCTGGCATATTTGGTTTATCATTAAGTTTAATAATTCGAATAGAGTTAAGTTCTTTAACATCTTTTATAGGTAATGATCAAATTTATAATAGAATTGTTACTATACATGCATTTATTATAATTTTTTTTATGGTTATACCAATTATAATTGGTGGTTTTGGAAATTGATTAATTCCATTAATATTAGGGGGTCCAGATATAGCATTTCCTCGTATAAATAATATAAGATTTTGACTATTAATTCCATCATTATTAATATTAATTATGAGAAGGTTAGTAAATGTTGGTGTTGGGACAGGATGAACAGTTTATCCTCCTTTATCTTTATTAATAGGTCATGGGGGAATTTCTGTAGATTTAAGAATTTTTTCTTTACATTTAGCGGGAGCTTCATCAATTATAGGTGCAATTAATTTTATTGTTACAATTATGAATATATGATTAGGTATAAAATATATAGATAAAATTTCTTTATTTAGATGATCAGTTTTAATTACTGCTATTTTATTATTATTATCATTACCTGTTTTAGCTGGGGCAATTACAATATTATTAACAGATCGAAATTTAAATACAAGATTTTTTGATCCGGCAGGAGGTGGGGATCCAATTTTGTATCAACATTTATTT.
Imaged specimen
♀: Guanacaste Province, Sector Cacao, Sendero Arenales, 10.92471, -85.46738, 1080 meters, University of Wyoming Insect Museum, Laramie, Wyoming.
Host data. None.
Voucher code of imaged specimen conspecific with the holotype. BIOUG33274-F09.
Other material
All Malaise-trapped: DHJPAR0045916, DHJPAR0045947, DHJPAR0046006, DHJPAR0046011, DHJPAR0046014, DHJPAR0046015, DHJPAR0046029, DHJPAR0047295, DHJPAR0047294, DHJPAR0047288, DHJPAR0047301, DHJPAR0047309, DHJPAR0047318, DHJPAR0047319, DHJPAR0047323, DHJPAR0047325, DHJPAR0047327, DHJPAR0047332, BIOUG36488-A02, BIOUG36495-H04.
Leptodrepana erasmocoronadoi , sp. nov.
Diagnostics
BOLD:ADA3192. Consensus barcode. TTTATTTTTGGAATATGATCAGGAATATTTGGATTATCATTAAGATTATTAATTCGTATAGAATTAAGTTCTATCACAACTTATATTGGAAATGATCAGATTTATAATAGAATTGTTACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTAATAATTGGGGGATTTGGAAATTGATTAGTACCTTTAATAATAGGTGGTCCTGACATATCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTATTACCTTCATTAATTTTATTAATTATAAGGAGTTTAGTAAATGTAGGTGTTGGTACTGGTTGAACAGTTTATCCTCCTTTATCTTTATTAATGGGTCATGGAGGTATTTCAGTAGATTTAAGAATTTTTTCTTTACATTTAGCTGGAGCTTCTTCAATTATAGGAGCTATTAATTTTATTGTAACAATTTTAAATATATGATTTAGTAAAAAATATATAGATAAGATTTCTTTATTTAGGTGATCAGTATTTATTACAGCAATTTTATTATTATTATCATTACCAGTATTAGCTGGAGCAATTACAATATTATTAACAGATCGAAATTTAAATACA------------------------------------.
Holotype ♀
Alajuela, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, 24‒31/iii/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28344-E11.
Paratypes
None.
Leptodrepana felipechavarriai , sp. nov.
Diagnostics
BOLD:ADB5898. Consensus barcode. ATTTTATATTTTATTTTTGGTTTATGATCAGGAATATTTGGGTTGTCATTGAGGTTAATGATTCGAATAGAATTAAGTTCAGTGGGTACTTATATAGGTAATGATCAAATTTATAATAGGATTGTAACAATACATGCATTTATCATAATTTTTTTTATAGTTATACCAGTTATAATTGGTGGATTTGGAAATTGATTGATTCCTTTGATGTTGGGGGGGCCTGATATATCTTTTCCTCGAATGAATAATATAAGATTTTGGTTGTTGGTACCATCATTATTTATATTAATTTTAAGAAGTTTAGTAAATACTGGGGTAGGGACTGGGTGAACAGTTTATCCTCCTTTATCTTTATTAATAGGTCATGGAGGAATTTCAGTTGATTTAAGAATTTTTTCATTACATTTAGCGGGGGCATCATCTATTATAGGGGCTATTAATTTTATTGTAACTATTATAAATATATGGTTAGGGTTTAAATTTTTAGATAAAATTTCTTTATTTAGGTGATCAGTGTTAATTACTGCAATTTTATTATTATTATCTTTACCAGTTTTAGCTGGAGCAATTACAATGTTATTGACA------------------------------------------.
Holotype ♂
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 03‒10/iv/2014, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher code. BIOUG29764-B11.
Paratypes
None.
Etymology
Leptodrepana felipechavarriai is named to honor Sr. Felipe Chavarria of Santa Rosa, ACG, for his decades of detail accurate handling of GDFCF accounting for GDFCF field projects, taking very high-quality images of ACG biodiversity, and being a core role in ACG and INBio Biodiversity Prospecting projects.
Leptodrepana freddyquesadai , sp. nov.
Diagnostics
BOLD:ADD4610. Consensus barcode. TTATATTTTATTTTTGGAATATGGTCTGGGATGTTTGGGTTATCATTAAGATTGTTAATTCGAATAGAATTAAGTTCATTAACTTCATATATGGGGAATGATCAAATTTATAATAGAGTTGTAACTATACATGCATTTATTATAATTTTTTTTATGGTGATACCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCATTAATATTAGGTGGTCCTGATATAGCGTTTCCACGAATAAATAATATAAGGTTTTGGTTATTAATTCCTTCATTATTTTTATTAATTTTAAGAAGATTAGTAAATGTAGGTGTTGGAACTGGATGAACAGTTTATCCGCCTTTATCTTTATTAATAGGGCATAGGGGAATTTCAGTGGATTTAAGAATTTTTTCTTTACATTTAGCTGGAGCATCTTCAATTATGGGAGCAATTAATTTTATTGTAACAATTATAAATATATGAATAGGTGTAAATTATATAGATAAAATTTCTTTATTTAGATGATCAGTATTAATTACAGCTATTTTATTATTATTATCTTTGCCAGTTTTAGCAGGTGCTATTACAATATTATTAACTGAT---------------------------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-2, 10.7634, -85.335, 824 meters, 10‒17/iv/2014, Malaise trap PL12-2A. Depository:
Host data. None.
Holotype voucher code. BIOUG30966-F04.
Paratypes
None.
Leptodrepana gilbertfuentesi , sp. nov.
Diagnostics
BOLD:ACF7385. Consensus barcode. AATTTTATATTTTATTTTTGGAATATGATCAGGTTTATTTGGTTTATCATTAAGATTATTAATTCGTATAGAATTAAGTTCAGTTTCA---AGATATTTAGGTAATGATCAAATTTATAATAGTGTAGTTACAATACATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGTTTTGGAAATTGATTAATTCCGTTAATGTTAGGAGGTCCAGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGGTTATTGGTTCCTTCTTTATTTTTATTGATTTTAAGAAGTTTAGTTAATGTTGGTGTTGGTACGGGTTGAACAGTTTATCCTCCTTTATCTTTGTTAATAGGTCATGGTGGAATTTCTGTAGATTTAAGAATTTTTTCTTTACATTTAGCTGGTGCTTCCTCTATTATAGGAGCTATTAATTTTATTGTAACAATTATTAATATATGAATAGGA---ATAAAATTTATAGATAAAATTTCTTTATTTAGATGATCTGTTTTAATTACTGCAATTTTATTATTATTATCTTTACCTGTTTTAGCAGGTGCTATTACTATATTGTTAACTGATCGAAATTTAAATACTAGATTTTTTGATCCTGCAGGTGGAGGGGATCCAATTTTGTATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 11‒18/vi/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG05181-B07.
Paratypes
None.
Leptodrepana manuelriosi , sp. nov.
Diagnostics
BOLD:ACR7934. Consensus barcode. ATAAAATTTATAGATAAAATTTCTTTATTTAGATGATCTGTTTTAATTACTGCAATTTTATTATTATTATCTTTACCTGTTTTAGCAGGTGCTATTACTATATTGTTAACTGATCGAAATTTAAATACTAGATTTTTTGATCCTGCAGGTGGAGGGGATCCAATTTTGTATCAACATTTATTTATTTTATATTTTATTTTTGGAATATGATCTGGGATATTTGGTTTATCATTAAGTTTAATTATTCGAATAGAATTAAGATCAGTAACATCTTTTTTAGGAAATGATCAAATTTATAATAGAGTAGTAACAATGCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGATTTGGAAATTGATTAATTCCTTTGATATTAGGAGGTCCTGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTGTGTATATTAATTTTAAGAAGATTAGTTAATATAGGAGTAGGGACAGGTTGAACAGTATATCCTCCTTTATCTTTATTAATAGGACATGGAGGGATTTCAGTAGATTTAGGGATTTTTTCATTACATTTAGCAGGAGCTTCATCAATTATAGGGGCAATTAATTTTATTGTAACAATTATAAATATATGATTAGGAATAAAATATATAGATAAGATTTCATTATTTAGATGATCAGTTTTAATTACTGCAATTCTATTATTATTATCTTTACCTGTATTAGCA------------------------------------------------------------------------.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 04‒11/vi/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG18005-B12.
Paratypes
None.
Phanerotoma Wesmael, 1838
There are 37 species recorded from the Neotropical region but none from Costa Rica (
Phanerotoma almasolisae , sp. nov.
Diagnostics
BOLD:ADB0692. Consensus barcode. ATATTATATTTTTTATTTGGAATATATTCAGGATTTTTTGGATTATCTTTTAGAATTATAATTCGATTAGAATTAAGAGTCTTAGGACAAATAATAGGAAATGATCAAATTTATAATATAATAGTAACAATTCATGCATTTGTTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTGGGAGGTCCTGATATAAGATTTCCTCGAATGAATAATATAAGATTTTGATTATTAATTCCTTCTTTATTATTATTAATTATATCAAGATTTATTAATATAGGGGTAGGTACAGGTTGAACAGTTTATCCTCCTTTATCATTAATTATAGGACATGGGGGTGTATCTGTAGATTTAGTAATTTTTTCTTTACATTTGGCTGGAATTTCTTCAATTTTAGGTGCAATTAATTTTATTAGAACTATTTTAAATATATGATATAATATAAAAGAACTAGAAAAATTATCTTTATTTAGATGATCTGTTGCTATTACTGCTTTATTGTTATTATTATCTTTACCAGTTTTAGCAGGTGCAATTACTATATTA---------------------------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, 20‒27/iii/2014, Malaise trap PL12-6A. Depository:
Host data. None.
Holotype voucher code. BIOUG29157-B02.
Paratypes
None.
Etymology
Phanerotoma almasolisae is named to honor Dr. Alma Solis of the Smithsonian and USDA for her decades of diligent and enthusiastic support for Costa Rican biodiversity development and taxonomy in INBio, ACG, and her home office in the USA, with special emphasis on Pyraloidea microlepidopteran identifications.
Phanerotoma alvaroherrerai , sp. nov.
Diagnostics
BOLD:AAL8247. Consensus barcode. GAGTTTTATATTTTTTATTTGGTATTTATTCAGGGTTTTTAGGTTTATCTTTAAGAATAATAATTCGTTTAGAATTAAGAGTGTTAGGGGRGATAYTAGGTAATGATCAAATTTATAATACAATTGTTACTATTCATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGRTTTGGRAATTGATTAGTRCCTTTAATATTAGGTGGTCCTGATATAAGRTTYCCTCGAATAAATAATATAAGATTTTGGTTATTAATTCCTTCATTAATTTTAATAATTTTATCTAGTTTTGTMAATGTTGGGGCTGGTACTGGATGAACARTYTAYCCTCCTTTRTCATTATTAGTTGGKCATGGGGGGGTTTCAGTTGATTTAGTAATTTTTTCTTTACATTTGGCAGGYRTTTCTTCAATTTTAGGGGCAATTAATTTTATTAGTACAGTTTTAAATATATGAGTTGGGTATAAAATGATAGATAAATTATCTTTRTTTATTTTTTCTGTAGTAATTACAGCATTATTATTATTATTATCTTTACCTGTTTTAGCTGGGGCYATTACAATRTTATTATTGGATCGAAATTTAAATACTAGATTTTTTGATCCTAGAGGMGGTGGGGATCCAGTTTTGTATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 21‒28/i/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG18290-F04.
Paratypes
All Malaise-trapped. BIOUG07918-F10, BIOUG09739-D11, BIOUG09739-C01, BIOUG08348-E03, BIOUG09739-H08, BIOUG10016-F02, BIOUG10017-G06, BIOUG10112-B02, BIOUG10112-B03, BIOUG10112-E11, BIOUG10247-E11, BIOUG17575-A10, BIOUG18289-H10, BIOUG18290-A04, BIOUG18290-D02, BIOUG18290-F04, BIOUG18290-F11, BIOUG18397-C11, BIOUG18398-B05, BIOUG18430-E07, BIOUG18499-D10, BIOUG18499-F05, BIOUG18499-F06. Depository:
Other material
Two specimens from the same BIN were found on BOLD; one from Oklahoma (BOLD voucher 09BBHYM-914) and the other from southern Florida (BOLD voucher 10BBHYM-1201). We doubt that they are conspecific with the Costa Rican specimens based on their distributions, but they were not examined and deserve closer scrutiny.
Phanerotoma anacordobae , sp. nov.
Diagnostics
BOLD:ACJ2167. Consensus barcode. GAGTTTTATATTTTTTATTTGGTATTTATTCTGGGKTTTTAGGTTTRTCTTTAAGAATAATAATTCGATTGGAATTAAGAGTWTTAGGGGAAATATTAGGTAATGAYCAAATTTATAATACAATTGTAACTATTCATGCTTTTATTATAATTTTTTTTATAGTYATACCAATTATAATTGGTGGTTTTGGTAATTGATTAGTWCCTTTAATATTAGGTGGACCTGATATAAGATTYCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATTTTAATAATTTTATCTAGTTTTGTAAATGTAGGGGCTGGAACTGGATGRACAGTTTATCCTCCTTTATCATTATTAGTTGGTCATGGTGGTATTTCAGTTGATTTAGTAATTTTTTCTTTACATTTGGCAGGTATTTCTTCAATTTTAGGTGCAATTAATTTTATTAGTACTGTWTTAAATATATGGGTTGRTTATAAATTGATAGATAAATTATCTTTATTTATTTTTTCTGTAGTAATTACAGCATTGTTATTGTTATTATCTTTACCTGTATTAGCTGGAGCAATTACAATGTTATTGTTGGATCGAAATTTAAATACAAGATTTTTTGACCCAAGAGGTGGAGGAGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 16‒23/iv/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG07616-D05.
Paratypes
All Malaise-trapped. BIOUG07616-F09, BIOUG07781-H06, BIOUG07918-H09, BIOUG08266-F05, BIOUG10016-G07, BIOUG10016-H06, BIOUG10017-H03, BIOUG10247-H02, BIOUG10753-B11, BIOUG17335-B07, BIOUG17335-E04, BIOUG17335-H08, BIOUG17346-E10, BIOUG17684-B01, BIOUG17684-B12, BIOUG17684-D11, BIOUG18290-C10, BIOUG18290-D04, BIOUG18290-E12, BIOUG18290-G07, BIOUG18290-H09, BIOUG18397-C12, BIOUG18398-B04, BIOUG18398-D11, BIOUG18398-F09, BIOUG18398-G07, BIOUG18498-H02, BIOUG18499-A12, BIOUG18499-H09, BIOUG18661-G06, BIOUG18664-E01.
Phanerotoma anamariamongeae , sp. nov.
Diagnostics
BOLD:ADA6322. Consensus barcode. AGTTTTATATTTTTTATTTGGTATTTATTCAGGATTTTTRGGDYTATCTTTAAGAATAATAATTCGTTTAGAATTAAGAGTATTAGGRGAAATATTAGGTAATGATCAAATTTATAATACAATTGTAACTATYCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGGTTTGGAAATTGATTAGTACCTTTAATATTAGGTGGTCCTGATATAAGGTTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAATTTTAATAATTTTGTCTAGRTTTGTAAATGTAGGAGCTGGAACTGGGTGAACAGTTTAYCCTCCTTTATCATTATTARTTGGRCATGGTGGTGTTTCAGTTGATTTGGTAATTTTTTCTTTACATTTAGCRGGTRTTTCTTCAATTTTAGGKGCAATTAATTTTATTAGAACGGTTTTAAATATATGAATTGGTTATAAAATAATGGATAAATTGTCTTTATTTATTTTTTCTGTAGTGATTACAGCATTATTRTTATTATTATCTTTACCTGTTTTAGCTGGGGCAATTACAATATTATTATTGGATCGAAATTTAAATACAAGATTTTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 06‒13/ii/2014, Malaise trap PL12-9A. Depository:
Host data. None.
Holotype voucher code. BIOUG28728-G08.
Paratypes
All Malaise-trapped. BIOUG28388-H02, BIOUG28667-D07, BIOUG28651-A02, BIOUG28674-H11, BIOUG28743-A02, BIOUG28678-G12, BIOUG28721-A12, BIOUG28770-H07, BIOUG28770-H10, BIOUG28770-H11, BIOUG28724-C04, BIOUG28728-A09, BIOUG28728-G09, BIOUG28728-G10, BIOUG28729-D07, BIOUG28846-B09, BIOUG28823-E08, BIOUG28825-B01, BIOUG28777-A04, BIOUG28779-B01. Depository:
Phanerotoma andydeansi , sp. nov.
Diagnostics
BOLD:ADA7613. Consensus barcode. GTATTATATTTTTTATTTGGTATTTATTCAGGATTTTTAGGTTTATCATTAAGAATAATAATTCGATTAGAATTAAGAGTTTTAGGTGAAATGTTAGGGAATGATCAGGTTTATAATACAATTGTTACAATACATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGTTTTGGGAATTGATTAGTACCTTTAATATTAGGTGGTCCTGATATAAGTTTTCCTCGAATAAATAATATAAGATTTTGATTGTTAATTCCTTCTTTAATTTTAATAATTTTATCAAGATTTGTAAATGTTGGTGCTGGTACAGGATGAACAGTTTATCCTCCTTTATCATTATTAATTGGTCATGGTGGTATTTCAGTTGATTTAGTAATTTTTTCTTTACATTTGGCAGGTATTTCATCAATTTTAGGTGCTATTAATTTTATTAGAACAGTTTTAAATATATGAATTAGTTTAAAAATAATAGATAAATTAACATTATTTATTTTTTCTGTAATAATTACAGCAATATTATTATTGTTGTCATTACCTGTWTTGGCTGGAGCAATTACAATATTATTATTAGATCGAAATATAAAT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, 01‒08/v/2014, Malaise trap PL12-6A. Depository:
Host data. None.
Holotype voucher code. BIOUG29322-E10.
Paratypes
All Malaise-trapped. BIOUG28394-F08, BIOUG28673-C05, BIOUG28764-F10, BIOUG28766-E08, BIOUG28722-F01, BIOUG28728-H01, BIOUG28823-E07, BIOUG28830-H02, BIOUG29059-D04, BIOUG29060-G09, BIOUG29064-E01, BIOUG29140-A12, BIOUG29146-E10, BIOUG29322-E08, BIOUG29322-E10, BIOUG29377-F12, BIOUG29403-F11, BIOUG29406-G10, BIOUG29388-H08, BIOUG29398-F04, BIOUG29530-G09, BIOUG29511-G04, BIOUG29725-C09, BIOUG29991-F01, BIOUG30959-G11. Depository:
Phanerotoma angelagonzalezae , sp. nov.
Diagnostics
BOLD:ADB3156. Consensus barcode. GTATTATATTTTTTGTTTGGAATTTATTCAGGAATTTTAGGTTTATCATTAAGTATAATAATTCGATTAGAATTAAGAGTTTTGGGGGAAATATTAGGTAATGATCAAGTTTATAATATAATAGTAACTATTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGATTTGGAAATTGGTTAGTACCTTTAATATTAGGAGGTCCTGATATAAGATTTCCTCGTATAAATAATATAAGATTTTGGTTATTAATTCCATCTTTAATATTATTAATTTTATCAAGATTTGTAAATATAGGAGCTGGAACAGGATGAACTATTTACCCTCCATTATCATTAATATTAGGTCATGGAGGAGTTTCTGTAGATTTAGTAATTTTTTCATTACATTTAGCAGGTATTTCATCAATTTTAGGTGCAATTAATTTTATTAGAACAGTTTTAAATATATGATTTATAAAAAGTATAATAGATAAATTGTCTTTATTTATTTTTTCAGTAGTTATTACTGCATTATTATTATTATTATCATTACCGGTTTTAGCTGGAGCTATTACTATATTATTAATAGATCGTAATTTAAAT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 08‒15/v/2014, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher code. BIOUG29845-C10.
Paratypes
All Malaise-trapped. BIOUG29320-A09, BIOUG29862-C07, BIOUG30984-H06. Depository:
Phanerotoma angelsolisi , sp. nov.
Diagnostics
BOLD:AAG8211. Consensus barcode. TGTATTRTATTTTTTWTTTGGAATTTATTCAGGTATTTTTGGTTTRTCATTAAGTATAATAATTCGATTAGAATTAAGAGTTTTAGGAGAARTRTTAGGTAATGATCAAGTTTATAATACAATTGTWACAATTCATGCTTTTATTATAATTTTTTTTATGGTTATACCARTTATAATTGGTGGTTTTGGWAATTGATTARTTCCTTTAATATTAGGAGGTCCYGATATAAGRTTTCCTCGAATAARTAATATAAGATTTTGRTTATTAATTCCTTCTTTATTAATATTAATTTTATCAAGTTTTRTTAATGTAGGTGYTGGAACAGGTTGAACTRTWTAYCCTCCTTTRTCTTTAYTRGTAGGTCATGGAGGTGTTTCRGTTGATATAGTAATTTTTTCTTTACATTTAGCAGGAATTTCWTCAATTTTRGGTGCTATTAATTTTATTAGAACAGTTTTAAATATATGGTTTGATTATRAAATAATAGATAAAYTGTCTTTATTTATTTTTTCTGTAKTTATTACAGCATTATTATTATTATYATCYTTACCTGTTTTAGCTGGTGCAATTACDATATTATTRTTAGATCGAAATTTAAAKACAAGATTTTTTGATCCAAGAGGAGGTGGGGATCCTATTTTATAYCAACATTTATTT. Restricted to Central America.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 12‒19/xii/2013, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher code. BIOUG29415-C09.
Paratype
BIOUG07751-A11. Depository:
Other material
BIOUG07751-A11 (Malaise-trapped) from Honduras was examined and considered conspecific, and it is included as a paratype. There are dozens of specimens in the same BIN that are from widespread locations across Canada and the USA. Several of these were examined and they differ significantly in color from the Mesoamerican specimens and are not considered conspecific. See BOLD for details.
Etymology
Phanerotoma angelsolisi is named to honor Mr. Angel Solis of BioAlfa for his decades of enthusiastic taxonomic and administrative support of INBio, parataxonomists, and all matters biodiversity of Costa Rica coupled with intensely developing BioAlfa of Costa Rica.
Phanerotoma barryhammeli , sp. nov.
Diagnostics
BOLD:ADG7956. Consensus barcode. AGTATTATATTTTTTATTTGGTATTTATTCAGGTGTATTAGGTTTATCTTTAAGTATAATAATTCGATTAGAATTGAGGGTTTTAGGAGAAATGTTAGGTAATGATCAAGTTTATAATACTATTGTTACTATTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGGTTTGGTAATTGGTTAGTTCCATTAATATTAGGGGGACCTGATATAAGATTCCCTCGTATGAATAATATAAGGTTTTGGTTATTAGTACCTTCATTAATTTTATTAGTTTTATCAAGATTTGTAAATATAGGGGTTGGTACAGGATGAACAGTTTACCCTCCATTATCTTTAACATTAGGACATGGAGGTATTTCAGTAGATTTAGTAATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTTTGGGGGCGATTAATTTTATTAGTACAGTATTAAATATATGATTTAATTATAAAATGATAGATAAATTATCTTTATTTATTTTTTCTGTAGTAATTACAGCATTATTATTATTATTATCTTTACCTGTTTTAGC.
Holotype ♂
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, 19‒26/ii/2015, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG32861-A06.
Paratypes
None.
Phanerotoma bernardoespinozai , sp. nov.
Diagnostics
BOLD:ACJ8069. Consensus barcode. AGTTTTATATTTTTTGTTAGGTATATATTCTGGAATAATAGGTTTATCATTAAGTTTATTAATTCGTTTGGAATTAAGTGTTTTAGGA------TGTATAATAGGTAATGATCAAGTTTACAATATAATTGTTACTATTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGATTAGTTCCATTAATATTAGGTGGACCTGATATAAGTTTTCCTCGAATAAATAATATAAGTTTTTGGTTATTAATTCCTTCTTTAATTTTATTAATAATATCAAGTTTTGTAAATGTTGGTGCTGGAACTGGTTGAACTGTTTATCCACCATTATCATTAATAATTGGTCATGGAGGAATTTCTGTTGATTTAGTAATTTTTTCTTTACATTTGGCTGGAATTTCTTCAATTTTAGGTGCTATTAATTTTATTTCTACTGTATTAAATATATGATTTAATTTAAAAATATTAGAAAAAATATCTTTATTTATTTGATCTGTTGTTATTACTGCTTTATTATTATTATTATCTTTACCTGTTTTAGCAGGTGCTATTACTATATTATTAATAGATCGAAATTTAAATACAAGATTTTTTGATCCAAGAGGAGGTGGTGATCCTGTGTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 04/vi/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher codes. BIOUG08356-H07.
Paratypes
None.
Etymology
Phanerotoma bernardoespinozai is named to honor Sr. Bernardo Espinoza for his decades of high-quality taxonomic actions for the curation and taxonomy of the INBio and now Museo Nacional insect collection, with special emphasis on BioAlfa and the Arctiidae of ACG and Costa Rica in overall.
Phanerotoma calixtomoragai , sp. nov.
Diagnostics
BOLD:ABV9880. Consensus barcode. AATTTTATATTTTTTATTTGGAATGTATTCAGGAATAATAGGTTTATCATTAAGTTTATTAATTCGTTTAGAATTAAGAGTTTTAGGAAATATATTAGGTAATGATCAAGTTTATAATATAATTGTTACTATTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGTTTTGGAAATTGGTTAATTCCTTTAATATTAGGAGGTCCTGATATAAGATTTCCTCGGATGAATAATATAAGGTTTTGATTATTAGTTCCTTCTTTAATTTTATTGAGTATGTCAAGATTTATTAATATAGGGGTTGGAACAGGTTGAACAGTTTATCCTCCTTTATCTTTAATAATTGGACATGGTGGGGTTTCCGTTGATTTAGTTATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTTTAGGGGCTATTAATTTTATTAGAACTATTTTAAATATATGAATTAATCTTAAAATAATAGATAAATTATCTTTATTTATTTGATCTGTTATGATTACTGTTTTATTATTGTTATTATCTTTACCAGTTTTAGCAGGAGCTATTACTATATTATTAATAGATCGAAATCTTAATACAAGATTTTTTGATCCAAGAGGAGGGGGGGATCCAGTATTATATCAGCATTTATTT.
Holotype ♀
Guanacaste, Sector Cacao, Sendero Cima, 10.933, -85.457, 1460 meters, 20/x/2008, Malaise trap. Depository:
Host data. None.
Holotype voucher code. DHJPAR0045952.
Paratypes
None.
Phanerotoma carolinacanoae , sp. nov.
Diagnostics
BOLD:ADB0776. Consensus barcode. TTATTATATTTTTTATTTGGTATATATTCTGGAATAATAGGATTATCTTTAAGATTATTAATTCGTTTAGAATTAAGGGTTTTAGGTAATATATTAGGGAATGATCAAATTTATAATATAGTTGTTACTATTCATGCTTTTATTATAATTTTTTTTATAGTAATACCAGTTATAATTGGTGGTTTTGGAAATTGATTAATTCCTTTAATGTTAGGAGGTCCTGATATAAGATTTCCTCGAATAAATAATATAAGTTTTTGATTGTTAATTCCTTCATTAATGTTATTAAGATTATCAAGATTTATTAATATAGGTGCTGGAACTGGTTGAACTATTTACCCTCCTTTATCCTTAATAATTGGTCATGGAGGATTTTCTGTTGATATAGTAATTTTTTCATTACATTTAGCTGGAATTTCATCAATTTTAGGAGCAATTAATTTTATCACTACAGTTTTAAATATATGGTTTAATATAAAAATAATAGATAAATTATCTTTATTTATTTGATCAGTAATTATTACTGCTTTATTATTATTATTATCTTTACCTGTATTAGCAGGAGCAATTACAATATTATTAATAGATCGTAAT---------------------------------------------.
Holotype ♂
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 16‒23/i/2014, Malaise trap PL12-9A. Depository:
Host data. None.
Holotype voucher code. BIOUG28722-E11.
Paratypes
None.
Phanerotoma christerhanssoni , sp. nov.
Diagnostics
BOLD:ACL3879. Consensus barcode. TGTTTTATATTTTTTATTTGGAATGTATTCAGGAATAATAGGTTTATCTTTAAGATTATTAATTCGTTTGGAATTAAGTGTATTAGGGTCAATATTAGGTAATGATCAAATTTATAATATAGTTGTTACAGTTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATGATTGGTGGTTTTGGAAATTGATTAGTTCCTTTAATATTAGGAGGACCTGATATAAGGTTTCCTCGAATAAATAATATAAGATTTTGATTGTTAATTCCTTCATTAATATTATTATGTTTATCTAGATTTATTAATATAGGTGCTGGTACAGGTTGAACTGTTTATCCTCCTTTATCTTTAATAATTGGTCATGGGGGAATTTCAGTAGATTTAGTAATTTTTTCATTACATTTAGCAGGAATTTCATCAATTTTAGGTGCTATTAATTTTATTAGAACTGTGTTGAATATGTGATTTAAATTGAAAATAATAGATAAATTATCATTATTTATTTGATCAGTTATAATTACTGCTTTATTGTTATTGTTATCTTTACCTGTATTGGCAGGTGCAATTACTATATTATTAATAGATCGTAATTTAAATACAAGTTTTTTTGATCCTAGAGGTGGTGGTGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 31/xii/2012‒07/i/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG09826-F06.
Paratypes
All Malaise-trapped. BIOUG10016-H08, BIOUG17462-A10, BIOUG17575-C11, BIOUG17616-B12, BIOUG18290-F03, BIOUG18397-B07. Depository:
Phanerotoma christhompsoni , sp. nov.
Diagnostics
BOLD:ACL5703. Consensus barcode. TGTTTTATATTTTTTATTTGGAATATATTCAGGTATGTTAGGTTTATCATTAAGTTTATTRATTCGTTTGGAATTAAGTGTTTTAGGATCTATATTAGGTAATGATCAAATTTATAATATAATTGTTACTRTTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGKTTTGGAAATTGATTAGTTCCTTTAATATTAGGGGGTCCTGATATAAGTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATATTATTATGTTTATCAAGATTTATTAATATAGGAGCTGGAACAGGATGAACAGTGTATCCACCTTTATCTTTAATAATTGGTCATGGAGGAATTTCAGTAGATTTAGTAATTTWTTCKTTACATTTGGCTGGAATTTCTTCTATTTTAGGTGCAATTAATTTTATTAGAACTGTTTTAAATATRTGATTTTTATTAAAAATAATAGATAAATTATCATTRTTTATTTGATCAGTAATWATTACTGCTTTATTATTATTATTATCTTTACCTGTATTAGCRGGTGCTATTACAATATTATTAATGGATCGAAATTTAAATACAAGRTTTTTTGATCCTAGAGGTGGTGGAGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 17‒24/xii/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG09739-B06.
Paratypes
All Malaise-trapped. BIOUG05346-A11, BIOUG08356-G03, BIOUG05422-D07, BIOUG08904-A11, BIOUG08905-F02, BIOUG08905-F11, BIOUG08911-A03, BIOUG09435-D11, BIOUG09435-D12, BIOUG09739-B06, BIOUG09739-F12,BIOUG09827-A02, BIOUG10017-D06, BIOUG17494-A10, BIOUG17494-C11, BIOUG17569-F01, BIOUG17569-F07, BIOUG17575-E05, BIOUG17575-E09, BIOUG18290-E11, BIOUG18290-F08, BIOUG18397-B01, BIOUG18551-A08. Depository:
Etymology
Phanerotoma christhompsoni is named to honor Dr. Chris Thompson of the Smithsonian Institution for his decades and continuing intense enthusiasm for the taxonomy of Costa Rican Syrphidae flies and subsequent development of the INBio collections and training of INBio curators, including their current financial support.
Phanerotoma davesmithi , sp. nov.
Diagnostics
BOLD:ADB4617. Consensus barcode. GTTTTATATTTTATATTTGGAATATATTCAGGAATGTTAGGTTTATCATTGAGATTATTAATTCGTTTAGAATTGAGTGTTTTAGGYTCTATATTGGGTAATGATCAAATTTATAATATAATTGTTACTATTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGTTTTGGAAATTGATTAGTGCCTTTAATATTAGGTGGTCCTGATATAAGTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATATTATTATGTTTATCTAGATTTATTAATATAGGAGCTGGAACAGGATGAACYGTTTATCCTCCATTATCTTTAATAATTGGACATGGTGGAATTTCTGTAGATTTAGTAATTTTTTCGTTACATTTAGCAGGAATTTCTTCAATTTTAGGTGCTATTAATTTTATTAGAACTGTTTTAAATATATGATTTAAATTAAAAATAATAGATAAATTATCTTTATTTATTTGATCAGTAATTATTACTGCTTTATTATTGTTATTATCTTTACCTGTATTAGCAGGTGCAATTACTATATTATTAATGGATCGAAATTTAAAT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, 05‒12/vi/2014, Malaise trap PL12-6A. Depository:
Host data. None.
Holotype voucher codes: BIOUG29436-A09.
Paratypes
All Malaise-trapped. BIOUG29437-F01, BIOUG29584-C07, BIOUG29964-B02, BIOUG29992-B03, BIOUG29999-D12, BIOUG30041-B07, BIOUG30497-D05, BIOUG30507-F05. Depository:
Phanerotoma davidduthiei , sp. nov.
Diagnostics
BOLD:ACL9696. Consensus barcode. TATTTTATATTTTATTTTTGGTATATATTCAGGTATATTAGGATTATCTTTAAGAATAATAATTCGTTTAGAATTAAGAGTTTTAGGATCATTATTAGGTAATGATCAAATTTATAATATAATAGTTACTAGACATGCTTTTATTATGATTTTTTTTATAGTTATACCAATTATGATTGGAGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGGGGTCCTGATATAAGTTTTCCTCGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCTTTATTTTTGTTAGTTTTATCAAGATATATTAATATAGGGGCAGGTACAGGTTGAACTGTTTATCCTCCATTATCTTTAATAATAGGTCATGGAGGTATTTCAGTTGATTTAGTAATTTTTTCTTTACATTTAGCTGGAGTTTCTTCAATTTTAGGAGCTATTAATTTTATTACAACAGTTTTAAATATGTGATTTAATATTAAAATATTAGATAAGTTAAGTTTATTTATTTGGTCTGTATTAATTACTGCATTATTATTATTATTATCTTTACCTGTATTAGCAGGGGCAATTACAATATTATTAATAGATCGTAATTTAAATACAAGATTTTTTGATCCTAGAGGAGGTGGGGATCCTGTATTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 11‒18/ii/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG10247-H11.
Paratypes
None.
Phanerotoma dirksteinkei , sp. nov.
Diagnostics
BOLD:ACA7259. Consensus barcode. GAATTTTRTATTTTTTGTTTGGTATATATTCAGGAATTTTAGGTTTATCTTTAAGRATAATAATTCGTTTAGAATTGAGGGTTTTRGGTTCAATATTAGGTAATGATCAGATTTATAATATGATAGTGACTAGGCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGGTTGGTTCCTTTAATATTGGGGGGTCCTGATATAAGATTYCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCATCTTTATTTTTATTAATTTTGTCTAGRTTTATTAATATAGGGGCAGGTACTGGTTGAACTRTTTATCCTCCATTATCTTTGATAATGGGTCATGGAGGAATTTCAGTTGATTTAGTAATTTTTTCTTTACATTTAGCTGGTATTTCATCTATTTTAGGAGCTATTAATTTTATTAGTACAGTTTTAAATATATGATTTAGAGTTAAAATATTGGATAAAATAAGTTTATTTATTTGGTCAGTGGTTATTACTGCTTTATTATTATTATTGTCATTACCTGTTTTGGCAGGGGCTATTACAATATTATTAATAGATCGAAATTTAAATACTAGATTTTTTGATCCTAGTGGTGGGGGAGATCCTGTTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438 -85.6138, 300 meters, 11‒18/iii/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher codes: BIOUG18661-F05.
Paratypes
All Malaise-trapped. BIOUG09442-B05, BIOUG09738-H03, BIOUG09740-A03, BIOUG09740-D01, BIOUG09826-A02, BIOUG09826-A08, BIOUG09827-A01, BIOUG09827-B03, BIOUG10017-A10, BIOUG13945-A09, BIOUG13945-B10, BIOUG13945-D01, BIOUG17520-A12, BIOUG17550-F01, BIOUG17566-G05, BIOUG17607-G02, BIOUG17608-B03, BIOUG17967-C02, BIOUG18290-D05, BIOUG18290-H03, BIOUG18330-D05, BIOUG18499-E11, BIOUG18660-H05, BIOUG18661-A05, BIOUG18661-B01, BIOUG18661-E02, BIOUG18661-F04, BIOUG18664-C06, BIOUG18664-H09, BIOUG18665-C07, BIOUG18665-F09, BIOUG18665-G01, BIOUG18748-E11, BIOUG19974-F05, BIOUG29209-A05, BIOUG29212-G12, BIOUG29320-A11, BIOUG29320-B09, BIOUG29322-E03, BIOUG29322-E04, BIOUG29322-F05, BIOUG29327-C06, BIOUG29327-C10, BIOUG29401-C04, BIOUG29401-C07, BIOUG29403-F09, BIOUG29403-F10, BIOUG29403-F12, BIOUG29490-E11, BIOUG29831-C08, BIOUG30887-G08, BIOUG02644-C08. Depository:
Other material
Two specimens, one from Munroe County in southern Florida and one from Honduras (BIOUG02644-C08 and BIOUG19974-F05), are in the same BIN in BOLD. They are morphologically similar to Costa Rican specimens and are included as paratypes.
Phanerotoma donquickei , sp. nov.
Diagnostics
BOLD:ACS0581. Consensus barcode. AATTTTATATTTTTTATTTGGTATGTATTCAGGAGTATTAGGGTTGTCATTAAGAATAATAATTCGTTTAGAATTGAGGGTTTTAGGATCTATATTGGGTAATGATCAAATTTATAATATAATGGTTACTAGACATGCTTTTATTATAATTTTTTTTATGGTTATACCAATTATAATTGGGGGTTTTGGGAATTGGTTAGTTCCTTTAATATTAGGAGGTCCTGATATAAGTTTTCCTCGAATAAATAATATAAGGTTTTGATTATTAATTCCATCATTATTTTTATTAGTTTTATCTAGATATATTAATATAGGAGCAGGAACTGGTTGAACTGTATACCCTCCTTTATCTTTAATAATGGGTCATGGTGGAATTTCGGTTGATTTAGTAATTTTTTCATTACATTTAGCTGGAATTTCTTCAATTTTAGGTGCTATTAATTTTATTAGAACAGTTTTGAATATATGATTTAGAATAAAAATGTTGGATAAATTAAGTTTATTTATTTGATCTGTAGTAATTACTGCTTTATTATTATTATTATCTTTACCTGTATTAGCAGGTGCTATTACTATATTATTAATGGATCGAAATTTAAATACTAGATTTTTTGA----------------.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 18‒25/ii/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG18510-C09.
Paratypes
None.
Phanerotoma duniagarciae , sp. nov.
Diagnostics
BOLD:ADB2375. Consensus barcode. ATTTTATATTTTTTATTTGGAATATATTCAGGCATTATAGGTTTATCTTTAAGAATAATAATTCGTTTGGAATTAAGAGTTTTAGGTTCTATGTTAGGAAATGATCAAATTTATAATATATTTGTTACAAGTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGGTTTGGGAATTGATTAGTTCCATTAATATTAGGTGGGCCTGATATAAGTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAGTTCCTTCTTTATTATTATTAATTTTGTCTAGATATACTAATATAGGAGCAGGGACTGGTTGAACAGTGTATCCTCCTTTATCTTTAATAATGGGTCATGGAGGAATTTCTGTTGATTTAGTAATTTTTTCTTTACATTTAGCTGGTATTTCTTCTATTATGGGGGCTATTAATTTTATTAGAACAATTTTAAATATATGATTTGATATTAAAATATTAGATAAATTGAGATTATTTATTTGATCTGTAATAATTACTGCTTTATTATTACTGTTATCATTACCTGTATTAGCTGGAGCTATTACAATATTATTAATGGAT------------------------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631 -85.3344, 820 meters, 19‒26/xii/2013, Malaise trap PL12-3A. Depository:
Host data. None.
Holotype voucher code. BIOUG29456-B07.
Paratypes
None.
Phanerotoma duvalierbricenoi , sp. nov.
Diagnostics
BOLD:ADD5784. Consensus barcode. ATTTTATATTTTTTATTTGGTATATATTCAGGCATTATAGGGTTGTCTTTAAGAATGATAATTCGTTTGGAATTAAGAGTATTAGGTTCAATATTAGGAAATGATCAAATTTATAATATATTTGTTACAAGACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGGTTAGTACCATTAATATTAGGTGGACCTGATATAAGTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTATTTTTATTAATTTTGTCTAGTTATACAAATATAGGTGCAGGTACTGGTTGAACAGTTTATCCACCTTTATCTTTAATAATAGGTCATGGGGGAATTTCTGTTGATTTAGTAATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATTATAGGGGCAATTAATTTTATTAGAACTGTTTTAAATATATGATTTAGAATAAAAATGTTAGATAAGTTAAGTTTATTTATTTGATCAGTTGTTATTACTGCTTTGTTATTGTTATTATCTTTACCTGTATTAGCTGGAGCTATTACAATATTATTA---------------------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-2, 10.7634, -85.335, 824 meters, 15‒22/v/2014, Malaise trap PL12-2A. Depository:
Host data. None.
Holotype voucher code. BIOUG30941-E12.
Paratypes
None.
Phanerotoma eddysanchezi , sp. nov.
Diagnostics
BOLD:AAW3563. Consensus barcode. AATTTTATATTTTTTATTTGGAATATATTCTGGRGTTATTGGTTTATCATTAAGTATAATAATTCGTTTAGAATTAAGAGTATTAGGATCTATATTAGGAAATGATCAAATTTATAATATATTTGTTACTAGACATGCTTTTATTATAATTTTTTTTATGGTTATACCTATTATAATTGGTGGTTTTGGAAATTGATTGGTTCCTTTAATGTTAGGGGGTCCTGATATAAGTTTCCCTCGAATAAATAATATRAGATTTTGRTTATTAGTTCCTTCTTTAATRTTATTAATTTTGTCTAGTTATACTAATATAGGGGCRGGTACTGGATGAACTGTTTATCCTCCTTTATCTTTAATAATAGGTCATGGGGGTATTTCTGTTGATTTAGTAATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTAGTACAGTTTTAAATATATGATTTAGAATTAAAATATTAGATAAATTAAGATTATTTATTTGGTCAGTTGTAATTACTGCTTTATTATTATTATTATCTTTACCTGTATTAGCTGGGGCTATTACAATACTATTAATAGATCGAAATTTAAATACTAGGTTTTTTGATCCAAGAGGTGGGGGTGATCCWG-TTTATATCAGCATTTATTT.
Holotype ♀
Guanacaste, Sector Mundo Nuevo, Estación La Perla, 10.76737, -85.43313, 325 meters, caterpillar collection date: 09/i/2011, wasp eclosion date: 23/ii/2011. Depository:
Host data. Gregarious parasitoid on Oryctometopia fossulatella (Pyralidae) feeding on Semialarium mexicanum (Celastraceae).
Caterpillar and holotype voucher codes. 11-SRNP-55189, DHJPAR0045461, two parasitoids, both with this voucher code, emerged from the host caterpillar.
Paratypes
Host = Oryctometopia fossulatella: BIOUG18005-A02, DHJPAR0034287. Depository:
Etymology
Phanerotoma eddysanchezi is named to honor Sr. Eddy Sanchez for his generous and supportive understanding of the integration of ACG conservation goals with the ICE goals of constructing a geothermal development site on the margin of ACG as a Natural World Heritage Site.
Phanerotoma eldarayae , sp. nov.
Diagnostics
BOLD:ACJ4087. Consensus barcode. TGTTTTATATTTTTTGTTTGGAATATATTCTGGAATTTTAGGTTTATCTTTAAGTATAATAATTCGTTTAGAATTA---AGAGTTTTAGGATCAATATTAGGTAATGATCAAATTTATAATATAATAGTTACTAGACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGGAATTGGTTAGTTCCTTTAATATTAGGTGGACCTGATATAAGATTTCCTCGAATAAATAATATAAGATTTTGGTTATTAGTTCCTTCTTTATTTTTATTAATTATATCTAGATATATTAATATAGGTGTAGGAACAGGATGAACTGTTTATCCTCCATTATCTTTAATAATAGGTCATGGTGGAATTTCAGTAGATTTAGTTATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTTTAGGTGCAATTAATTTTATTAGAACAGTAATAAATATATGATTTAGAATAAAAATATTAGATAAATTAAGATTATTTATTTGATCTGTAGTTATTACTGCTTTATTGTTATTATTATCTTTACCTGTTTTAGCAGGTGCTATTACTATATTATTAATAGATCGAAATTTAAATACAAGATTTTTTGATCCAAGAGGAGGTGGAGATCCTGTATTGTATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 07/v/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG07919-D03.
Paratypes
None.
Phanerotoma eliethcantillanoae , sp. nov.
Diagnostics
BOLD:ADE1674. Consensus barcode. ---------------------------TTCTGGAGTTTTAGGTTTGTCATTAAGAATAATAATTCGTTTAGAATTAAGTGTTTTAGGATCAATATTAGGTAATGATCAAATTTATAATATAATAGTTACTAGACATGCTTTTGTAATGATTTTTTTTATAGTAATACCTATTATAATTGGAGGTTTTGGGAATTGGTTGGTTCCTTTAATATTAGGTGGACCTGATATAAGATTTCCTCGGATAAATAATATAAGATTTTGATTATTGATTCCTTCTTTATTTTTATTAATTTTGTCTAGTTATATTAATATAGGGGCAGGTACAGGATGGACTGTTTATCCTCCATTATCTTTAATAATAGGTCATGGTGGAATTTCTGTAGATTTAGTTATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATTTTAGGTGCTATTAATTTTATTAGAACAGTTTTTAATATATGATTTAGAATAAAAATATTAGATAAATTAAGATTGTTTATTTGATCTGTAGTAATTACTGCTTTATTATTATTATTATCTTTACCTGTTTTGGCTGGTGCAATTACTATATTATTAATAGATCGAAATTTAAATACAAGGTTTTTTGATCCAAGAGGGGGGGGGGATCCTGTATTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Rincon Rain Forest, Camino Rio Francia, 10.90425, -85.28651, 410 meters, caterpillar collection date: 05/iv/2016, wasp eclosion date: 02/v/2016. Depository:
Host data. phyBioLep01 BioLep17 (Pyralidae) feeding on Stryphnodendron microstachyum (Fabaceae).
Caterpillar and holotype voucher codes. 16-SRNP-40256, DHJPAR0059458.
Paratypes
None.
Phanerotoma jenopappi , sp. nov.
Diagnostics
BOLD:ADY2296. Consensus barcode. TTTATATTTTATATTTGGTTTATATTCTGGATTTTTAGGATTATCTTTAAGTTTAATAATTCGTTTGGAATTAAGAGTTTTAGGAGTTATATTAGGTAATGATCAAATTTATAATATAATAGTTACAGTTCATGCATTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCATTAATGTTAGGAGGTCCTGATATAAGATTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATTTTAATAATTATTTCAAGATTTATTAATATAGGAGCAGGAACAGGTTGAACAGTTTATCCTCCATTATCGTTAATAATGGGACATGGTGGAATTTCTGTGGATTTAGTAATTTTTTCTTTACATTTGGCAGGAATTTCATCTATTTTAGGGGCAATTAATTTTATTAGTACTGTTATAAATATATGGTTTATAATTAAAATATTAGAAAAATTATCTTTATTTATTTGGTCTGTATTAATTACTGCTTTATTATTATTATTATCTTTACCAGTTTTAGCTGGTGCTATTACAATATTATTAATAGATCGTAATTTAAATACTAGTTTTTTTGATCCTAGTGGAGGTGGGGATCCAATTTTATATCAACATTTATT.
Holotype ♂
Guanacaste, Sector Pailas Dos, PL12-3, 10.7637, -85.3331, 853 meters, Malaise trap, 19/iii/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG46564-D07.
Paratypes
None.
Pseudophanerotoma Zettel, 1990
The genus is restricted to the New World and contains 14 described species. Eight species are recorded from the Neotropical region (
Pseudophanerotoma alanflemingi , sp. nov.
Diagnostics
BOLD:ACL3198. Consensus barcode. TATTTTATATTTTATATTTGGTATATATTCTGGTGTGTTAGGTTTGTCTTTAAGTTTATTAATTCGAATAGAATTAAGATGTTTAGGTAGATTATTAGGTAATGATCAAATTTATAATATAGTAGTAACAATTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAGTTATAATTGGTGGATTTGGTAATTGATTAATTCCTTTAATAATTGGTGGGCCTGATATATCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTATTTTTATTACTTATATCAAGTTTTGTAAATATAGGAGTAGGAACTGGTTGAACAGTTTATCCTCCTTTATCTTTAATGATAGGTCATGGTGGTATATCWGTAGATTTAGCTATTTATTCATTACATTTAGCTGGAATTTCATCAATTATAGGAGCAGTAAATTTTATTACAACAATTTTAAATATATGAAATTTAAATTTTATAGATAAATTATCTTTATTTAGTTGATCTGTTATTATTACAGCTTTATTATTATTATTATCATTACCTGTATTAGCAGGTGCTATTACTATATTATTAAGAGATCGTAATTTAAATACTAGATTTTTTGATCCAAGTGGAGGGGGGGATCCTGTTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 16‒23/i/2014, Malaise trap PL12-9A. Depository:
Host data. None.
Holotype voucher code. BIOUG28722-F02.
Paratype
Malaise-trapped. BIOUG09826-G03. Depository:
Pseudophanerotoma albanjimenezi , sp. nov.
Diagnostics
BOLD:AAV8843. Consensus barcode. AGTTTTATATTTTATATTTGGTATATATTCAGGTTTTATTGGTTTATCATTAAGATTGTTAATTCGGATAGAATTAAGATGTTTAGGGGGATTAATAGGTAATGATCAAATTTATAATATAATTGTAACAATTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATAATTGGTGGACCTGATATATCTTTCCCACGAATAAATAATATAAGATTTTGATTATTAGTTCCTTCCTTATTTTTATTAATATTTTCAAGTTTTGTTAATATAGGGGTTGGGACAGGTTGAACAGTTTAYCCTCCATTATCATCAATTACTGGACATAGGGGTATATCTGTAGATATAGCTATTTATTCTTTACATTTAGCTGGTGCGTCTTCAATTATAGGGGCAGTAAATTTTATTACAACTATTTTAAATATATGAAATTTTAAATTATTAGATAAAATAYCATTATTTGTATGATCAGTTTTTATTACAGCTTTATTATTATTATTATCTTTACCTGTCTTAGCTGGTGCAATTACAATGTTATTAAGAGATCGAAATTTGAATACAAGATTTTTTGATCCAAGAGGTGGAGGTGACCCTATTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal, Tajo Angeles, 10.86472, -85.41531, 540 meters, caterpillar collection date: 22/viii/2011, wasp eclosion date: 09/ix/2011. Depository:
Host data. Episimus ortygia (Tortricidae) feeding on Vismia baccifera (Hypericaceae).
Caterpillar and holotype voucher codes. 11-SRNP-3257, DHJPAR0048279.
Paratypes
Host = Episimus ortygia: DHJPAR0040083, DHJPAR0055207. Depository:
Pseudophanerotoma alejandromarini , sp. nov.
Diagnostics
BOLD:ACE1984. Consensus barcode. AATTTTATATTTTATATTTGGAATATATTCTGGWATRTTAGGRTTATCATTAAGTTTATTAATTCGAATAGAGTTAAGATGTTTAGGAAGATTATTAGGAAATGATCAAATTTATAATATAATTGTAACAATTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAGTAATAATTGGTGGRTTTGGAAATTGATTAGTACCTTTAATAATAGGTGGACCAGATATATCTTTYCCACGAATAAATAATATAAGATTTTGATTATTRATTCCTTCTTTAATATTATTAATTTTATCAAGTTTYGTTAATATAGGAGTTGGGACAGGWTGAACAGTTTAYCCWCCTTTATCATTAGTATTAGGRCATGGGGGTATATCAGTTGATATAGCAATTTATTCATTACATTTAGCTGGAATTTCATCAATTATAGGTGCTATTAATTTTATTTCTACAATTATYAATATATGAAGTTTAAAATTATTTGATAAAATATCTTTATTTAGYTGATCAGTAATTATTACAGCTTTTTTATTATTAATGTCATTACCAGTATTAGCTGGTGCAATTACRATATTATTAAGGGATCGAAATTTAAATACAAGATTTTTTGATCCAAGTGGAGGAGGGGATCCAATTTTATACCAACATTTATTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 05‒12/xii/2013, Malaise trap PL12-9A. Depository:
Host data. None.
Holotype voucher code. BIOUG28669-E05.
Paratypes
All Malaise-trapped. BIOUG04583-E05, BIOUG25658-C12, BIOUG28388-B02, BIOUG29507-E10, BIOUG29577-D11, BIOUG04583-E05, BIOUG25658-C12. Depository:
Note
Two specimens from Honduras, BIOUG04583-E05 and BIOUG25658-C12, are in the same BIN and are likely conspecific, though they were not examined.
Pseudophanerotoma alexsmithi , sp. nov.
Diagnostics
BOLD:ACB1516. Consensus barcode. TATTTTATATTTTATATTTGGTATATATTCTGGTATATTAGGGTTATCATTAAGTTTATTAATTCGTATAGAATTAAGGTGTTTAGGTAGGTTATTAGGRAATGATCAAATTTATAATATAATTGTTACAATTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTTATAATTGGAGGATTCGGAAATTGATTGGTACCTTTAATAATTGGTGGACCAGATATATCTTTTCCTCGAATAAATAATATAAGATTTTGACTATTAYTACCTTCTTTATTTTTATTAATTATATCAAGATTTGTAAATATAGGAGTTGGTACAGGATGAACTGTTTATCCTCCATTGTCATTAATAATTGGTCATGGTGGTATATCAGTTGATATAGCTATTTATTCATTACATTTAGCTGGTATTTCTTCTATTATAGGAGCAATTAATTTTATTACAACAATTACAAATATATGGAATTTTAAATTGTTTGATAAAATATCYTTGTTTAGATGGTCAGTAATTATTACAGCTTTATTACTTTTATTATCATTACCAGTTTTGGCTGGTGCTATTACAATATTATTAAGAGATCGAAATTTAAATACAAGATTTTTTGATCCAAGGGGTGGTGGGGATCCAATTTTATACCAACATTTGTTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Palomo, 10.96187, -85.28045, 96 meters, caterpillar collection date: 05/iii/2012, wasp eclosion date: 20/iii/2012. Depository:
Host data. Cosmorrhyncha albistrigulana (Tortricidae) feeding on Dialium guianense (Fabaceae).
Caterpillar and holotype voucher codes. 12-SRNP-67407, DHJPAR0049445.
Paratypes
Hosts = Cosmorrhyncha albistrigulana, Cosmorrhyncha brown001. DHJPAR0049430, DHJPAR0049441, DHJPAR0049443, DHJPAR0049444, DHJPAR0049450, DHJPAR0049452, DHJPAR0049453, DHJPAR0049456, DHJPAR0049458, DHJPAR0049459, DHJPAR0049460, DHJPAR0049461, DHJPAR0049462, DHJPAR0049463, DHJPAR0049466, DHJPAR0049473, DHJPAR0055194, DHJPAR0055300. Depository:
Etymology
Pseudophanerotoma alexsmithi is named to honor Dr. Alex Smith of the University of Guelph in recognition of his dedicated logistic, taxonomic, and ecological support for all things ACG, with special emphasis on the biology of the volcano elevational gradients in biodiversity.
Pseudophanerotoma allisonbrownae , sp. nov.
Diagnostics
BOLD:ACJ2201. Consensus barcode. AGTATTATATTTTATTTTTGGTATTTATGCAGGAGTAGTAGGTTTGTCAATAAGAATAATAATTCGATTAGAATTAAGAGTTTTAGGATCATTAATAGGAAATGATCAAATTTATAATGTATTAGTGACAAGACATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAAGTTTTCCTCGAATAAATAATATAAGATTTTGATTATTGGTACCTTCATTATATTTTTTAATAATATCAATATTTATTAATATAGGAGCAGGAACTGGATGAACAGTATATCCTCCTTTGTCAATAACTATAAGGCATAGAGGGGTTTGTGTAGATTTAGTAATTTTTTCTTTACATTTAGCTGGGATTTCATCAATTTTAGGTTCAATTAATTTTATTAGAACTATTTTTAATATATGAATTGATTATAAATTAATAGATAAATTGAGTTTATTTATTATTTCAGTATTAGTAACTGCTTTTTTATTGTTATTGTCTTTACCAGTATTAGCAGGTGCAATTACAATATTATTAAGAGATCGAAATTTGAATACAAGATTTTTTGATCCTAGAGGAGGTGGTGATCCTGTTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 16‒23/iv/2012, Malaise Trap, DH Janzen and W Hallwachs. Depository:
Host data. None.
Holotype voucher code. BIOUG07616-H01.
Paratype
BIOUG07918-H11.
Etymology
Pseudophanerotoma allisonbrownae is named in honor of Allison Brown, formerly of the Centre for Biodiversity Genomics, for her intensely accurate specimen processing of ACG insects for international taxonomists and for her tolerance of their quirks.
Pseudophanerotoma bobrobbinsi , sp. nov.
Diagnostics
BOLD:ADB6487. Consensus barcode. TTTTTTTTTGGTATTTATTGTGGAATAATAGGTTTATCATTAAGAATATTAATTCGATTAGAATTAAGAGTTTTAGGGTCTTTATTAGGAAATGATCAAATTTATAATGTAATAGTTACTAGTCATGCATTTATTATAATTTTTTTTATGGTTATACCCATTATGATTGGTGGGTTTGGTAATTGATTAGTACCATTAATATTGGGTGGYCCAGATATAAGATTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATATTATTAATTATATCAAGGTTTGTTAATATAGGTGCAGGAACTGGTTGAACTGTTTATCCACCATTATCATTAATAGTTGGACATAGAGGAATTTCTGTTGATTTAGTAATTTTTTCTTTACATTTAGCTGGTATATCTTCTATTTTAGGGGCTATTAATTTTATTAGGACAGTTTTAAATATATGAATTAGTATAAAAATATTGGATAAATTATCTTTATTTATTTGATCAGTTATAATTACTGCTTTATTATTATTATTATCTTTACCAGTTTTAGCAGGTGCTATTACTATATTATTAAGAGATCGAAATTTAAATACA.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 13‒20/ii/2014, Malaise trap PL12-3A. Depository:
Host data. none.
Holotype voucher code. BIOUG29681-H06.
Paratypes
BIOUG33275-D10, BIOUG33411-D01. Depository:
Etymology
Pseudophanerotoma bobrobbinsi is named in honor of Bob Robbins of the Smithsonian Institution for his decades of taxonomic support for ACG Lycaenidae and the inclusion of ACG voucher specimens in the National Museum of Natural History in Washington, D.C.
Chapter 5: Homolobinae
Homolobinae is a small subfamily containing three genera, two of which occur in the New World. The subfamily is cosmopolitan with several widespread species. The world fauna was revised by van
Key to the New World genera of Homolobinae
Exasticolus van Achterberg, 1979
Exasticolus is restricted to the New World, and the most recent key to species is found in
Exasticolus jennyphillipsae , sp. nov.
Diagnostics
BOLD:AAW1602. Consensus barcode. GTTTTATATTTTTTATTGGGAATTTGATCAGGAATTTTAGGTATATCAATAAGATTTTTAATTCGAATAGAATTAATAATACCAGGTAGATTTTTAAGAAATGATCAAATTTATAATAGTATCGTTACTAGACATGCTTTTATTATAATTTTTTTTTTAGTTATACCAATAATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGATGTCCTGATATGGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATTTTATTATTATTAAGAAGTATGGTAAATTTAGGAGTAGGTACTGGTTGAACAGTTTATCCTCCTTTGTCATTAAATTTAAGTCATAGGGGTATATCTGTAGATATGGCTATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATTATAGGAGCAATTAATTTTATTACAACAATTTTAAATATACGTTCTAATATAGTTTTTATGGATAAAATTCCTTTATTTGTTTGATCAGTATTTATTACTGTAATTTTACTATTATTGTCTTTACCTGTTTTAGCAGGAGCTATTACTATGTTATTGACTGATCGAAATTTAAATACATCTTTTTTTGATCCTTGTGGTGGAGGGG--------------------------.
Similar to E. xmatkuilensis López-Martínez, Delfin-González and van Achterberg 2011 but differing in the ratio of forewing veins r/3RSa, which is 0.9‒1.1 in E. xmatkuilensis and 1.3 in this species.
Holotype ♀
Guanacaste, Sector Cacao, Sendero Cima, 10.93328, -85.45729, 1460 meters, collection date: 30/vi/2008, wasp eclosion date: 29/vii/2008. Depository:
Host data. noto 08-SRNP-35960 (Notodontidae) feeding on Vaccinium poasanum (Ericaceae).
Caterpillar and holotype voucher codes. 08-SRNP-35960, DHJPAR0028118.
Paratypes
None.
Etymology
Exasticolus jennyphillipsae is named in honor of Jenny (Eugenie) Phillips of San José, Costa Rica in recognition of her extreme patience in dealing with the foundational bureaucracy of the startup of the BioAlfa process to facilitate bioliteracy in Costa Rica.
Exasticolus jennyphillipsae, cocoon. A snow-white cocoon of Exasticolus jennyphillipsae was discovered in the silk and leaf pupal chamber of 08-SRNP-35960, a medium large Notodontidae caterpillar. However, without an associated adult moth, the identity of the host is unknown at present. As the cocoon ages, it turns golden brown in color, but long after the wasp has emerged through the circular hole cut by the wasp in the end of the cocoon.
Exasticolus randallgarciai , sp. nov.
Diagnostics
BOLD:ACM4427. Consensus barcode. TATTTTATATTTTTTATTAGGTATTTGAGCAGGGATTTTAGGAATATCTATGAGAATTTTAATTCGTATAGAATTAATGATACCTGGGAGATTTTTAAGTAATGATCAGATTTATAATAGGATAGTAACTAGACATGCTTTTATTATAATTTTTTTTTTAGTTATACCAATAATAATTGGGGGATTTGGAAATTGATTAATTCCTTTAATGTTAGGATGTCCAGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATTTTATTAATATTAAGAAATATAGTAAATTTAGGAGTAGGTACTGGTTGAACTGTATACCCTCCTTTATCTTTAAATTTAAGTCATAGGGGGATATCTGTAGATATGGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTACAACAATTTTAAATATACGATCAGAAGGAATTTTTATAGATAAAATTCCTTTATTTGTTTGATCTGTTTTTATTACAGTAATTTTATTATTATTATCTTTACCTGTTTTAGCTGGGGCCATTACTATATTATTGACTGATCGAAATTTAAATACATCATTTTTTGATCCTTGTGGTGGGGGAGATCCAATTTTATATCAACATTTATTT. Similar to E. xmatkuilensis López-Martínez, Delfin-González & van Achterberg, 2011 but differing in the degree of sculpture on the frons, which is relatively smooth in E. xmatkuilensis and with more carinae in this species. The ocelli are much smaller in E. xmatkuilensis, and the black color restricted to the stemmaticum in E. xmatkuilensis is more extensive in E. randallgarciai.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Nacho, 10.98445, -85.42481, 710 meters, caterpillar collection date: 11/xi/2013, wasp eclosion date: 15/i/2014. Depository:
Host data. Leptostales angulataDHJ01 (Geometridae) feeding on Sabicea panamensis (Rubiaceae).
Caterpillar and holotype voucher codes. 13-SRNP-31747, DHJPAR0054461.
Paratypes
None.
Exasticolus robertofernandezi , sp. nov.
Diagnostics
BOLD:AAA5869. Consensus barcode. AATTTTATATTTTATTTTGGGTATTTGATCAGGTATTTTAGGTATATCAATAAGATTATTAGTACGAATRGARTTGATAGTTCCTGGAAAATTTTTAGGTAAYGATCAAATTTATAATAGAATTGTTACTGGCATGCATTTTTATAATTTTTTTTTTAGTTATACCAATAATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGATGTCCAGATATAGCTTTTCCTCGTATAAATAATATAAGGTTTTGGTTATTAATTCCTTCTTTATTATTATTAATTATTAGAAGATTAATAAATGTTGGGTTGGAACTGGGTGAACAGTTTATCCTCCGTTATCTTTAAATATAAGTCATAGAGGAATATCTGTAGATATAGCAATTTTTTCATTACATTTGGCAGGAATTTCTTCTATTATAGGAGCAATTAATTTTATTACAACTATTTTAAATATACGTTCAAGTTTAATTTATATAGATAAAATTTCTTTATTAACTTGATCAGTATTTATTACAGTRATTTTATTATTATTATCTTTACCAATTTTAGCAGGAGCTATTACTATATTATTAACTGATCGTAATTTAAATACTTCTTTTTTTGATCCGTGKGGAGGTGGAGATCCAATTTTATATCAACATTTATTT. Close to Exasticolus nigriceps (Enderlein, 1920) but differing in the much larger ocelli of Exasticolus robertofernandezi.
Holotype ♀
Guanacaste, Sector Horizontes, Vado Esteron, 10.763, -85.56, 95 meters, caterpillar collection date 20/vii/1999, wasp eclosion 21/viii/1999. Depository:
Host data. Gregarious parasitoid of Ormetica sicilia (Erebidae, Arctiinae) feeding on leaves of Inga vera (Fabaceae); 6 wasps eclosed 21/viii/1999 and 4 eclosed 23/viii/1999 from 10 beige wasp cocoons spun next to the cadaver inside the moth cocoon.
Caterpillar and holotype voucher codes. 03-SRNP-9821, DHJPAR0029331.
Paratypes
As recorded for each of the vouchers below, all are hairy, medium-sized Arctiinae (Erebidae) caterpillars. All 15 caterpillar cocoons have gregarious parasitoids that spin their white cocoons inside the caterpillar cocoon after emerging from the prepupal caterpillar. Hosts = Lophocampa modestaDHJ01, Lophocampa maroniensis, Ormetica ataenia, Ormetica sicilia, Viviennea tegyra. DHJPAR0021125, DHJPAR0022194, DHJPAR0029333, DHJPAR0029334, DHJPAR0029335, DHJPAR0021107, DHJPAR0021108, DHJPAR0021124, DHJPAR0021126, DHJPAR0021127, DHJPAR0021128, DHJPAR0021133, DHJPAR0016437, DHJPAR0016438, DHJPAR0016440, DHJPAR0016441, DHJPAR0022099, DHJPAR0022100, DHJPAR0022101, DHJPAR0022102, DHJPAR0022103, DHJPAR0022104, DHJPAR0022105, DHJPAR0022107, DHJPAR0022108, DHJPAR0022109, DHJPAR0022114, DHJPAR0023536, DHJPAR0023537, DHJPAR0016439, DHJPAR0023538, DHJPAR0023539, DHJPAR0023540, DHJPAR0023541, DHJPAR0023542, DHJPAR0023543, DHJPAR0023544, DHJPAR0023545, DHJPAR0023546, DHJPAR0029394, DHJPAR0029395, DHJPAR0029396, DHJPAR0029397, DHJPAR0029399, DHJPAR0037963, DHJPAR0023343, DHJPAR0023344, DHJPAR0023345, DHJPAR0023346, DHJPAR0023347, DHJPAR0023348, DHJPAR0023349, DHJPAR0023350, DHJPAR0036312, DHJPAR0036313, DHJPAR0036314, DHJPAR0036315, DHJPAR0036316, DHJPAR0029330, DHJPAR0029331, DHJPAR0029401, DHJPAR0029402, DHJPAR0055433, DHJPAR0029185, DHJPAR0029327, DHJPAR0029328, DHJPAR0029329. DHJPAR0029400. Depository:
Etymology
Exasticolus robertofernandezi is named in honor of Roberto Fernandez Ugalde of Cartago, Costa Rica, in recognition of his persistent efforts to plan minimal environmental impacts for geothermal and hydroelectric projects by Costa Rica’s National Electric Company (ICE).
Exasticolus sigifredomarini , sp. nov.
Diagnostics
BOLD:ACM4428. Consensus barcode. AATTTTATATTTTTTATTGGGAATTTGATCAGGAATTTTAGGTATGTCTATAAGAGTTTTAATTCGTATAGAATTAATAATACCAGGTAGATTTTTAAGTAATGATCAAATTTATAATAGTATTGTTACTAGACATGCTTTTATTATAATTTTTTTTTTAGTTATACCAATAATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGATGTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATTTTATTATTATTAAGAAGTATAGTAAATTTGGGTGTAGGGACAGGTTGAACAGTTTAYCCTCCTTTGTCATTAAATTTAAGTCATAGAGGAATATCTGTGGATATAGCTATTTTTTCTTTACATTTGGCTGGAATTTCATCTATTATAGGAGCAATTAATTTTATTACAACAATTTTAAATATACGTTCTAATATAATTTTTATAGATAAAATTCCTTTATTTGTTTGATCAGTATTTATTACTGTAATTTTATTGTTATTATCTTTACCTGTTTTAGCAGGGGCTATTACTATATTATTGACTGATCGAAATTTAAATACATCTTTTTTTGATCCTTGTGGTGGGGGGGACCCAATTTTGTATCAACATTTATTT. Similar to E. fuscicornis (Cameron, 1887) but differing in the ratio of forewing veins r/3RSa, which is 1.4–1.5 in E. fuscicornis and 1.1 in Exasticolus sigifredomarini.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Sendero Aura, 10.96536, -85.32386, 432 meters, caterpillar collection date: 02/vi/2017, wasp eclosion date: 29/vi/2017. Depository:
Host data. Malocampa matralis (Notodontidae) feeding on Pseudolmedia mollis (Moraceae).
Caterpillar and holotype voucher codes. 17-SRNP-26903 DHJPAR0061471.
Paratypes
Hosts = Malocampa matralis and Sericochroa felderi (Notodontidae). DHJPAR0029336, DHJPAR0054460. Depository:
Etymology
Exasticolus sigifredomarini is named to honor Sigifredo Marin of Liberia, Costa Rica, in emphatic recognition of his decades of effort to support the germination and growth of Area de Conservación Guanacaste and the projects of the Guanacaste Dry Forest Conservation Fund.
Exasticolus tomlewinsoni , sp. nov.
Diagnostics
BOLD:ADB0948. Consensus barcode. ATTTTGTATTTTTTATTAGGAATTTGATCAGGAATTTTAGGAATATCTATAAGAATTTTAATTCGAATAGAGTTAATAATACCAGGTAGGTTTTTAAGTAATGATCAAATTTATAATAGTATAGTTACTAGACATGCTTTTATTATAATTTTTTTTTTAGTTATACCTATAATAATTGGGGGGTTTGGTAATTGATTAATTCCTTTAATATTAGGATGTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGGTTATTAATTCCTTCATTAATTTTATTATTATTAAGAAGTATAGTAAATTTAGGAGTAGGAACTGGTTGAACGGTTTATCCTCCTTTATCATTAAATTTAAATCATGGRGGGATGTCTGTAGATATAGCTATTTTTTCTTTACATTTGGCTGGAATTTCTTCTATTATAGGTGCTATTAATTTTATTACAACAATTTTAAATATACGATCTGATATAATTTTTATAGATAAAATTCCTTTATTTGTTTGATCAGTATTTATTACAGTAATTTTATTATTATTATCTTTACCYGTATTAGCTGGTGCAATTACTATATTATTAACTGATCGA. Similar to E. xmatkuilensis López-Martínez, Delfin-González & van Achterberg (2011) but differing in that the basal antennal flagellomeres and the area lateral to the ocelli are black in specimens of Exasticolus tomlewinsoni and pale in E. xmatkuilensis.
Holotype ♀
Guanacaste, Sector Cacao, Pailas Dos, 10.7612, -85.3353, 791 meters, Malaise trap, 5/xii/2013. Depository:
Host data. None.
Holotype voucher code. BIOUG28731-E04.
Paratypes
BIOUG28713-H02, BIOUG28408-A02, BIOUG29043-D07, BIOUG28817-F06, BIOUG28731-F03. Depository:
Etymology
Exasticolus tomlewinsoni is named to acknowledge the participation and guidance of Tom Lewinson at the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Chapter 6: Hormiinae
There are only two New World genera of Hormiinae as strictly defined, Hormius and Parahormius. These can be differentiated with the key below. The subfamily may be distinguished from all other New World braconids by the possession of the following suite of character states: oral cavity cyclostome (= concave dorsally); metasomal terga, except first tergum, membranous medially and weakly sclerotized laterally; occipital carina complete; second submarginal cell 5-sided; stigma normal, not linear; RS vein of forewing reaching margin of wing; and Cu vein of forewing straight, not bent towards the posterior margin of the wing. For the Hormiinae NJ tree, see Suppl. material
Key to the New World genera of Hormiinae
Hormius Nees, 1819
No species of Hormius was previously recorded from Costa Rica. The few described Neotropical species are mostly from Brazil, Paraguay, and Cuba. Based on distribution, the only likely candidate to be conspecific with any of the species described here is Hormius deletus Wharton, 1993, and it is distinctive from all listed species here in having the r-m crossvein of the forewing lacking. The few New World species with recorded life-histories are gregarious ectoparasitoids of Gelechiidae, Tortricidae, and possibly Coleophoridae. Here we describe species that are parasitoids of the following families: Hesperiidae, Crambidae, Depressariidae, Gelechiidae, Pyralidae, and Tortricidae. In some of the cases below, there is just one parasite from a given caterpillar host. In these instances, the wasp may be solitary, or it may be that only one of multiple offspring survived.
Hormius anamariamongeae , sp. nov.
Diagnostics
BOLD:AAW4506. Consensus barcode. AATTTTATATTTTTTATTTGGAATATGGGCTGGTATAGTTGGTTTATCAATAAGATTAATTATTCGTTTGGAATTAGGAATACCAGGGAGTTTATTAGGTAATGATCAGATTTATAATAGAATAGTGACTGCTCATGCATTTATTATGATTTTTTTTATAGTTATACCAATTATAATTGGGGGGTTTGGAAATTGATTAATTCCTTTAATATTAGGGTCTCCTGATATAGCCTTTCCTCGAATAAATAATATAAGATTTTGATTATTAGTTCCTTCATTAATATTATTAGTTTTTAGTGGTGTTTTAAATATTGGGGTTGGTACAGGGTGGACTATATATCCTCCTTTATCTTCTTTAATTGGTCATAGAGGAATTTCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATAGGGGCTATTAATTTTATTTCTACAATTTTTAATATAAGTTTAAATTATATGAAAATAGATCAAATTAATTTATTAATTTGATCTATTTTAATTACTGCAATTTTATTATTATTATCTCTTCCTGTTTTAGCAGGGGCTATTACTATATTATTAACTGATCGTAATTTAAATACAACATTTTTTGATTTTTCTGGAGGTGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Jacobo, 10.94076, -85.3177, 461 meters, caterpillar collection date: 09/i/2011, wasp eclosion date: 23/i/2011, one of seven wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Rhectocraspeda Solis05 (Crambidae) feeding on Solanum jamaicense (Solanaceae).
Caterpillar and holotype voucher codes. 11-SRNP-69067, DHJPAR0042073.
Paratypes
Hosts = Rhectocraspeda Solis05, and Rhectocraspedia periusalis. 6 specimens, same data as holotype, and DHJPAR0056686. Depository:
Hormius angelsolisi , sp. nov.
Diagnostics
BOLD:AAA5364. Consensus barcode. ATTATTATATTTTTTATTTGGGATATGATCTGGTATAATTGGTTTATCTATAAGTTTAATTATTCGTTTAGAATTAGGAATACCTGGTAGATTATTGAGTAATGATCAAATTTATAATAGTATAGTAACCGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGATCTCCTGATATGGCTTTTCCTCGAATAAATAATATAAGGTTTTGATTATTAATCCCTTCTTTAATATTATTAATTTTTAGAAGAATTTTAAATGTAGGAGTAGGGACGGGTTGAACTATATATCCACCTTTATCTTCTTTAATTGGTCATGGGGGAATTTCTGTTGATTTAGCTATTTTTTCTTTACATTTGGCTGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTTCAACTATTTTAAATATAAATTTATATAATATAAAATTGGATCAAATTAATTTATTAATTTGATCTATTTTAATTACTGCTATTTTATTATTATTATCTTTACCTGTTTTAGCAGGGGCTATTACTATATTATTAACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGTGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste Sector Pitilla, Pasmompa, 11.01926, -85.40997, 440 meters, caterpillar collection date: 10/viii/2005, wasp eclosion date: 28/viii/2005, one of 27 wasps that emerged from the same host caterpillar. Depository:
Host data. Parphorus decora (Hesperiidae) feeding on leaves of Olyra latifolia (Poaceae).
Caterpillar and holotype voucher codes. 05-SRNP-33249, DHJPAR0029016.
Paratypes
Host = Parphorus decora: 26 specimens with same data as holotype, and DHJPAR0029009, DHJPAR0038938, DHJPAR0029007, DHJPAR0060215. Depository:
Hormius anniapicadoae , sp. nov.
Diagnostics
BOLD:AAU2092. Consensus barcode. TGGTCTGGGATAATTGGGCTTTCAATAAGATTAATTATTCGAATAGAATTAGGTATACCTGGAAGTTTATTAGGTAATGATCAAATTTATAATAGTATTGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGGTCTCCTGATATAGCTTTTCCACGAATAAATAATATAAGGTTTTGATTATTGATTCCTTCTTTATTTTTATTAATTTTTAGGGGTTTATTAAATGTTGGGGTAGGAACGGGTTGAACAATATATCCCCCCTTGTCTTCTTTAATTGGTCATAATGGTGTATCTGTTGATTTAGCTATTTTTTCTTTGCATTTAGCTGGTATTTCTTCTATTATAGGTGCTGTAAATTTTATTTCAACTATTTTTAATATAAATTTATATAATATAAAATTTGATCAAATTAATTTATTAATTTGATCAATTTTGATTACTGCATTATTATTATTATTGTCTTTACCTGTATTAGCTGGAGCTATTACAATATTATTAACTGATCGTAATTTAAATACAACATTTTTTGATTTTTCTGGTGGTGGGGACCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal. Sendero Huerta, 10.93050, -85.37223, 527 meters, caterpillar collection date: 16/viii/2009, wasp eclosion date: 29/viii/2009, one of five wasps that emerged from the host caterpillar. Depository:
Host data. Gregarious parasitoid of Amorbia productana (Tortricidae) feeding on leaves of Hyptis obtusiflora (Lamiaceae).
Caterpillar and holotype voucher codes. 09-SRNP-4249, DHJPAR0040024.
Paratypes
4 specimens, same data as holotype. Depository:
Etymology
Hormius anniapicadoae is named in honor of Annia Picado of San José, Costa Rica for her long years sorting Malaise trap specimens, identifying Diptera, and now dissecting genitalia of Microlepidoptera in the former INBio collections.
Five tightly packed elongate cocoons of Hormius anniapicadoae (DHJPAR0040024).
Hormius arthurchapmani , sp. nov.
Diagnostics
BOLD:ADA7345. Consensus barcode. ATTTTATATTTTTTATTTGGGATATGGTCAGGGATATTAGGTTTATCAATAAGGATAATTGTTCGTTTAGAATTAGGGATACCCGGTAGATTGTTAGGTAATGATCAGATTTATAATAGAATAGTAACTGCTCATGCATTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGGTTTGGAAATTGATTAGTTCCTTTAATATTAGGGTCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGGTTTTGATTATTAATTCCTTCATTGATATTATTAATTTTTAGAGGGGTGTTGAATGTTGGAGTCGGGACAGGATGAACTATTTATCCTCCATTGTCTTCTTTAATTGGTCATAGAGGAATTTCAGTTGATTTGGCTATTTTTTCTTTACATTTAGCAGGTGCTTCTTCAATTATAGGGGCAATTAATTTTATTACTACTATTTTAAATATAAATTTATATATAAAAATAGATCAAATTAGTTTATTAATTTGATCTATTATGATTACGGCAATTTTATTATTATTATCTTTGCCAGTTTTAGCTGGAGCTATTACAATACTTTTAACTGATCGAAATTTGAATACT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 8/v/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29831-H04.
Paratypes
BIOUG29863-E06, BIOUG29862-H08. Depository:
Etymology
Hormius arthurchapmani recognizes the participation of Arthur Chapman at the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and the contribution of his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius barryhammeli , sp. nov.
Diagnostics
BOLD:AAM1049. Consensus barcode. AGTATTATATTTTTTATTTGGGATATGAGCTGGTATAATTGGTTTATCAATAAGATTAATTATTCGTTTAGAGTTAGGTATACCAGGTAGTTTATTAGGTAATGATCAAATTTATAATAGTATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATGATTGGGGGTTTTGGTAATTGATTAGTTCCTTTAATATTAGGTTCACCTGATATAGCATTTCCACGAATAAATAAYATAAGATTTTGATTATTAGTTCCTTCTTTAATATTATTAATTTTTAGTGGATTATTAAATATTGGGGTAGGTACTGGATGAACTATGTATCCACCTTTATCTTCTTTAATTGGTCATGGGGGAATTTCAGTTGATTTAGCAATTTTTTCATTACATTTGGCTGGTATTTCTTCAATTATAGGGGCTATTAATTTTATTTCAACTATTTTAAATATAAATTTATATTATATAAAATTTGATCAAATTAGTTTATTGATTTGATCAATTTTAATTACTGCTGTATTATTATTATTATCTTTGCCTGTTTTAGCTGGGGCAATTACTATATTGTTGACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGAGGTGACCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Jacobo, 10.94076, -85.3177, 461 meters, caterpillar collection date: 07/v/2013, wasp eclosion date: 17/v/2013, one of eight wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Monoloxis flavicintalisDHJ02 (Pyralidae) feeding on upper surface of leaves of Lacistema aggregatum (Lacistemataceae).
Caterpillar and holotype voucher codes. 13-SRNP-69652, DHJPAR0052245.
Paratypes
Hosts = Monoloxis flavicintalisDHJ02 and Cordia panamensis (Cordiaceae). 7 specimens, same data as holotype, and DHJPAR0038037. Depository:
Etymology
Hormius barryhammeli is named in honor of Barry Hammel in recognition of his persistent high quality and essential taxonomic efforts for the flora of Costa Rica, working in tandem with Nelson Zamora.
Multiple white cocoons (09-SRNP-23352-DHJ495931.jpg) of Hormius barryhammeli (DHJPAR0038037) glued to the silk spun on the leaf surface of the nest of the host caterpillar, Monoloxis flavicintalisDHJ04 (Pyralidae). The hole through the leaf on the left is the escape hole used by the caterpillar when disturbed.
Hormius carloswalkeri , sp. nov.
Diagnostics
BOLD:ACK8132. Consensus barcode. ATTATTATATTTTTTATTTGGGATGTGATCAGGAATAATTGGTTTATCAATAAGATTGATTATTCGTTTAGAATTAGGGATACCTGGAAGATTATTAGGTAATGATCAAATTTATAATAGAATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGTTTTGGAAATTGATTAATTCCTTTAATATTGGGATCTCCCGATATAGCTTTTCCTCGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAATAATATTAATTTTTAGAGGGATTTTAAATGTTGGAGTTGGTACAGGTTGAACAATATATCCTCCTTTATCTTCACTTATTGGACATAGAGGAATTTCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATAGGTGCTATTAATTTTATTTCTACTATTTTAAATATAAATTTATATTCATTAAAATTAGATCAAATTAATTTATTAATTTGATCAATTTTAATTACTGCTATTTTATTATTATTATCTTTACCTGTTTTAGCAGGAGCAATTACTATATTATTAACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCAGGAGGAGGAGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Pasmompa, 11.01926, -85.40997, 440 meters, caterpillar collection date: 27/iii/2006, wasp eclosion date: 13/iv/2006, one of 32 wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Vettius aurelius (Hesperiidae) feeding on Lasiacis procerrima (Poaceae).
Caterpillar and holotype voucher codes. 06-SRNP-31447, DHJPAR0029027.
Paratypes
Hosts = Joanna joanna, Vettius aurelius, Cynea Irma, Vehilius vetula, Justinia norda, Tigasis simplex (all Hesperiidae). 31 specimens, same data as holotype and DHJPAR0029010, DHJPAR0057466, DHJPAR0047098, DHJPAR0047119, DHJPAR0047127, DHJPAR0049061, DHJPAR0049084, DHJPAR0053764. Depository:
Etymology
Hormius carloswalkeri is named in recognition of Carlos Walker’s many years of mentoring the financial arrangement between ACG, GDFCF, and IEDO (formerly Mesoamerica) for the power line crossing Sector Mundo Nuevo of ACG.
Hormius carmenretanae , sp. nov.
Diagnostics
BOLD:ABA7266. Consensus barcode. AGTTTTATATTTTTTATTTGGAATATGGGCTGGTATAGTTGGTTTATCAATAAGTTTAATTATTCGATTAGAATTAGGAATACCAGGTAGATTATTGGGTAATGATCAAATTTATAATAGAATAGTTACTGCACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTTTAATAATAGGAGCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATATTATTGATTTTTAGGGGAGTATTAAATATTGGGGTTGGAACTGGTTGAACTATATATCCTCCTTTATCTTCTTTAATTGGTCATGGTGGTATTTCTGTTGATTTAACAATTTTTTCTTTACATTTGGCTGGAATTTCTTCAATTATAGGAGCTATTAATTTTATTTCTACAATTTTAAATATAAATTTATATTATATAAAATTTGATCAAATTAGTTTATTAATTTGATCAATTTTAATTACTGCTATTTTATTATTATTATCTTTACCTGTTTTAGCTGGGGCTATTACAATATTATTAACAGATCGTAATTTGAATACTACTTTTTTTGATTTTTCTGGGGGAGGGGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Camino Albergue Oscar, 10.87741, -85.32363, 560 meters, caterpillar collection date: 06/iii/2011, wasp eclosion date: 28/iii/2011, 1 wasp eclosed. Depository:
Host data. Apparently solitary parasitoid of phyBioLep01 BioLep757 (Pyralidae) feeding on Croton billbergianus (Euphorbiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-954, DHJPAR0042983.
Paratypes
None.
Etymology
Hormius carmenretanae is named for Carmen Retana in recognition of her long years of guiding the Minister of Environment’s administrative office of the Ministerio del Ambiente y Energia.
Single cocoon of Hormius carmenretanae of wasp DHJPAR0042983 spun on leaf surface of nest of phyBioLep01 BioLep757.
Hormius cesarsuarezi , sp. nov.
Diagnostics
BOLD:ACK7796. Consensus barcode. AATTTTGTATTTTTTATTTGGAATATGAGCAGGTATAGTTGGGTTATCAATAAGATTAATTATTCGGTTAGAATTAGGGATACCAGGAAGATTATTAGGTAATGATCAAATTTATAATAGRATAGTTACTGCACATGCATTTGTTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGTTCACCTGATATAGCTTTYCCTCGTATAAATAATATAAGATTTTGATTATTAGTTCCYTCATTAATATTATTAATTTTTAGTGGTTTATTAAATATTGGRGTGGGTACAGGATGGACAATGTATCCTCCATTATCTTCATTAATTGGACATGGAGGTATTTCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATGGGGGCTATTAATTTTATTTCAACAATTTTTAATATAAATTTGTATTATTTAAAATTGGATCAGATTAATTTATTAATTTGATCAATTTTAATTACTGCTGTTTTATTAYTATTATCTCTACCTGTTTTAGCTGGAGCAATTACAATATTATTAACTGATCGTAATTTAAATACAACATTTTTTGATTTTTCMGGAGGRGGTGAYCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Sector Pitilla, Ingas, 11.00311, -85.42041, 580 meters, caterpillar collection date: 15/ii/2012, wasp eclosion date: 29/ii/2012, one of three wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Herpetogramma salbialis (Crambidae) feeding on Zexmenia virgulta (Asteraceae).
Caterpillar and holotype voucher codes. 12-SRNP-30368, DHJPAR0049093.
Paratypes
Hosts = Herpetogramma salbialis. 2 specimens, same data as holotype and DHJPAR0047041, DHJPAR0047116, DHJPAR0039420, DHJPAR0039421, DHJPAR0031136, DHJPAR0038115, DHJPAR0038262, DHJPAR0053699, DHJPAR0029000, DHJPAR0029006, DHJPAR0029013, DHJPAR0035321, DHJPAR0035390, DHJPAR0041698, DHJPAR0041709, DHJPAR0041724. Depository:
Etymology
Hormius cesarsuarezi is named in recognition of Cesar Suarez’s continued management support for the IEDO powerline crossing Sector Mundo Nuevo of ACG.
Hormius danbrooksi , sp. nov.
Diagnostics
BOLD:ADY0841. Consensus barcode. TTTATATTTTTTATTTGGAATATGATCTGGAATAATTGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAGGTATACCAGGGAGTTTATTAGGTAATGATCAAATTTATAATACAATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATCGGAGGTTTTGGAAATTGATTAGTTCCTTTAATATTAGGTTCTCCAGATATAGCTTTTCCTCGGATAAATAATATAAGTTTTTGGTTATTAGTTCCTTCTTTAATATTATTAATTTTTAGTGGTTTATTGAATATTGGTGTTGGAACAGGTTGAACAATATATCCTCCATTATCTTCTTTAATTGGACATAGAGGAATTTCTGTTGATTTGGCTATTTTTTCATTACATTTGGCTGGTATTTCATCAATTATAGGTGCAATTAATTTTATTTCAACTATTTTAAATATAAATTTATATTATATAAAATTAGATCAGATTAGTTTATTAATTTGATCAATTTTAATTACTGCTATTTTATTATTATTGTCTTTACCCGTTTTAGCTGGTGCAATTACTATATTATTAACTGATCGTAATTTAAATACTACATTTTTTGATTTTTCTGGTGGAGGTGATCCAATTTTATTTCAACATTTAT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-4, 10.7629, -85.3341, 820 meters, Malaise trap, 20/xi/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG46727-F07.
Paratypes
None.
Etymology
Hormius danbrooksi is named in recognition of the attendance and guidance of Dan Brooks at the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius eddysanchezi , sp. nov.
Diagnostics
BOLD:ACC0955. Consensus barcode. AATTTTATATTTTTTTTTTGGAATATGGGCTGGTATAGTTGGCTTATCTATAAGTTTAATTATTCGTTTAGAATTAGGGATACCTGGTAGATTATTAGGTAATGATCAAATTTATAATAGAATAGTTACTGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGTTTYGGAAATTGATTAATTCCTTTAATATTAGGATCTCCTGATATAGCTTTYCCTCGAATAAATAACATAAGATTTTGATTATTAGTACCTTCTTTAATATTATTAGTTTTTAGAGGAATTTTAAATATTGGGGTAGGAACAGGTTGAACAATATACCCACCATTATCTTCATTAATTGGTCATAGAGGAATTTCAGTTGATATAGCTATTTTTTCATTACATTTAGCTGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTTCTACTATTTTTAATATAAAATTAAATTATATAAAAATAGATCAAATTAATTTATTAATTTGATCTATTTTAATTACAGCAATTTTATTATTATTATCTTTRCCCGTTTTAGCCGGTGCAATTACTATATTATTAACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGAGGAGGGG.
Holotype ♀
Rincon Rain Forest, Jacobo, 10.94076, -85.3177, 461 meters, caterpillar collection date: 14/i/2013, wasp eclosion date: 30/i/2013, one of two wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of crambidBioLep01 BioLep77 (Crambidae) feeding on Lepidaploa tortuosa (Asteraceae).
Caterpillar and holotype voucher codes. 13-SRNP-69106, DHJPAR0051034.
Paratypes
Hosts = Herpetogramma salbialis crambidBioLep01 BioLep77 (both Crambidae). DHJPAR0056927, DHJPAR0050031. Depository:
Etymology
Hormius eddysanchezi is named in honor of Eddy Sanchez in recognition of his bureaucratic bravery of allowing the government’s PL12 geothermal project to be biomonitored by ACG.
Hormius erikframstadi , sp. nov.
Diagnostics
BOLD:ACG5362. Consensus barcode. TATTTTATATTTTTTATTTGGAATATGATCAGGAATATTAGGTTTATCAATAAGATTAATTATTCGTTTAGAATTA---GGGATACCTGGTAGATTATTAGGTAATGATCAAATTTATAATAGGATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATGATTGGTGGATTTGGGAATTGGTTAATTCCTTTAATGTTAGGGTCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGGTTTTGATTATTAATTCCTTCATTAATATTATTAATTTTTAGGGGGTTATTAAATATTGGGGTTGGGACAGGATGAACTGTTTATCCCCCATTATCTTCTTTAATTGGGCATGGGGGGATTTCTGTGGATTTAGCAATTTTTTCTTTACATTTGGCTGGTGTTTCATCAATTATGGGGGCAATTAATTTTATTACTACTATTTTAAATATAAATTTATATATA---AAAATAGATCAAATTAATTTATTAATTTGATCTATTATAATTACGGTAATTTTGTTATTATTATCATTACCAGTTTTGGCTGGTGCTATTACTATACTTTTAACTGATCGAAATTTAAATACAACTTTTTT-------------------------------------------------------------------.
Holotype ♀
Guanacaste, Sector Santa Rosa Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 30/vii/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG05883-A02.
Paratypes
None.
Etymology
Hormius erikframstadi is named in honor of Erik Framstad for his participation in the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius georgedavisi , sp. nov.
Diagnostics
BOLD:ACG5375. Consensus barcode. TGTTTTATATTTTTTATTTGGAATATGATCAGGTATATTAGGTTTATCAATAAGATTGATTGTTCGTTTAGARTTAGGGATACCTGGTAGTTTATTAGGTAATGATCAAATTTATAATTCTATAGTTACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATGATTGGAGGTTTTGGTAATTGATTAATTCCTTTAATATTAGGATCTCCTGATATAGCTTTTCCTCGAATAAATAATATGAGGTTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTTAGAGGTTTATTAAATATTGGGGTTGGTACAGGTTGAACTATTTATCCTCCTTTATCTTCATTAATTGGTCATGGAGGTATTTCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCTATTATAGGGGCTATTAATTTTATTACTACTATTTTTAATATGAATTTATATATAAAAATAGATCAAATTAGTTTATTAATTTGATCTATTATGATTACAGCTATTTTATTATTATTGTCTTTACCTGTATTAGCTGGAGCTATTACTATATTATTAACTGATCGAAATTTAAATACAACTTTTTT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, Malaise trap, 20/ii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29020-B06.
Paratypes
BIOUG29015-D12, BIOUG05883-C01. Depository:
Etymology
Hormius georgedavisi is named in honor of George Davis attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius grettelvegae , sp. nov.
Diagnostics
BOLD:AAM1094. Consensus barcode. AGTTTTATATTTTTTATTTGGAATATGAGCTGGTATAATTGGTTTATCAATAAGTTTAATTATTCGTTTAGAATTAGGAATACCGGGTAGGTTATTAGGAAATGATCAAATTTATAATAGTATAGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGTTTTGGAAATTGATTAGTTCCTTTAATATTAGGTTCTCCTGATATGGCTTTTCCACGTATAAATAATATAAGGTTTTGATTATTAATTCCTTCTTTGATATTATTAATTTTTAGAGGAGTATTAAATATTGGRGTAGGAACTGGATGAACTATATATCCTCCATTATCTTCATTGATTGGACATAGAGGTATTTCAGTTGATTTAGCAATTTTCTCTTTACATTTAGCTGGAATTTCATCAATTATAGGAGCAATTAATTTTATTTCTACAATTTTAAATATAAATTTGTATAATATAAAATTTGATCAAATTAGATTATTAATTTGATCTATTTTAATTACTGCAATTTTACTTTTATTATCTTTACCTGTTTTRGCGGGTGCAATTACTATATTATTAACAGATCGAAATTTAAATACAACTTTTTTTGAYTTTTCGGGGGGAGGTGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Tamarindo, 10.94259, -85.31333, 478 meters, caterpillar collection date: 10/iii/2010, wasp eclosion date: 28/iii/2010, one wasp eclosed from two cocoons in the host caterpillar. Depository:
Host data. Solitary parasitoid of Antaeotricha ribbei (Depressariidae) feeding on Guarea rhopalocarpa (Meliaceae).
Caterpillar and holotype voucher codes. 10-SRNP-69369, DHJPAR0038964.
Paratypes
DHJPAR0058966 reared from Gonionota Janzen116 (Depressariidae). Depository:
Hormius gustavoinduni , sp. nov.
Diagnostics
BOLD:ABA7247. Consensus barcode. AGTTTTATATTTTTTATTTGGAATATGATCTGGAATGGTTGGGTTATCTATAAGTTTAATTATTCGATTAGAGTTAGGGATACCAGGGAGATTATTGGGGAATGATCAGATTTATAATAGTATAGTAACCGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGTAATTGATTAATTCCTTTAATAATAGGATCTCCTGATATAGCTTTTCCTCGAATAAACAATATAAGCTTTTGGTTATTGATTCCTTCATTGATATTATTAATTTTTAGTGGTATATTAAATATTGGTGTTGGAACTGGTTGAACAATATATCCTCCTTTATCTTCTTTAATTGGGCATGGAGGTATTTCTGTTGATTTGGCAATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTACAACAATTATAAATATAAATTTATATTATATTAAATTTGACCAAATTAGTTTATTAATTTGGTCAATTTTAATTACTGCTATTTTATTATTATTATCTTTACCTGTTTTAGCTGGGGCTATTACAATATTGTTAACAGATCGTAATTTAAATACTACCTTTTTTGATTTTTCTGGAGGAGGGGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Estación Quica, 10.99697, -85.39666, 470 meters, caterpillar collection date: 22/i/2010, wasp eclosion date: 10/ii/2010 one of two wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of elachJanzen01 Janzen764 (Depressariidae) feeding on Microgramma percussa (Polypodiaceae).
Caterpillar and holotype voucher codes. 10-SRNP-70422, DHJPAR0038344.
Paratypes
Hosts = elachJanzen01 Janzen764. 1 specimen same data as holotype and DHJPAR0043056. Depository:
Hormius hartmanguidoi , sp. nov.
Diagnostics
BOLD:ACJ2468. Consensus barcode. TGTTTTATATTTTATATTTGGAATATGATCTGGAATAGTTGGTTTATCAATGAGATTAATTATTCGTTTAGAGTTGGGGATACCTGGAAGTTTATTAGGAAATGATCAAATTTATAATAGTATGGTTACATCTCATGCTTTTATTATAATTTTTTTTATGGTTATACCTATTATGATTGGAGGGTTTGGAAATTGGTTAGTTCCTTTAATATTAGGGTCTCCTGATATGGCTTTTCCTCGAATAAATAATATGAGATTTTGATTATTGGTTCCTTCTTTAATATTATTAATTTTTAGTGGATTATTAAATATTGGGGTTGGAACAGGTTGAACTATATATCCACCTTTATCTTCATTAATTGGTCATGGAGGAATTTCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTTCAACAATTTTAAATATAAATTTGTATTATATAAAATTGGATCAGATTAATTTATTAATTTGATCTATTTTAATTACTGCTATTTTATTATTATTATCTTTACCTGTATTAGCAGGGGCTATTACTATATTATTAACTGATCGTAATTTAAATACAACATTTTTTGATTTTTCAGGTGGAGGAGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Sector Rincon Rain Forest, Camino Rio Francia, 10.90425, -85.28651, 410 meters, caterpillar collection date: 14/xii/2012, wasp eclosion date: 11/i/2013, one of seven wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Dichomeris Janzen703 (Gelechiidae) feeding on leaves of Lepidaploa tortuosa (Asteraceae).
Caterpillar and holotype voucher codes. 12-SRNP-87112, DHJPAR0050996.
Paratypes
6 specimens, same data as holotype. Depository:
Etymology
Hormius hartmanguidoi is named in honor of Hartman Guido in recognition of his long-standing support for the biomonitoring of ICE’s biomonitoring of PL12 geothermal project.
Mass of tightly clumped cocoons of Hormius hartmanguidoi (DHJPAR0050996) inside the lightly silked leaves of the cocoon of Dichomerus Janzen703.
Hormius hectoraritai , sp. nov.
Diagnostics
BOLD:ADY6062. Consensus barcode. ATTATATTTTATCTTTGGAATATGGTCAGGTATATTAGGGTTATCAATAAGTTTAATTATTCGAATAGAGTTGGGGATACCAGGAAGGTTGTTAGGTAATGATCAAATTTATAATACGATGGTGACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATGATTGGGGGGTTTGGAAATTTTTTAATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGGTTTTGATTATTAATTCCTTCTTTAATTTTATTAATTTTAAGAAGGTTATTAAATACAGGGGTAGGTACAGGTTGAACAATATATCCTCCTTTATCTTTATTATTAGGACATGGAGGTATTTCTGTAGATTTATCAATTTTTTCTTTACATTTAGCAGGAATTTCATCAATTATGGGGGCAATTAATTTTATTACAACTATTTTTAATATAAATTTATTTACAATTAAAATAGATCAAATTATATTATTAGTTTGATCTGTAATAATTACTGCTTTTTTATTATTATTATCATTACCTGTATTAGCGGGGGCTATCACAATATTATTAACAGATCGTAATTTAAATACAAGATTTTTTGATTTTTCAGGAGGGGGGGATCCTATTTTATTTCAACATTTATT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 5/ii/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG44643-B02.
Paratypes
None.
Etymology
Hormius hectoraritai is named in honor of Hector Arita attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius hesiquiobenitezi , sp. nov.
Diagnostics
BOLD:ACJ1489. Consensus barcode. AGTTTTATATTTTTTATTTGGTATATGATCTGGAATAATTGGATTATCAATAAGTTTAATTATTCGATTAGAATTAGGTATACCAGGTAGTTTATTAGGAAATGACCAAATTTATAATAGAATAGTAACCGCTCATGCTTTTATTATAATTTTTTTTATAGTCATACCAATTATAATTGGAGGTTTTGGAAATTGATTAATTCCTTTAATAATAGGATCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGGTTTTGATTATTAATTCCTTCATTAATAATATTAGTTTTTAGGGGTTTATTAAATATTGGAGTTGGAACTGGTTGAACTATATATCCTCCTTTGTCTTCATTAATTGGACAYAGAGGTATTTCTGTTGATTTGGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATAGGAGCTATTAATTTTATTTCTACAATTATAAATATAAATTTATATTATATAAAATTTGATCAGATTAGATTATTAATTTGATCAATTTTAATTACTGCTATTTTATTATTGTTATCATTACCTGTTTTAGCTGGGGCTATTACAATATTATTAACAGATCGTAATTTGAATACAACTTTTTTTGATTTTTCTGGAGGAGGTGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 23/iv/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG07617-B11.
Paratypes
BIOUG07616-G03, BIOUG07458-G07. Depository:
Etymology
Hormius hesiquiobenitezi is named in honor of Hesiquio Benitez attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius irenecanasae , sp. nov.
Diagnostics
BOLD:ACB2774. Consensus barcode. AGTTTTATATTTTTTATTTGGAATGTGAGCTGGAATAGTTGGTTTATCAATGAGTTTAATTATTCGTTTAGAATTAGGTATACCAGGTAGATTATTAGGTAATGATCAAATTTATAATAGTATAGTYACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGWTTTGGAAATTGATTAGTTCCATTAATAATAGGTGCYCCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTTAGGGGATTATTAAATATTGGRGTTGGTACTGGTTGAACAATATATCCTCCTTTATCTTCTTTAATTGGRCATGGAGGAATTTCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATAGGAGCTATTAATTTTATTTCAACAATTTTAAATATAAATTTATATTATATAAAATTTGATCAGATTAGATTATTAATTTGATCAATTTTAATTACTGCTATTTTATTATTATTATCWTTACCTGTTTTRGCTGGGGCTATTACAATATTATTAACAGATCGTAATTTAAATACTACTTTTTTTGATTTTTCAGGAGGGGGGGACCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Sector Rincon Rain Forest, Rio Francia Arriba, 10.89666, -85.29003, 400 meters, caterpillar collection date: 04/iii/2012, wasp eclosion date: 17/iii/2012, 1 wasp eclosed.
Host data. Apparently solitary parasitoid of gelJanzen01 Janzen235 (Gelechiidae) feeding on Croton schiedeanus (Euphorbiaceae). Depository:
Caterpillar and holotype voucher codes. 12-SRNP-40926, DHJPAR0049383.
Paratypes
Host = Asturodes fimbriauralisDHJ02 (Crambidae): DHJPAR0052293. Depository:
Etymology
Hormius irenecanasae is named for Irene Cañas in recognition of her broad-minded attitude about integrating Costa Rican wild biodiversity conservation with industrial geothermia.
Single cocoon of Hormius irenecanasae of wasp DHJPAR0049383 glued to the leaf nest of its host caterpillar.
Hormius isidrochaconi , sp. nov.
Diagnostics
BOLD:AAT8859. Consensus barcode. ATTATTATATTTTTTATTTGGGATATGAGCTGGAATAGTTGGTTTATCAATAAGTTTAATTATTCGTTTAGAATTAGGTATACCTGGGAGATTATTAGGTAATGATCAAATTTATAATAGAATCGTTACTGCACATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGTTTTGGAAATTGATTAGTTCCTTTAATATTAGGTTCACCTGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGGTTATTAGTTCCTTCTTTATTTATATTAATTTTTAGAGGTTTATTAAATGTGGGAGTTGGTACGGGTTGAACTATATATCCTCCTTTATCTTCATTGATTGGTCATGGGGGAGTGTCTGTTGATTTGGCTATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATTATGGGGGCAGTAAATTTTATTTCAACAATTTTAAATATAAAATTATTTAATATAAAATTTGATCAAATTAATTTATTAATTTGATCAATTTTAATTACTGCTATTTTATTATTATTATCTTTACCTGTTTTGGCAGGGGCTATTACTATATTATTAACTGATCGAAATTTAAATACAACATTTTTTGATTTTTCTGGTGGTGGTGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector El Hacha, Sendero Bejuquilla, 11.03004, -85.52699, 280 meters, caterpillar collection date: 18/vi/2009, wasp eclosion date: 30/vi/2009. Depository:
Host data. Gregarious parasitoid of Antaeotricha BioLep38 (Depressariidae) feeding on Arrabidaea chica (Bignoniaceae).
Caterpillar and holotype voucher codes. 09-SRNP-21577, DHJPAR0041628.
Paratypes
None.
Etymology
Hormius isidrochaconi is named to honor Sr. Isidro Chacon of San José, Costa Rica in recognition of his 40 years of cataloguing and taxonomizing the Lepidoptera of Costa Rica, and supporting the founding and development of INBio, the National Biodiversity Institute of Costa Rica.
Hormius jaygallegosi , sp. nov.
Diagnostics
BOLD:ACK6504. Consensus barcode. AGTTTTATATTTTTTATTTGGTATGTGATCTGGAATAGTTGGTTTGTCAATAAGTTTAATTATTCGATTGGAATTAGGTATACCAGGTAGATTATTAGGTAATGATCAAATTTATAATAGAATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGAAATTGGTTAGTTCCTTTAATAATAGGGTCTCCTGATATAGCTTTTCCACGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAATATTATTAATTTTTAGTGGTTTATTAAATATTGGGGTTGGAACTGGGTGAACAATATATCCTCCTTTATCTTCTCTTATTGGTCATAGAGGAATTTCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTATAGGAGCAATTAATTTTATTTCTACAATTTTAAATATAAATTTATATTATATAAAGTTTGATCAAATTAGTTTATTAATTTGGTCTATTTTAATTACTGCTATTTTATTATTATTATCTTTACCTGTTTTAGCGGGAGCTATTACTATATTATTAACAGATCGAAATTTAAATACTACTTTTTTTGATTTTTCTGGAGGTGGGGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste Sector El Hacha, Sendero Bejuquilla, 11.03004, -85.52699, 280 meters, caterpillar collection date: 17/xii/2003, wasp eclosion date: 08/i/2004, one of 11 wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Cliniodes opalalis (Crambidae) feeding on Daphnopsis americana (Thymelaeaceae).
Caterpillar and holotype voucher codes. 03-SRNP-38276, DHJPAR0029001.
Paratypes
Host = Cliniodes opalalis: 10 specimens, same data as holotype and DHJPAR0062198, DHJPAR0029014, DHJPAR0029015, DHJPAR0029023, DHJPAR0029025. Depository:
Etymology
Hormius jaygallegosi is named for Jay Gallegos in recognition of Mesoamerica (IEDO) moral and financial support for the IEDO electric power line highly positive interaction with Sector Mundo Nuevo of ACG.
Hormius jimbeachi , sp. nov.
Diagnostics
BOLD:ADB3326. Consensus barcode. TTTTTATTTGGTATATGGTCTGGAATATTAGGGTTATCAATAAGATTAATTATTCGTTTAGAGTTAGGTATGCCTGGGAGATTATTAGGTAATGATCAAATTTATAATAGAATAGTAACAGCTCATGCATTTGTTATAATTTTTTTTATAGTGATACCAATTATAATTGGAGGATTTGGAAATTGGTTAATTCCTTTAATATTAGGGTCACCTGATATGGCTTTTCCTCGAATAAATAATATAAGGTTTTGATTATTAGTTCCTTCTTTAATATTATTAATTTTTAGGGGATTATTAAATATTGGGGTTGGTACAGGTTGAACTATTTATCCTCCTTTATCTTCTTTAATTGGTCATAGAGGAATTTCAGTTGATTTAGCAATTTTTTCTTTACACTTAGCAGGGGCATCTTCTATTATAGGGGCAATTAATTTTATTACAACTATTTTGAATATAAATTTATATATAAAAATAGATCAAATTAGTTTATTAATTTGATCAATTATAATTACGGCTATTTTATTATTATTATCATTACCTGTATTAGCT------------------------------------------------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-7, 10.7612, -85.3353, 791 meters, Malaise trap, 5/vi/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29355-F10.
Paratypes
None.
Etymology
Hormius jimbeachi is named in honor of Jim Beach attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius jimlewisi , sp. nov.
Diagnostics
BOLD:AAK5352. Consensus barcode. RRKTTTATATTTTTTATTTGGAATGTGAGCTGGRATAATTGGTTTATCAATGAGTTTAATTATTCGTTTAGAATTAGGAATACCAGGAAGATTRTTGGGTAATGATCAAATTTATAATAGTATAGTAACTGCYCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGATTTGGTAATTGATTGGTYCCTTTAATAATAGGRGCYCCTGATATGGCTTTYCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTTAGAGGTTTATTGAATATTGGGGTTGGTACTGGTTGAACAATATATCCACCTTTATCTTCATTGATTGGTCATGGAGGAATTTCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATRGGAGCAATTAATTTTATTTCAACAATTTTAAATATAAATTTATATTATATAAAATTTGATCAAATTAGATTATTAATTTGATCAATTTTAATTACTACTATTTTATTATTATTATCATTACCTGTTTTAGCTGGAGCTATTACAATATTATTAACAGATCGTAATTTAAATACTACTTTTTTTGATTTTTCAGGAGGGGGGGATCCAATTTTATTTCAGCATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Area Administrativa, 10.83764, -85.61871, 295 meters, caterpillar collection date: 03/xi/1995, wasp eclosion date: 18/xi/1995, one of two wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Salbia cassidalis (Crambidae) feeding on Lasiacis sorghoidea (Poaceae).
Caterpillar and holotype voucher codes. 95-SRNP-10855, DHJPAR0030617.
Paratypes
Hosts = Salbia cassidalis, Orphanostigma haemorrhoidalis (Crambidae). 1 specimen same data as holotype, and DHJPAR0030615, DHJPAR0038122, DHJPAR0042956, DHJPAR0047155, DHJPAR0051229. Depository:
Etymology
Hormius jimlewisi is named in recognition of Jim Lewis’ long years of curating the Hemiptera and other insects of the former INBio national insect inventory.
Hormius joelcracrafti , sp. nov.
Diagnostics
BOLD:ACX9208. Consensus barcode. ATTTTATATTTTTTATTAGGTATATGATCTGGTATATTAGGTTTATCAATAAGATTAATTATTCGTTTAGAGTTAGGAATACCTGGAAGATTGTTAGGGAATGATCAGATTTATAATAGAATAGTTACTGCACATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGGAATTGGTTAGTTCCTTTAATATTAGGTTCACCTGATATAGCTTTTCCTCGAATAAATAATATAAGGTTTTGATTATTGATTCCTTCATTAATATTATTAATTTTTAGTAGTTTATTAAATATTGGGGTTGGTACAGGTTGAACTATTTATCCTCCTTTGTCTTCATTAATTGGTCATAGGGGGATTTCAGTTGATATAGCAATTTTTTCTTTACATTTAGCGGGTGTATCTTCTATTATAGGGGCAATTAATTTTATTACTACTATTTTAAATATAAATTTATATATAAAAATAGATCAGATTAGTTTATTAATTTGATCTATTATAATTACAGCAATTTTATTATTATTATCTTTACCAGTTTTGGCTGGAGCTATTACAATACTTTTAACTGATCGGAAT---------------------------------------------.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, Malaise trap, 24/ii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG24700-H07.
Paratypes
None.
Etymology
Hormius joelcracrafti is named in honor of the participation of Joel Cracraft in the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius johanvalerioi , sp. nov.
Diagnostics
BOLD:ACC1525. Consensus barcode. TGTTTTATATTTTTTATTTGGAATATGATCTGGTATAATTGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAGGGATACCGGGAAGATTATTGGGGAATGATCAAATTTATAATAGTATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGGTTTGGGAATTGGTTAATTCCATTGATATTAGGATCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGACTATTAATTCCTTCTTTAATATTATTAGTTTTTAGAGGTATTTTAAATATTGGTGTTGGTACAGGTTGAACAATATATCCTCCTTTGTCTTCATTAATTGGTCATGGTGGAATTTCTGTTGATTTAGCTATTTTTTCATTACATTTAGCTGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTTCAACTATTTTAAATATAAATTTATATTATATAAAGTTAGATCAAATTAGATTATTAATTTGATCTATTTTAATTACTGCTATTTTATTATTATTGTCTTTACCTGTTTTAGCTGGTGCAATTACTATATTATTAACTGATCGTAATTTAAATACAACATTTTTCGATTTTTCTGGTGGGGGGGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Laguna, 10.9888, -85.42336, 680 meters, caterpillar collection date: 22/ix/2012, wasp eclosion date: 06/x/2012, one of five wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Mictopsichia Janzen340 (Tortricidae) feeding on Marcgravia nervosa (Marcgraviaceae).
Caterpillar and holotype voucher codes. 12-SRNP-31369, DHJPAR0050059.
Paratypes
Four specimens, same data as holotype. Depository:
Hormius johnburleyi , sp. nov.
Diagnostics
BOLD:ADA5377. Consensus barcode. TATTTTATATTTTTTATTTGGAATGTGATCAGGAATATTAGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAGGGATACCTGGTAGATTATTAGGTAATGATCAAATTTATAATAGGATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCAATTATGATTGGGGGATTTGGAAATTGATTAATTCCTTTAATATTAGGATCACCTGATATAGCTTTCCCTCGAATAAATAATATAAGGTTTTGATTATTAATTCCTTCATTAATATTATTAATTTTTAGAGGATTATTAAATATTGGGGTTGGGACGGGATGAACTGTTTATCCTCCATTATCTTCTTTAATTGGTCATAGGGGGATTTCTGTAGATTTAGCAATTTTTTCTTTGCATTTGGCTGGTGCTTCATCAATTATAGGGGCAATTAATTTTATTACTACTATTTTAAATATGAATTTATATATAAAAATAGATCAAATTAATTTATTAATTTGATCTATTATAATTACGGCAATTTTATTATTATTATCATTACCAGTTTTGGCTGGGGCTATTACTATACTTTTAACTGATCGAAATTTAAATACAACTTTTTTTGATTTTTCTGGGGGGGGAGATCCTATTTTGTTTCAACATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal. Sendero Huerta, 10.88, -85.389, 575 meters, Malaise trap, 28/iv/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG28377-A03.
Paratype
BIOUG28377-D02. Depository:
Etymology
Hormius johnburleyi is named in honor of John Burley attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius joncoddingtoni , sp. nov.
Diagnostics
BOLD:ADA9980. Consensus barcode. AATTTTATATTTTTTATTTGGTATATGATCAGGTATATTAGGTTTATCAATAAGTTTGATTGTTCGTTTAGAGTTAGGAATACCTGGAAGAATATTAGGGAATGATCAAATTTATAATAATATAGTTACTGCTCATGCTTTTATYATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGGTTTGGAAATTGATTAATTCCTTTAATATTAGGGTCTCCTGATATGGCTTTTCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATATTATTAATTTTTAGTGGRTTATTAAATATTGGGGTTGGGACAGGATGAACTATTTATCCTCCATTATCTTCTTTAATTGGACATAGAGGTATTTCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTGCTTCTTCAATTATAGGGGCAATTAATTTTATTACTACTATTTTAAATATAAATTTATATATAAAAATAGATCAAATTAATTTATTAATTTGATCTATTATAATTACAGCAATTTTATTATTGTTGTCTTTACCAGTTTTAGCTGGTGCYATTACTATACTTTTAACTGATCGAAATTTAAAT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, Malaise trap, 16/x/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29696-G02.
Paratypes
BIOUG29696-F02, BIOUG29696-E09. Depository:
Etymology
Hormius joncoddingtoni is named in honor of Jon Coddington for his participation in the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius jorgecarvajali , sp. nov.
Diagnostics
BOLD:AAW4502. Consensus barcode. AGTTTTATATTTTTTATTTGGTATATGATCTGGAATATTAGGGTTATCAATAAGATTAATTATTCGATTAGAATTAGGTATACCTGGAAGATTATTAGGTAATGATCAAATTTATAATAGTATAGTAACTGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGGTCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGTTTTTGATTATTAGTTCCTTCTTTAATATTATTAATTTTTAGAGGAATTTTAAATGTAGGGGTTGGTACTGGATGAACTATATATCCTCCTTTATCATCTCTTATTGGTCATAGGGGTATTTCAGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATATCTTCTATTATAGGAGCTATTAATTTTATTTCAACTATTTTAAATATAAAATTATATTATRTAAAATTAGATCAAATTAATTTATTAATTTGATCAATTTTAATTACTGCAATTTTATTATTRTTATCTTTACCTGTATTAGCTGGAGCTATTACAATATTATTAACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGAGGAGACCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Quebrada Escondida, 10.89928, -85.27486, 420 meters, caterpillar collection date: 19/vii/2011, wasp emergence date: 29/vii/2011, one of two wasps that emerged from the same host caterpillar. Depository:
Host data. Sometimes gregarious parasitoid of Desmia octomaculalis (Crambidae) feeding on Notoplura uliginosa (Rubiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-43374, DHJPAR0055451.
Paratypes
Hosts = Desmia ploralis, Desmia octomaculalis. 1 specimen same data as holotype, and DHJPAR0041689, DHJPAR0051039, DHJPAR0054838, DHJPAR0064146, DHJPAR0064159, DHJPAR0051039, DHJPAR0038269, DHJPAR0038357, DHJPAR0064159 (four specimens do not show in the BOLD BIN because they have relatively short barcodes, but by their host caterpillars and tight clustering in the NJ tree, they are unambiguously Hormius jorgecarvajali). Depository:
Etymology
Hormius jorgecarvajali is named in honor of Jorge Carvajal, the decades-long logistics facilitator for INBio and now, BioAlfa in its temporary residence in the INBio facilities.
Hormius juanmatai , sp. nov.
Diagnostics
BOLD:AAT9025. Consensus barcode. AGTTTTATATTTTTTATTTGGAATRTGATCTGGAATAATTGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAGGAATACCTGGAAGTTTATTAGGTAATGATCAAATTTATAATAGAATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATRATTGGGGGATTTGGAAATTGATTAGTTCCATTAATATTAGGATCACCTGATATRGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAGTTCCTTCWTTAATGTTATTAATTTTTAGAGGTATATTAAATATTGGGGTTGGAACAGGTTGAACAATATATCCTCCTTTATCTTCATTAATTGGTCATGGAGGAATTTCTGTTGATTTAGCTATTTTTTCATTACATTTAGCTGGTATTTCTTCTATTATAGGGGCTATTAATTTTATTTCAACAATTTTAAATATAAATTTATATTATATAAAGTTAGATCAAATTAGTTTATTAATTTGATCTATTTTAATTACTGCTATTTTRTTATTATTATCTTTACCAGTTTTAGCTGGAGCTATTACTATATTATTAACTGATCGAAATTTAAATACAACATTTTTTGATTTTTCTGGAGGAGGAGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Mundo Nuevo, Mamones, 10.77074, -85.42874, 365 meters, caterpillar collection date: 10/xii/2010, wasp eclosion date: 28/xii/2010, one of five wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Palpusia Solis25 (Crambidae) feeding on Guettarda macrosperma (Rubiaceae).
Caterpillar and holotype voucher codes. 10-SRNP-57322, DHJPAR0041552.
Paratypes
Hosts = Palpusia Solis25 and Asturodes fimbriauralis (Crambidae). 4 specimens, same data as holotype, and DHJPAR0050993. Depository:
Etymology
Hormius juanmatai is named to honor Juan Mata of San José, Costa Rica and the former INBio insect collection in Santo Domingo de Heredia, and for his diligent skill in creating images for publications from the INBio collections.
Two separated cocoons of Hormius juanmatai (DHJPAR0041552) that were tightly but irregularly clustered in the pupal chamber of the ultimate instar caterpillar.
Hormius manuelzumbadoi , sp. nov.
Diagnostics
BOLD:AAD0121. Consensus barcode. TGTTTTATATTTTTTATTTGGAATGTGGTCTGGAATAATTGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAGGAATACCTGGAAGTTTATTAGGTAATGATCAAATTTATAATAGTATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATGATTGGAGGTTTTGGAAATTGATTAGTTCCTTTAATATTAGGGTCTCCTGATATGGCTTTTCCTCGAATAAATAATATAAGTTTTTGGTTATTAATTCCTTCTTTAATATTATTGATTTTTAGAGGTATATTAAATATTGGAGTTGGAACAGGTTGAACAATATATCCTCCTTTATCTTCATTAATTGGTCATGGTGGAATTTCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTTCAACTATTTTAAATATAAATTTATATAATATAAAATTGGATCAAATTAGTTTATTGATTTGATCTATTTTAATTACTGCTATTTTATTATTATTATCTTTACCAGTGTTAGCTGGAGCTATTACTATATTATTAACTGATCGTAATTTAAATACTACTTTTTTTGATTTTTCTGGTGGAGGAGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Camino Porvenir, 10.90383, -85.25964, 383 meters, caterpillar collection date: 04/v/2012, wasp eclosion date: 24/v/2012, one of 18 wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Stenoma Janzen20 (Depressariidae) feeding on leaves of Miconia xalapensis (Melastomataceae).
Caterpillar and holotype voucher codes. 12-SRNP-41874, DHJPAR0049109.
Paratypes
Hosts = elachJanzen01 Janzen01 (Depressariidae) and Stenoma Janzen20. 17 specimens, same data as holotype, and DHJPAR0035338, DHJPAR0049128, DHJPAR0049476. Depository:
Etymology
Hormius manuelzumbadoi is named in honor of Manuel Zumbado in recognition of his decades of curating Diptera in the Costa Rican national insect collection.
Four white scattered cocoons of Hormius manuelzumbadoi (DHJPAR0049109) glued to the wall of the caterpillar rearing container.
Hormius mercedesfosterae , sp. nov.
Diagnostics
BOLD:ACG3568. Consensus barcode. TATTTTATATTTTTTATTTGGAATATGAGCAGGAATATTGGGTTTATCAATAAGATTAATTGTTCGTTTAGAGTTAGGAATACCTGGTAGATTATTAGGTAATGATCAAATTTATAATAGAATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGATCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTATTATTATTAATTTTTAGAGGATTATTAAATATTGGGGTTGGAACAGGATGAACTGTTTATCCTCCATTGTCTTCTTTAATTGGTCATAGTGGAATTTCTGTTGATTTAGCAATTTTTTCTTTRCATTTAGCCGGTGCTTCTTCAATTATAGGAGCAATTAATTTTATTACTACTATTTTGAATATAAATTTATATATAAAGATAGATCAAATTAATTTATTGATTTGATCTATTATAATTACGGCGATTTTATTATTATTATCATTACCGGTTTTGGCTGGTGCTATTACTATACTTTTAACTGATCGAAATTTAAATACAACTTTTTTTGATTTTTCGGGAGGAGGAGATCCAATTTTGTTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 29/x/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG09074-A02.
Paratypes
BIOUG09074-A10, BIOUG09074-A08. Depository:
Etymology
Hormius mercedesfosterae is named in honor of Mercedes Foster for her participation in the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius modonnellyae , sp. nov.
Diagnostics
BOLD:ADA9991. Consensus barcode. TATTTTATATTTTTTATTTGGAATATGAGCAGGAATATTGGGTTTATCAATAAGATTAATTGTTCGTTTAGAGTTAGGAATACCTGGTAGATTATTAGGTAATGATCAAATTTATAATAGGATAATTACTGCTCATGCTTTCATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGGTCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAGTATTATTAATTTTTAGAGGATTATTAAATATTGGAGGTGGAACAGGATGAACTATTTATCCTCCATTGTCTTCTTTAATTGGTCATACTGGAATTTCTATTGATTTAGCAATTTTTTCTTTACATTTAGCTGGTGCTTCTTCGATTATAGGAGCAATTAATTTTATTACTACTATTTTGAATATAAATTTATATATAAAAATAGATCAAATCAATTTATTGATTTGATCTATTATAATTACGGCGATTTTATTATTATTGTCATTACCGGTTTTAGCTGGTGCTATTACTATACTTTTAACTGATCGAAATTTAAATACA.
Holotype ♂
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 23/x/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG30201-B10.
Paratypes
BIOUG30201-B07, BIOUG30201-A12. Depository:
Etymology
Hormius modonnellyae is named in honor of Mo Donnelly attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius nelsonzamorai , sp. nov.
Diagnostics
BOLD:AAM1047 Consensus barcode. TTATCAATAAGTTTAATTATTCGATTAGAATTAGGAATACCAGGTAGATTATTAGGTAATGATCAGATTTATAATAGTATAGTAACCGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGATTTGGAAATTGATTAGTACCTTTAATAATAGGTTCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGTTTTTGATTATTAATCCCCTCTCTTATATTGTTGATTTTTAGAGGTTTATTAAATATTGGGGTTGGGACTGGTTGGACAATTTATCCTCCTTTGTCTTCATTAATTGGTCATAGAGGAATTTCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATAGGAGCTATTAATTTTATTTCAACAATTTTAAATATAAATTTATATTATATAAAATTTGATCAGATTAGATTATTAATTTGATCTATTTTAATTACTGCTATTTTATTATTATTGTCTTTACCTGTTTTAGCTGGAGCTATTACAATATTATTGACGGATCGAAATTTAAATACAACTTTTTTTGA-------------------------------------------.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Estación Llanura, 10.93332, -85.25331, 135 meters, caterpillar collection date: 12/vi/2009, wasp eclosion date: 25/vii/2009, one of nine wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Stenoma Janzen07feeding on Vismia billbergiana (Hypericaceae).
Caterpillar and holotype voucher codes. 09-SRNP-44582, DHJPAR0035445.
Paratypes
Eight specimens, same data as holotype. Depository:
Etymology
Hormius nelsonzamorai is named in recognition of Nelson Zamora’s decades of cheerfully and energetically providing high quality identifications for the foods of the caterpillar hosts of the parasitoids of ACG.
Nine neatly arrayed white cocoons of Hormius nelsonzamorai (DHJPAR0035445) that were glued to the leaf wall of the larval nest, over the cadaver, revealed by stripping off one wall of the collectively spun cover of the cocoons.
Hormius pamelacastilloae , sp. nov.
Diagnostics
BOLD:AAM1076. Consensus barcode. AATTCTTTATTTTTTATTTGGTATATGAGCAGGAATATTAGGTTTATCAATAAGATTAATTATTCGTTTAGAACTAGGAATACCAGGTAGATTATTAGGGAATGACCAAATTTATAATAGAATAGTTACAGCTCATGCTTTTGTTATAATTTTTTTTATAGTTATACCAGTAATAGTTGGAGGGTTTGGGAATTGATTATTACCTTTAATATTAAGGGCCCCTGATATAGCTTTCCCACGTTTAAATAATATAAGATTTTGGTTATTAATTCCTTCTTTATTTTTATTATTAATAAGAAGAGTATTAAATGTAGGTGTTGGTACTGGATGAACAATATATCCTCCTTTGTCTTCTTCTTTAGGTCATAGGGGTATATCAGTTGATTTAGCTATTTTTTCTTTACATATTGCGGGTATTTCATCAATTTTAGGGGCTATAAATTTTATTTCAACTATTTTTAATATACATTTATTAACTTTAAAATTAGATCAATTAACTTTATTTATTTGATCAATTTTTATTACAACTTTGTTATTATTATTATCTTTACCAGTATTAGCTGGGGCTATTACCATATTATTAACTGATCGAAATTTAAATACTTCTTTTTTTGATTTTTCTGGTGGAGGTGACCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Estación Llanura, 10.93332, -85.25331, 135 meters, caterpillar collection date: 11/i/2010, wasp eclosion date: 22/i/2010, one of three wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Omiodes humeralisDHJ07 (Crambidae) feeding on leaves of Inga oerstediana (Fabaceae).
Caterpillar and holotype voucher codes. 10-SRNP-75038, DHJPAR0038272.
Paratypes
Two specimens, same data as holotype. Depository:
Etymology
Hormius pamelacastilloae is named to honor Pamela Castillo in recognition of her direct support of the integration of the BioAlfa process into National System of Conservation Areas (SINAC) of the government of Costa Rica.
Three separate cocoons of Hormius pamelacastilloae (DHJPAR0038272) that were glued between the overlapping leaves of the nest of the host caterpillar Omiodes humeralisDHJ07 (10-SRNP-75038).
Hormius raycypessi , sp. nov.
Diagnostics
BOLD:ADB7824. Consensus barcode. TTATTATATTTTATTTTTGGAATATGATCAGGTATAGTTGGGTTAGCTATAAGTTTAATTGTACGTTTAGAATTAGGAATACCTGGTAGTTTATTAGGTAATGATCAAATTTATAATAGTATAGTTACAGCGCATGCATTTATTATAATTTTTTTTATAGTGATACCTATTATAATTGGAGGGTTTGGAAATTTTTTAGTTCCTTTAATATTAGGGTCTCCTGATATAGCATTTCCTCGTATAAACAATATAAGGTTTTGATTATTAATTCCTTCATTAATTTTATTAATTTTGAGAGGGATTTTGAATGTAGGAGTGGGGACTGGATGAACTATATATCCTCCATTATCTTCATTAGTAGGTCATAGAGGTATTTCTGTAGATTTATCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTACAACTATTTTTAATATAAGTTTATTTTCAATTAAAATAGATCAAATTATGTTATTGATTTGATCTGTGATAATTACTGCTTTTTTATTATTATTATCTTTACCCGTATTAGCGGGAGCAATTACAATATTATTAACTGAT---------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 30/i/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29646-F01.
Paratypes
None.
Etymology
Hormius raucypessi is named in honor of Ray Cypess attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius ritacolwellae , sp. nov.
Diagnostics
BOLD:ADB4713. Consensus barcode. GTTTTATATTTTTTATTTGGTATATGATCTGGAATAATTGGTTTATCAATAAGTTTAATTATTCGTTTAGAATTAGGGATACCTGGTAGATTATTAGGGAATGATCAAATTTATAATAGAATAGTGACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGGTTGATTCCATTAATATTGGGATCTCCCGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTATTATTATTAATTTTTAGAAGAATTTTAAATATTGGTGTTGGAACAGGTTGAACAATATATCCTCCTTTATCTTCATTAATTGGTCATGGTGGAATTTCTGTTGATTTAGCTATTTTTTCATTACATTTGGCTGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTTCAACTATTTTAAATATAAATTTATATTATATAAAATTAGATCAAATTAGATTATTAATTTGATCTATTTTAATTACCGCTATTTTATTGTTATTATCTTTACCTGTTTTGGCTGGTGCAATTACTATATTATTAACTGAT------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 9/i/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29536-F03.
Paratypes
None.
Etymology
Hormius ritacolwellae is named in honor of Rita Colwell attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius robcolwelli , sp. nov.
Diagnostics
BOLD:ADZ9590. Consensus barcode. TATTTTTTATTTGGAATATGATCAGGAATATTAGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAGGGATACCTGGTAGATTATTAGGTAATGATCAAATTTATAATAGGATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATGATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGGGCACCTGATATAGCTTTTCCTCGAATAAATAATATAAGGTTTTGATTATTAATTCCTTCATTAATATTATTAATTTTTAGGGGGTTATTAAATATTGGGGTTGGGACAGGATGAACTGTTTATCCTCCATTATCTTCTTTAATTGGACATAGGGGGATTTCTGTAGATTTAGCAATTTTTTCTTTACATTTAGCTGGTGCTTCATCAATTATAGGAGCAATTAATTTTATTACTACTATTTTAAATATAAATTTATATATAAAAATAGATCAAATTAATTTATTAATTTGATCTATTATAATTACGGCAATTTTATTATTATTATCATTACCAGTTTTGGCTGGT------------------------------------------------------------------.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, Malaise trap, 25/viii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG27759-A09.
Paratypes
None.
Etymology
Hormius robcolwelli is named in honor of Rob Cowell attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius rogerblancosegurai , sp. nov.
Diagnostics
BOLD:ADA9724. Consensus barcode. GTATTATATTTTATTTTTGGTATATGGGCTGGAATAGTGGGKTTATCAATGAGGTTAATTGTTCGTATAGAATTAGGTATGCCTGGGAGGTTATTAGGYAATGATCAAATTTATAATAGAATAGTTACTGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATGATTGGGGGATTTGGTAAYTTTTTAATTCCTTTAATATTAGGGGCTCCTGATATGGCTTTCCCTCGTATAAATAATATAAGTTTTTGATTGTTGATTCCTTCRTTAATTTTATTAATTTTAAGGGGAATTTTAAATACTGGGGTGGGGACAGGGTGAACTATATATCCYCCTTTGTCTTCATTATTGGGSCATGGGGGRGTTTCTGTAGATTTAGCAATTTTYTCTTTACATTTAGCTGGGGTYTCTTCAATCATAGGGGCTATTAATTTTATTACAACTATTTTTAATATAAATTTATTTTCAATTAAAATAGATCAAATTATATTGTTAATTTGGTCAGTGATGATTACTGCTTTTTTATTAYTGTTATCTTTACCAGTATTGGCAGGGGCTATTACAATATTATTAACAGATCGTAATTTAAATACTAGGTTTTTT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 26/xii/2013. Depository:
Host data. None.
Holotype voucher code. BIOUG29456-G12.
Paratypes
BIOUG28748-C02, BIOUG29537-H06. Depository:
Etymology
Hormius rogerblancosegurai is named in honor of Roger Blanco Segura attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius ronaldzunigai , sp. nov.
Diagnostics
BOLD:AAA5366. Consensus barcode. AATTTTATATTTTTTATTTGGTATATGATCTGGTATAGTTGGTTTATCAATAAGATTAATTATTCGGTTAGAATTAGGAATACCAGGAAGATTATTAGGTAATGATCAAATTTATAACAGTATAGTAACCGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGCGGATTCGGAAATTGATTAATTCCTTTAATAATAGGGTCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGTTTTTGGTTATTAGTTCCTTCATTAATGTTATTAATTTTTAGTGGGTTATTAAATATTGGAGTGGGGACTGGGTGAACTATATATCCTCCATTATCTTCATTAATTGGACATAGTGGGATTTCAGTTGATTTAGCTATTTTTTCTTTACATTTAGCAGGGGTTTCATCAATTATGGGGGCTATTAATTTTATTTCTACAATTTTAAATATAAATTTATATAATATGAAATTTGATCAAATTAGTTTATTAATTTGGTCTATTTTAATTACTGCAATTTTACTTTTATTATCTCTTCCCGTTTTAGCAGGTGCAATTACTATGTTATTAACAGATCGGAATTTAAATACAACTTTTTTTGATTTTTCTGGAGGGGGTGACCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.86472, -85.41531, 540 meters, caterpillar collection date: 19/x/2009, wasp eclosion date: 11/xi/2009, one of three wasps that emerged from the same host caterpillar. Depository:
Host data. Gregarious parasitoid of Antaeotricha fascicularis (Depressariidae) feeding on Pouteria viridis (Sapotaceae)
Caterpillar and holotype voucher codes. 09-SRNP-5487, DHJPAR0038086.
Paratypes
Host = Chlamydastis tryphon (Depressariidae), 2 wasps, same data as holotype, and DHJPAR0029024. Depository:
Etymology
Hormius ronaldzunigai is named for Ronald Zuñiga in recognition of his decades of curating Hymenoptera of Costa Rica.
Dispersed white cocoons of Hormius ronaldzunigai (DHJPAR0038086) in the remains of the caterpillar nest of leaves spun together with light silk.
Hormius russchapmani , sp. nov.
Diagnostics
BOLD:ACG4356. Consensus barcode. GTATTTTATATTTTTTATTAGGAATATGGTCTGGAATATTAGGATTATCAATAAGTTTAATTGTTCGGTTAGAATTAGGTATACCAGGAAGATTATTAGGGAATGATCAAATTTATAATAGAATAGTAACTGCACATGCATTTATTATAATTTTTTTTATAGTGATACCAATTATAATTGGTGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGGGTGCCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAATGTTATTGATTTTTAGAAGTTTATTAAATATTGGGGTTGGTACTGGTTGAACTATTTATCCTCCTTTATCTTCTTTAATTGGTCATAGAGGAATTTCAGTTGATTTAGCAATTTTTTCATTACATTTAGCAGGTGCTTCATCAATTATAGGAGCAATTAATTTTATTACTACTATTTTAAATATAAATTTATATATAAAAATAGATCAAATTAGTTTATTAATTTGATCTATTATAATTACAGCTATTTTATTATTGTTATCTTTACCAGTTTTAGCTGGAGCTATTACTATACTTTTAACTGATCGAAATTTGAATACTACTTTTTTTGATTTTTCTGGGGGAGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 25/vi/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG05195-E11.
Paratype
BIOUG10677-B04. Depository:
Etymology
Hormius russchapmani is named in honor of Russ Chapman attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius virginiaferrisae , sp. nov.
Diagnostics
BOLD:ADB9401. Consensus barcode. GTATTATATTTTTTGTTTGGAATATGAGCTGGTATATTAGGGTTATCAATAAGTTTAATTGTTCGGTTAGAATTAGGTATACCTGGGAGTTTATTAGGTAATGATCAAATTTATAATAGAATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTGATACCAATTATAATTGGTGGGTTTGGAAATTGATTGGTTCCTTTAATATTGGGGTCTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGTTTTTGGTTATTGATTCCTTCATTAATATTATTAATTTTTAGGGGTGTTTTAAATACGGGCGTCGGCACTGGATGAACAGTTTATCCTCCATTATTTCATTAATTGGTCACAGAGGTATTTCAGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGTGCTTCATTATTATAGGGGCTATTAATTTTATTACTACTATTTTAAATATAAATTTATATATAAAAATAGATCAAATTAGTTTATTAATTTGGTCAATTATGATTACGGCTGTTTTATTATTATTATCTTTACCTGTGTTAGCAGGAGCTATCACGATACTTTTAACTGAT------------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 23/x/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG30103-A11.
Paratypes
None.
Etymology
Hormius virginiaferrisae is named in honor of Virginia Ferris attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius warrenbrighami , sp. nov.
Diagnostics
BOLD:ACS0289. Consensus barcode. TTTTTATTTGGGATGTGATCTGGAATGTTAGGTTTATCAATAAGTTTAATTGTTCGTTTAGAATTAGGAATACCAGGAAGTTTATTAGGTAATGATCAAATTTATAATAGAATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATGCCAATTATAATTGGGGGGTTTGGAAATTGATTAATTCCTTTAATATTAGGTTCACCTGATATAGCTTTTCCTCGAATAAATAATATGAGGTTTTGACTTTTAGTTCCTTCTTTAATATTATTAATTTTTAGAGGATTTTTAAATACTGGAGTAGGTACAGGTTGAACAATTTATCCTCCTTTATCTTCTTTAATTGGTCATGGTGGAATTTCTGTAGATTTGGCAATTTTTTCTTTACATCTTGCTGGAATTTCTTCAATTATAGGTGCAATTAATTTTATTACAACTATTTTGAATATAAATTTATATTTAAAAATAGATCAAATTAGTTTATTAATTTGATCAATTATAATTACAGCTATTTTATTATTATTATCTTTACCTGTTTTAGCAGGTGCTATTACTATATTATTGACAGATCGT------------------------------.
Holotype ♂
Guanacaste, Sector Santa Rosa Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 28/v/2012. Depository:
Host data. None.
Holotype voucher code. BIOUG18764-G08.
Paratypes
None.
Etymology
Hormius warrenbrighami is named in honor of Warren Brigham attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Hormius willsflowersi , sp. nov.
Diagnostics
BOLD:ADB9850. Consensus barcode. TTTTTATTTGGAATATGATCTGGAATAGTTGGTTTATCTATAAGTTTAATTATTCGTTTGGAATTAGGAATACCAGGTAGATTATTAGGAAATGATCAAATTTATAATAGTATAGTAACCGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGGGGATTTGGAAATTGATTAATTCCTTTAATAATAGGGGCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCTTTAATATTATTAGTTTTTAGTGGTTTATTAAATATTGGTGTTGGAACTGGTTGAACAATATATCCTCCTTTATCATCTTTAATTGGACATGGAGGAATTTCTGTTGATTTAGCAATTTTTTCTTTACATTTGGCTGGTATTTCTTCAATTATGGGAGCAATTAATTTTATTTCTACAATTTTAAATATAAATTTATATTATATAAAATTTGATCAAATTAGTTTATTAATTTGATCAATTTTAATTACTGCTATTTTATTATTATTATCTTTACCTGTTTTAGCTGGG---------------------------------------------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 28/viii/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG29991-F05.
Paratypes
None.
Etymology
Hormius willsflowersi is named in honor of Wills Flowers attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, Philadelphia, USA, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Chapter 7: Ichneutinae
The subfamily Ichneutinae is treated here as comprising the tribes Ichneutini Foerster, 1862 and Muesbeckiini Mason, 1969. The tribe Proteropini van Achterberg, 1976 was removed from Ichneutinae and elevated to subfamily rank by Chen and van Achterberg (2019). This separation is the result of numerous phylogenetic analyses that do not resolve the Ichneutinae s. l. as monophyletic, e.g.,
Key to the New World genera of Ichneutinae
Oligoneurus Szépligeti, 1902
Members of the genus are recorded from the Holarctic, Oriental, and Neotropical realms. There are 13 described species of which two are Neotropical (Brazil, Peru). The two published host records are on Cosmopterigidae and Gracillariidae (both Lepidoptera). Here we report a new host family, Crambidae.
Oligoneurus jorgejimenezi , sp. nov.
Diagnostics
BOLD:ADB9923. Consensus barcode. ATATTATATTTTATTTTCGGTATATGATCAGGTATATTTGGTTTATCTTTAAGATTATTAATTCGATTAGAATTAGGAAATTTAGGAAATTTTATTGGAAATGATCAAATTTATAATAGAATTGTTACTTCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGGGGACCAGATATATGTTTTCCTCGTTTAAATAATATAAGATTTTGATTATTAATTCCTTCAATAATATTTATAATTATTAGAAGATTTGTTGGAAGTGGATGTGGCACAGGATGAACTGTATATCCTCCATTATCTTCATTGATTGGTCATTCTAGTTTATCGGTAGATTTTGGAATTTTTTCTTTACATTTAGYAGGAGTTTCTTCTATTTTAGGATCAATAAATTTTATTTCAACAATTTTAAATATACGATCTTTAAAATTTACTATAGAAAAAATTTCTTTATTTTGTTGAGCTGTATTAATTACAACAATTTTATTATTATTATCATTACCTGTATTAGCTGGTGCAATTACTATATTATTAACAGAT.
Holotype ♂
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, Malaise trap, 25/ix/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG30059-F11.
Paratype
BIOUG30154-F02. Depository:
Etymology
Oligoneurus jorgejimenezi is named in honor of Jorge Jimenez attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Oligoneurus kriskrishtalkai , sp. nov.
Diagnostics
BOLD:ABA7267. Consensus barcode. TATTTTATATTTTATTTTTGGTTTATGATCAGGAATAATGGGATTATCTTTAAGAATTTTAATCCGTATAGAATTAAGTGCATTAGGACAATTAATTGGTAATGATCAAATTTATAATAGAATTGTAACTTCACATGCTTTTGTGATAATTTTTTTTATAGTAATACCTGTAATAATTGGGGGATTTGGAAATTGGTTGATTCCTTTAATATTAGGAAGACCTGATATATGTTTCCCTCGATTAAATAATATAAGGTTTTGACTATTAGTTCCTTCTGTCCTCTTTATAATCATTAGAAGATTCGTAAGAAATGGATGTGGTACAGGATGAACAGTTTATCCACCTTTATCATTATTAATTGGACATTCAGGCATCTCTGTAGATTTTAGAATTTTTTCTTTACATTTAGCTGGTGTTTCTTCTATTTTAGGATCAATAAATTTTATTTCAACAATTTTGAATATACGATCTATAAAATTTAAAATAGAAAATATTTCATTATTATGCTGATCTGTAATAATTACTACAATTTTACTTTTATTATCATTGCCTGTTTTAGCGGGAGCAATTACGATATTATTAACAGATCGAAATTTAAATACAACATTTTTTGATCCTTCTGGAGGCGGAGACCCAGTTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.86472, -85.41531, 540 meters, caterpillar collection date: 8/ii/2011, wasp eclosion date: 20/iii/2011. Depository:
Host data. Desmia benealisDHJ02 (Crambidae) feeding on Drymonia macrophylla (Gesneriaceae).
Caterpillar and holotype voucher codes. 11-SRNP-521, DHJPAR0043136.
Paratypes
None.
Etymology
Oligoneurus kriskrishtalkai is named in honor of Kris Krishtalka attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Paroligoneurus Muesebeck, 1931
Members of the genus are found worldwide. There are 18 described species of which six are described from Mexico and one from Ecuador. The sole published host record is on Nepticulidae (Lepidoptera) (
Paroligoneurus elainehoaglandae , sp. nov.
Diagnostics
BOLD:ADA8821. Consensus barcode. ATTTTATATTTTTTATTTGGTATATGAGCTGGAATATTTGGTTTGTCAATAAGTTTAATTATTCGTTTAGAGTTGGGTATGTTGGGGTCTTTTATTGGTAGAGATCAAATTTATAATAGAATAGTTACTTCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGCGGATTTGGAAATTGATTAATTCCTTTAATATTATCAGCTCCAGATATAGCCTTTCCTCGTATGAATAATATGAGATTTTGATTATTAATTCCTTCTTTAATTATATTAATTTTAAGGGGGTTAATAAATAGGGGAGCAGGTACAGGGTGAACAGTTTATCCTCCTTTATCTTTATTAATTGGTCATGGGGGCATATCAGTTGATATATGTATTTTTTCTTTACATTTAGCAGGTGCTTCTTCAATTATAGGTGCAATTAATTTTATTACTACTATTTTAAATATACGTTCTATTTATTTAACTATAGATAAAATTTCTTTATTAAGATGATCAGTTTTAATTACTGCAATTTTGTTATTATTATCATTACCTGTTTTAGCTGGGGCTATTACTATACTTTTAACA---------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-1, 10.7642, -85.335, 828 meters, Malaise trap, 2/x/2013. Depository:
Host data . None.
Holotype voucher code. BIOUG28589-E03.
Paratypes
None.
Etymology
Paroligoneurus elainehoaglandae is named in honor of Elaine Hoagland attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Paroligoneurus julianhumphriesi , sp. nov.
Diagnostics
BOLD:ADF4538. Consensus barcode. AATTTTATATTTTTTATTTGGTATATGAGCTGGAATATTTGGTTTGTCAATAAGTTTAATTATTCGTTTAGAGTTGGGTATGTTGGGGTCTTTTATTGGTAGAGATCAGATTTATAATAGAATAGTTACTTCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTATCAGCTCCAGATATAGCTTTTCCTCGTATAAATAATATGAGATTTTGATTATTAATTCCTTCTTTAATTATATTAATTTTAAGGGGGCTAATGAATAGGGGAGCAGGYACAGGGTGAACAGTTTATCCTCCTTTATCTTTATTAATTGGTCATGGGGGTATATCAGTTGATATATGTATTTTTTCTTTACATTTAGCAGGTGCTTCTTCGATTATAGGTGCAATTAATTTTATTACTACTATTTTAAATATACGTTCTATTTATTTAACTATAGATAAAATTTCTTTATTAAGATGATCAGTTTTAATTACTGCAATTTTGTTATTATTATCATTACCTGTTTTAGCTGGGGCTATTACTATACTTTTAACAGATCGTAATTTGAATACT.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 30/x/2014. Depository:
Host data. None.
Holotype voucher code. BIOUG31504-H06.
Paratypes
BIOUG31533-C04, BIOUG31533-E09. Depository:
Etymology
Paroligoneurus julianhumphriesi is named in honor of Julian Humphries attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Paroligoneurus mikeiviei , sp. nov.
Diagnostics
BOLD:ADB7695. Consensus barcode. GGAATTTTATATTTTTTATTTGGTATATGGGCTGGAATATTTGGTTTGTCAATAAGTTTAATTATTCGTTTAGAGTTGGGTATGTTGGGATCTTTTATTGGTAGAGATCAAATTTATAATAGAATAGTTACTTCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTATCAGCTCCAGATATAGCATTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATTATATTAATTTTAAGGGGATTAATAAATAGGGGTGCAGGTACGGGGTGAACAGTTTATCCYCCCTTATCTTTATTAATTGGTCATGGGGGTATATCAGTTGATATATGTATTTTTTCTTTACATTTAGCAGGKGCKTCTTCAATTATAGGTGCTATTAATTTTATTACTACTATTTTTAATATACGCTCTATTTATTTAACTATAGATAAAATTTCTTTATTAAGATGATCAGTTTTAATTACTGCAATTTTATTATTATTATCATTACCTGTTTTAGCTGGGGCTATTACTATACTTTTAACAGATCGTAATTTGAATACTAGTTTTTTTGATCCTGC-GGAGGGG--.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, Malaise trap, 5/ii/2015. Depository:
Host data. None.
Holotype voucher code. BIOUG33120-D06.
Paratypes
BIOUG31723-D02, BIOUG32722-B01, BIOUG32747-H05, BIOUG32722-C04, BIOUG31532-E01, BIOUG32722-B01. Depository:
Etymology
Paroligoneurus mikeiviei is named in honor of Mike Ivie attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Chapter 8: Macrocentrinae
The New World genera of Macrocentrinae may be distinguished using the key that follows. Interestingly, no species have been recorded from Costa Rica (
Key to the New World genera of Macrocentrinae
Austrozele Roman, 1910
Worldwide in distribution, but most species found in the Palaeotropical region. Recorded hosts are Geometridae and Noctuidae; Notodontidae is recorded as a host for the first time in this work. No species have been recorded from Costa Rica. The types of all five Austrozele species previously recorded from the Neotropical region (Colombia, Mexico, Brazil, and Guyana) under Austrozele were described in Paniscozele Enderlein, 1920, a junior synonym of Austrozele Roman, 1911. These were examined by CvA and all belong to Dolichozele Viereck. For details and new combinations see the latter genus. As a result, we report here the genus as new for the Neotropical region.
Austrozele jorgecampabadali , sp. nov.
Diagnostics
BOLD:AAE4116. Consensus barcode. AATTTTATATTTTTTATTTGGGATATGATCAGGAATAATTGGATTATCAATAAGAATTATTATTCGAATAGAATTAAGTCAATCTGGATCAATAATTGGAAATGATCAAATTTACAATAGATTTGTTACTACCCATGCTTTCATTATAATTTTTTTTATAGTAATACCTACTATATTAGGGGGGTTTGGAAATTGATTAATYCCATTAATATTAGGAAGAGTTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTATYATTATTATTATTAAGAGGATTTATAAATATTGGAGTTGGTACTGGATGAACTGTTTAYCCTCCATTATCTTTAAATATTAGACATATAGGAATATCTGTTGATATAGCTATTTTTTCTTTACATTTAGCAGGAATTTCTTCTATTATAGGATCAATTAATTTTATTGTTACTATTATAAATATACGTAATTATGGAGTAATTATAGAAAAAATTAGTTTATTATGTTGATCAATTTTAATTACAGTAATTTTATTATTATTATCATTGCCTGTTTTAGCAGGAGCAATTACTATATTATTAACTGATCGAAATTTAAATACTACTTTTTTTGATCCTGCAGGAGGGGGAGATCCAATTTTATATCAACATTTATTT. Morphologically very similar to A. jorgesoberoni because of posteriorly slightly widened first metasomal tergite, glabrous apical half of subbasal cell of forewing combined with a faint sclerome and dark brown posterior half of metasoma. Differs by having the diameter of the lateral ocellus ca. equal to its distance from the eye (vs. 1.2–1.3 × in A. jorgesoberoni), vein cu-a of forewing straight and vertical (bent posteriorly and reclivous) and base of first tergite wider in lateral view (vs. slenderer).
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Palo Alto, 570 meters, 10.88186, -85.38221, caterpillar collection date: 20/viii/2004, wasp eclosion date: 14/ix/2004. Depository:
Host data. Dunama janewaldronae (Notodontidae) feeding on Chamaedorea dammeriana (Arecaceae).
Caterpillar and holotype voucher codes. 04-SRNP-4200, DHJPAR0009366.
Paratypes
Host = Dunama janewaldronae: DHJPAR0009367, DHJPAR0009370, DHJPAR0029346, DHJPAR0030311, DHJPAR0030381, DHJPAR0061211. Depository:
Austrozele jorgesoberoni , sp. nov.
Diagnostics
BOLD:AAF0520. Consensus barcode. AATTTTATATTTTTTATTTGGAATATGAGCGGGAATAATTGGGTTATCAATAAGAATTATTATTCGTATAGAATTAAGTCAACCTGGATCATTAATTGGAAATGATCAAATTTATAATAGATTTGTTACCACCCATGCTTTCATTATAATTTTTTTTATAGTAATGCCTATTATATTGGGGGGATTTGGTAATTGATTAATTCCATTAATATTAGGAAGAGTTGATATAGCTTTYCCACGAATGAATAATATAAGATTTTGATTACTAATTCCTTCTTTATTATTATTATTATTAAGAGGATTTATAAATATTGGAGTTGGTACTGGATGAACTGTTTACCCTCCCTTATCTTTAAATATTAGACATATAGGGATATCAGTTGATATAGCTATTTTTTCTTTACATTTAGCAGGAATTTCTTCTATTATAGGGTCAATTAATTTTATTGTTACAATTTTGAATATACGTAATTACGGARTAATTATAGAAAAAATTAGTTTATTATGTTGATCAATTTTAGTTACAGTAATTTTATTATTATTATCATTACCTGTTTTAGCAGGAGCTATTACTATATTATTAACTGATCGAAATTTAAATACTACTTTTTTTGACCCTGCGGGAGGGGGGGATCCAGTTTTATATCAACATTTATTT. For morphological differences see diagnosis of A. jorgecampabadali.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Naciente, 700 meters, 10.98705, -85.42816, caterpillar collection date: 04/xi/2013, wasp eclosion date: 18/xii/2013. Depository:
Host data. Acrotomodes bolaDHJ01 (Geometridae) feeding on Pimenta guatemalensis (Myrtaceae).
Caterpillar and holotype voucher codes. 13-SRNP-31637, DHJPAR0054444.
Paratypes
Host = Pyrinia Janzen02: DHJPAR0009365, DHJPAR0029343, DHJPAR0029348, DHJPAR0042749, DHJPAR0054443, DHJPAR0054445, DHJPAR0054446, DHJPAR0054447, DHJPAR0054453, DHJPAR0063839. Depository:
Dolichozele Viereck, 1911
Dolichozele is exclusively New World in distribution. Of the twelve recorded species only D. gravitarsis has host data. It is reported as a parasitoid of Lophocampa argentata and Lophocampa ingens (Erebidae, as Arctiidae; Marsh 1979). Here we report a new host family, Noctuidae. There are ten named Neotropical species; the species named by
Austrozele citreitarsis (Enderlein, 1920) Paniscozele. Valid species in Dolichozele Viereck, 1911, comb nov.
Austrozele carinifrons (Enderlein, 1920) Paniscozele. New junior synonym of Dolichozele fuscivertex (Enderlein, 1920).
Austrozele fuscivertex (Enderlein, 1920) Paniscozele. Valid species in Dolichozele Viereck, 1911, comb nov.
Austrozele griseipes (Enderlein, 1920) Paniscozele. New junior synonym of Dolichozele koebelei Viereck, 1911.
Austrozele nigricauda (Enderlein,1920) Paniscozele. New junior synonym of Dolichozele quaestor (Fabricius, 1804). (originally described as Ophion quaestor Fabricius, 1804).
Dolichozele josefernandeztrianai , sp. nov.
Diagnostics
BOLD:ACK6060. Consensus barcode. ATTTTATATTTTTTATTCGGAATATGAGCAGGTATAATTGGTTTATCAATAAGAATCATTATTCGAATCGAATTAAGTCAATCAGGATCATTTATTAGTAATGATCAAATTTATAATAGATTTGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGAAATTGATTAATTCCTTTAATATTAGGAAGAGTTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTATTATTATTATTAATAAGAGGATTTATAAATATTGGAGTGGGAACTGGATGAACAGTTTATCCTCCTTTATCATTAAATATTAGTCATAGAGGAATATCAGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGTATATCATCAATTATAGGATCTATTAATTTTATTGTAACTATTTTAAATATACGAAATTATGGAGTTATTTTAGAAAAAATTAGATTATTAAGATGATCAATTTTAATTACTGTAATTTTATTATTATTATCTTTACCTGTTTTAGCAGGAGCAATTACAATATTATTAACAGATCGAAATTTAAATACTACTTTTTTTGATCCTTCTGGGGGGGGAGATCCTGTATTATACCAACATTTATTT. Differs from other species by its brown hind tarsus, vein cu-a of forewing bent posteriorly and subbasal cell of forewing without a faint sclerome.
Holotype ♀
Guanacaste, Sector Pitilla, Colocho, 375 meters, 11.02367, -85.41884, caterpillar collection date: 25/vii/2005, wasp eclosion date: 02/x/2005. Depository:
Host data. Goniohelia Poole02 (Erebidae) feeding on Senegalia multipinnata (Fabaceae).
Caterpillar and holotype voucher codes. 05-SRNP-33496, DHJPAR0028979.
Paratypes
None.
Dolichozele josephinerodriguezae , sp. nov.
Diagnostics
BOLD:AAF3799. Consensus barcode. AATTTTATATTTTATATTTGGAATATGATCAGGAATAATTGGTTTATCGATAAGAATTATTATTCGAATAGAATTAAGACAATCAGGATCATTTATTTGTAATGATCAAATTTATAATAGATTTGTAACTGCACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATGATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGTAGAGTTGATATAGCTTTTCCTCGAATAAATAATATAAGGTTTTGATTATTAATTCCTTCATTATTATTATTATTAATGAGTGGATTTATAAATATTGGAGTAGGAACAGGTTGAACTGTTTATCCTCCTTTATCTTTAAATATTAGTCAAAGAGGAATATCAGTTGATATGGCTATTTTTTCTTTACATTTAGCTGGAGTATCATCAATTATAGGGTCAATTAATTTTATTGTAACTATTTTAAATATACGAAATTATGGAATTATTATAGAAAAAATTAGATTATTATGTTGATCAATTTTAATTACTGTAATTTTATTATTAATATCATTACCTGTATTAGCAGGAGCAATTACTATATTATTAACAGATCGAAATTTAAATACTACTTTTTTTGATCCTGCAGGAGGAGGAGATCCTGTATTATATCAACATTTATTT. The new species shares with D. fuscivertex (Enderlein) the dark brown band on the vertex, but differs by the comparatively robust first tergite (distinctly widened posteriorly and approx. 3 × longer than its apical width (parallel-sided in D. fuscivertex and approx. 5 × longer than its apical width), inner hind claw of female different from outer hind claw (similar in D. fuscivertex) and subbasal cell of forewing with faint nearly round sclerome (absent in D. fuscivertex).
Holotype ♀
Guanacaste, Sector Pailas, Estación Pailas, 785 meters, 10.76969, -85.34080, caterpillar collection date: 01/viii/1998, wasp eclosion date: 3/ix/1998. Depository:
Host data. Bagisara albicosta (Noctuidae) feeding on Croton morifolius (Euphorbiaceae).
Caterpillar and holotype voucher codes. 98-SRNP-11256, DHJPAR0029357.
Paratypes
Host = Bagisara albicosta: DHJPAR0029416, DHJPAR0029417, DHJPAR0029415, DHJPAR0029356, DHJPAR0021564, DHJPAR0021114. Depository:
Note
males and females differ strongly in color.
Etymology
Dolichozele josephinerodriguezae is named to honor Dr. Josephine Rodriguez of the University of Virginia’s College at Wise, for her intense collaboration and taxonomy of ACG Microgastrinae as part of the ACG ongoing Biodiversity Inventory.
Dolichozele gravitarsis
Diagnostics
BOLD:AAI9864. Consensus barcode. ATTTTATATTTTTTATTTGGAATATGATCAGGAATAATTGGTTTATCAATAAGAATTATTATTCGAATAGAATTRAGACAATCAGGAATATTTATTGGTAATGATCAAATTTATAATAGATTTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTCATACCAATTATAATTGGTGGATTTGGAAATTGAYTAATTCCTTTAATATTAGGAAGWRTTGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGATTATTAATTCCATCATTATTTTTATTATTAATAAGAGGATTYATAAATATTGGTRTTGGAACTGGTTGAACTGTATATCCTCCTTTATCATTAAATGTAAGTCATAGAGGAATATCTGTTGATATAGCTATTTTTTCTCTTCATTTAGCWGGAATTTCTTCAATYATAGGATCAATTAATTTTATTGTTACTATTTTAAATATACGTAATTATGGAATCATTATAGAAAAAATTAGATTATTATGTTGATCAATTTTAATTACTGTTATTTTATTATTAATRTCTTTACCAGTTTTRGCTGGAGCAATTACTATATTATTAACAGATCGAAATTTAAATACTACATTTTTTGATCCTGCTGGAGGAGGTGATCCTGTATTATAYCAACATTTATTT. Both sexes have a strongly bent forewing vein M+Cu1 and comparatively short hind tibial spurs; the females have a robust and yellowish hind tarsus, which is ivory or whitish and slenderer in males.
Figured specimen
♂, Alajuela, Sector Rincon Rain Forest, Camino Albergue Oscar, 10.877, -85.324, 560 meters, caterpillar collection date 3/iii/2008, wasp eclosion date 26/v/2008.
Host data. Turuptiana obliqua (Erebidae) feeding on Nectandra hihua (Lauraceae).
Caterpillar and figured male voucher codes. DHJPAR0027736.
Additional specimens
Host = Hypocrisias Espinoza01 DHJPAR0027736, DHJPAR0029351, DHJPAR0029352, DHJPAR0029353, DHJPAR0029355. Depository:
Notes
The species is widespread from British Columbia, Canada, through the western USA, Mexico, and Meso America. One specimen from British Columbia (BIOUG22381-E09) is in the same BIN.
Hymenochaonia Dalla Torre, 1901
Hymenochaonia is exclusively New World in distribution although H. delicata (Cresson, 1872) has been introduced (apparently unsuccessfully) to Italy for biocontrol purposes. Hymenochaonia delicata has been recorded as a parasitoid of many caterpillar species including members of Depressariidae, Gelichiidae, Noctuidae, Pyralidae, and Tortricidae. It cannot be ruled out that more than one species of parasitoid were treated in these published reports (see
Hymenochaonia kalevikulli , sp. nov.
Diagnostics
BOLD:AAP9411. Consensus barcode. AATATTATATTTTATTTTTGGAATATGGGCAGGTATAATTGGTTTATCAATAAGATTAATTATTCGWATAGAATTAGGTCAATCAGGRAGTTTAATTGCTAATGATCAAATTTAYAATAGRGTAGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGRGGATTTGGTAATTGATTAATTCCTTTAATATTAGGGRGGGTTGATATAGCTTTCCCACGTATAAATAATATAAGATTTTGGTTATTAATTCCTTCAATTATTTTATTATTAATAAGAGGATTTTTGAATATTGGGGTTGGTACTGGTTGAACTATTTATCCTCCTTTATCTTTAAATATTAGACATTTAGGAGTTTCTTTAGATATAACTATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATTATAGGGGCTATAAATTTTATTGTAACAATTTTAAATATACGTAATTCTGGAGTATTAATAGATAAAATTAGATTATTATGTTGATCAATTTTAATTACTGCTATTTTATTATTATTGTCTTTACCAGTGTTAGCAGGGGCAATTACAATATTATTAACTGAYCGAAATTTAAATACTTCTTTTTTTGATCCTTCAGGAGGGGGGGACCCAATTTTATAYCAGCATTTATTT. This species and H. keithwillmotti sp. n. share a long and widened vein 1Cua of forewing, antenna (except blackish base) pale yellowish or whitish and distinctly curved vein cu-a of forewing. Hymenochaonia kalevikulli differs by having vein 1Cua of forewing longer than vein cu-a, tarsal claws simple, hind tibia white basally and remainder black, subbasal cell of forewing largely glabrous and apical half of metasoma black.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Sendero Albergue Crater, 10.84886, -85.3281, 980 meters, caterpillar collection date: 06/xii/2010, wasp eclosion date: 7/x/2010. Depository:
Host data. Antaeotricha caprimulgaEPR01 (Depressariidae) feeding on Gonocalyx pterocarpus (Fabaceae).
Caterpillar and holotype voucher codes. 10-SRNP-3034, DHJPAR0040587.
Paratypes
Hosts = Antaeotricha caprimulgaEPR01 and Antaeotricha caprimulgaEPR02. DHJPAR0040484, DHJPAR0040567, DHJPAR0040571 DHJPAR0040572, DHJPAR0040574, DHJPAR0040578, DHJPAR0040579, DHJPAR0040580, DHJPAR0040584, DHJPAR0040585, DHJPAR0040591, DHJPAR0040595. Depository:
Hymenochaonia kateperezae , sp. nov.
Diagnostics
BOLD:AAA7061. Consensus barcode. GGTTTTATATTTTATATTTGGGTTATGGGCGGGTATAATTGGKATATCAATAAGATTAGTTATTCGTTTAGAATTAGGTCAATCAGGGGTATTAATTGGTAATGAYCAAATTTATAATAGATTTGTAACTGCCCATGCTTTTGTTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGGTTTGGTAATTGATTAATTCCTTTAATATTAGGAAGGGTTGATATAGCATTTCCTCGGATAAATAATATGAGATTTTGATTATTAATTCCTTCAATTAATTTATTAGTTTTTAGGGGATTTATAAATATTGGGGTAGGTACTGGGTGAACTGTTTATCCCCCTTTATCATTAAATATTAGACATATAGGTATTTCTGTGGATATAGCAATTTTTTCTTTACATTTAGCAGGGGTTTCTTCTATTATAGGAGCTGTAAATTTTATTGTAACTATTATAAATATACGAAATAATAAAGTATTTATAGATAAAATTAGATTATTATGTTGATCTATTTTAATTACTGYAATTTTATTATTATTGTCTTTACCAGTTTTAGCYGGGGCAATTACAATATTATTAACTGATCGTAATTTAAATACATCTTTTTTTGACCCAGCAGGTGGAGGGGATCCTGTTTTATACCAGCATTTATTT. The long vein 2-SR+M of forewing (ca. as long as vein m-cu), ovipositor sheath ca. 1.2 × as long as body and simple tarsal claws are shared with the H. texana (Muesebeck, 1932) from USA and Mexico. However, the new species has vein 1Cua distinctly wider than vein 2-Cu1 (similar in H. texana), POL ca. equal to diameter of posterior ocellus (ca. 2 × diameter of posterior ocellus in H. texana), vertex laterally pale yellowish (dark brown in H. texana) and vein cu-a of forewing inclivous (vertical in H. texana).
Holotype ♀
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.86472, -85.41531, 540 meters, caterpillar collection date: 23/vii/2011, wasp eclosion date: 15/viii/2011. Depository:
Host data. elachBioLep01 BioLep754 (Depressariidae) feeding on Tapirira brenesii (Anacardiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-2902, DHJPAR0045062.
Paratypes
Host = Stenoma BioLep86: DHJPAR0035531, DHJPAR0035533. Depository:
Etymology
Hymenochaonia kateperezae is named to honor Ms. Kate Perez for her many years laboring in execution and administration of the field and laboratory actions performed by the Centre for Biodiversity Genomics to rapidly and accurately DNA barcode tens of thousands of Costa Rican insects from Malaise traps installed by ACG and BioAlfa.
Hymenochaonia katherinebaillieae , sp. nov.
Diagnostics
BOLD:ACK7901. Consensus barcode. WATATTATATTTTTTATTTGGRATTTGATCWGGGATAATTGGTTTATCAATAAGTTTAATTATTCGTTTAGAGTTAGGTCAATCTGGTTCATTAATTGGTAATGATCAAATTTATAATAGTATAGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTWATACCAATTATAATTGGAGGATTTGGRAATTGATTAATTCCTTTAATATTAGGTAGAATTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGRTTAYTRATTCCTTCAATTAAATTATTAATTTTAAGGGGATTTATAAATATTGGRGTWGGTACTGGTTGAACAGTTTAYCCYCCTTTATCATTAAATATAAGACATATAGGYATTTCTGTAGATATATCWATTTTTTCTTTACATTTGGCGGGTATTTCTTCTATTATAGGKTCAATTAATTTTATTGTWACTATTTTAAATATACGAAATTATGGARTTTTAATAGATAAAATTAGTTTATTATGTTGATCTATTTTAATTACTGTTATTTTATTATTATTKTCATTRCCTGTRTTGGCTGGTGCAATYACAATATTATTRACTGATCGTAATTTAAATACATCTTTTTTTGATCCTTCAGGRGGTGGRGAYCCTGTTTTATATCAACATTTATTT. Similar to H. kateperezae sp. nov. because of the long vein 2-SR+M of forewing (ca. as long as vein m-cu) and moderately widened and medium-sized vein 1Cua of forewing, but differs by having a brownish stripe in the subbasal cell of forewing (absent in H. kateperezae), vein m-cu of forewing much longer than vein 3-Cu1 (slightly longer in H. kateperezae), hind tarsus brownish yellow (ivory in H. kateperezae), tarsal claws with lamella (absent in H. kateperezae) and vein cu-a of forewing subvertical (inclivous in H. kateperezae). The long vein 2-SR+M of forewing (ca. as long as vein m-cu), simple tarsal claws and distinctly widened marginal cell of hind wing are shared with the H. texana (Muesebeck, 1932) from USA and Mexico. However, the new species has vein 1Cua distinctly wider than vein 2-Cu1 (similar in H. texana), claws with ventral lamella (with no lamella in H. texana), ovipositor sheath ca. as long as body (distinctly longer than body in H. texana) and first metasomal tergite hardly widened posteriorly (distinctly widened in H. texana).
Holotype ♀
Alajuela, Sector San Cristobal, Corrales Viejos, 10.89974, -85.38085, 495 meters, caterpillar collection date: 8/xi/2003, wasp eclosion date: 18/xii/2003. Depository:
Host data. Quadraforma obliqualis (Pyralidae) feeding on Myrcia splendens (Myrtaceae)
Caterpillar and holotype voucher codes. 03-SRNP-34090, DHJPAR0029337.
Paratypes
Hosts = Quadraforma obliqualis, Pococera sadotha, Carthara abruptaDHJ01, Carthara abruptaDHJ02, Chloropaschia pegalis, Carthara abrupta, Chloropaschia granitalis, Accinctapubes albifasciataDHJ03, Accinctapubes albifasciataDHJ01, Chloropaschia mennusalis, Chloropaschia Janzen3596, Tancoa crinite, Deuterollyta majuscule, Chloropaschia Janzen05 (all Pyralidae). DHJPAR0009297, DHJPAR0009298, DHJPAR0029180, DHJPAR0028980, DHJPAR0009658, DHJPAR0009767, DHJPAR0028996, DHJPAR0029369, DHJPAR0030262, DHJPAR0030263, DHJPAR0030264, DHJPAR0030265, DHJPAR0037792, DHJPAR0037865, DHJPAR0037866, DHJPAR0037867, DHJPAR0037868, DHJPAR0037869, DHJPAR0037870, DHJPAR0037871, DHJPAR0010512, DHJPAR0040229, DHJPAR0040230, DHJPAR0040231, DHJPAR0040232, DHJPAR0040235, DHJPAR0040236, DHJPAR0040237, DHJPAR0040238, DHJPAR0040240, DHJPAR0040609, DHJPAR0041130, DHJPAR0041131, DHJPAR0041133, DHJPAR0041134, DHJPAR0041135, DHJPAR0041136, DHJPAR0041137, DHJPAR0041209, DHJPAR0041210, DHJPAR0041211, DHJPAR0041212, DHJPAR0041213, DHJPAR0041214, DHJPAR0041216, DHJPAR0041217, DHJPAR0041218, DHJPAR0041219, DHJPAR0041220, DHJPAR0041221, DHJPAR0042391, DHJPAR0042392, DHJPAR0045055, DHJPAR0045072, DHJPAR0045378, DHJPAR0045813, DHJPAR0045819, DHJPAR0045821, DHJPAR0045829, DHJPAR0046725, DHJPAR0046726, DHJPAR0046727, DHJPAR0046728, DHJPAR0046729, DHJPAR0048080, DHJPAR0048082, DHJPAR0048084, DHJPAR0048085, DHJPAR0050913, DHJPAR0050918, DHJPAR0054448, DHJPAR0054449, DHJPAR0054450, DHJPAR0054451, DHJPAR0054452, DHJPAR0055123, DHJPAR0062983, DHJPAR0048088. Depository:
Hymenochaonia katherineellisonae , sp. nov.
Diagnostics
BOLD:ADF5149. Consensus barcode. GTATACTTTATTTTTTATTTGGTATGTGATCAGGAATATTGGGGYTATCTATAAGTGTGTTAATTCGTTTAGAATTAGGATGTTCTGGCACTCTTATTAATAATGATCAAATTTATAATAGATTTGTTACTGCTCATGCTTTTATTATAATTTTTTTTATGGTTATACCTATTATAATTGGGGGGTTTGGTAATTGATTAGTTCCTTTAATGTTGGGGAGTGTTGATATAGCTTTCCCTCGAATAAATAATATGAGATTTTGATTATTAATTCCTTCTATTAATATATTGTTATTAAGGGGTTTTATTAATATTGGGGTAGGTACTGGTTGAACAGTTTATCCTCCTTTATCTTTAAATGTAAGTCATATAGGTATTTCTGTTGATATAGCTATTTTTTCTTTACATTTAGCGGGTATTTCTTCAATTATAGGGGCTATTAATTTTATTGTTAGAATTTTAAATATACGAAATTATGGAATTTTAATAGAAAAAATTAGGTTAATATGTTGATCTATTTTAATTACTGCTATTTTATTATTACTTTCATTGCCTGTATTGGCTGGTGCTATTACAATGTTGTTAACTGAT. An easily recognizable species because of the robust (ca. 2 × as long as wide apically) and black first metasomal tergite, the simple tarsal claws and the dark brown stigma.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, 20/xi/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG31549-B03.
Paratypes
All Malaise-trapped: BIOUG33120-C06, BIOUG36548-H01. Depository:
Hymenochaonia katyvandusenae , sp. nov.
Diagnostics
BOLD:AAL5547. Consensus barcode. AATATTATATTTTTTATTTGGGATATGATCAGGAATAATTGGTTTGTCAATAAGTTTAATTATTCGATTGGAATTAGGTCAATCAGGTACATTAATTGGTAATGATCAAATTTATAATAGGTTTGTTACTTCACATGCATTTATTATAATTTTTTTCATAGTTATACCAATTATAATTGGAGGGTTTGGGAATTGATTAATCCCTTTAATATTAGGAAGAGTTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGGTTATTAATTCCTTCTTTAATAATGTTAATTTTAAGAGGATTTGTAAATATTGGGGTGGGTACTGGTTGAACAGTTTACCCCCCTTTATCATTAAATATTAGACATATAGGAATTTCTGTAGATATAGCTATTTTTTCTTTACATTTAGCAGGAATTTCATCAATTATAGGGGCTATTAATTTTATTGTAACAATTATAAATATACGAAATTATGGAGTAATAATAGATAAAATTAGATTATTAAGTTGATCAATTTTAATCACTACTATTTTGTTATTATTATCATTACCTGTATTGGCTGGTGCTATTACTATATTATTAACTGATCGAAATTTAAATACGTCTTTTTTTGACCCTTCAGGAGGTGGGGATCCAATTTTATATCAACATTTATTT. An easily recognizable species because of the unique oval shape of vein cu-a of forewing; in addition, the vein is shorter than normal (subequal to length of vein 1Cua), the first metasomal tergite is very slender and the face is bicolored.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Sendero Albergue Crater, 10.84886, -85.3281, 980 meters, caterpillar collection date: 1/iii/2010, wasp eclosion date: 29/iii/2010. Depository:
Host data. Pilocrocis purpurascens (Crambidae) feeding on Pentagonia donnell-smithii (Rubiaceae).
Caterpillar and holotype voucher codes. 10-SRNP-1080, DHJPAR0038863.
Paratypes
Host = Pilocrocis purpurascens (Crambidae): DHJPAR0036780, DHJPAR0037835, DHJPAR0039536, DHJPAR0040227, DHJPAR0041215, DHJPAR0042732, DHJPAR0052091, DHJPAR0052092, DHJPAR0052093, DHJPAR0057264, DHJPAR0057303. Depository:
Hymenochaonia kazumifukunagae , sp. nov.
Diagnostics
BOLD:AAX3464. Consensus barcode. GTTTTATATTTTTTATTTGGGATATGATCAGGAATAATTGGTTTATCTATAAGAATAATTATTCGTTTAGAATTAGGGCAATC---AGGGACTTTAATTAATAATGATCAAATTTATAATAGATTTGTAACTTCACATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATCGGGGGGTTTGGAAATTGGTTAATTCCTTTAATATTAGGTAGGGTAGATATAGCTTTCCCTCGGATGAATAATATAAGATTTTGATTATTAATTCCATCATTAATAATATTGATTTTAAGGGGATTTATAAATATTGGGGTAGGTACGGGATGAACAGTATATCCTCCTTTATCTTTGAATGTAAGGCATATAGGGGTTTCAGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGTATTTCCTCTATTATGGGGGCTATAAATTTTATTATTACTATTTTAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATGTTGATCAGTTTTAATTACTGCAATTTTATTATTATTATCATTGCCAGTTTTAGCAGGAGCTATTACAATATTATTAACTGATCGAAATTTAAATACATCTTTTTTTGATCCATCAGGAGGGGGGGATCCAATTTTGTATCAACATTTATTT. Both H. keithlangdoni and H. kazumifukunagae have the ovipositor sheath and antenna of ♀ medially white or ivory combined with a strongly widened vein 1Cua of forewing which is shorter than curved vein cu-a. Differs from H. keithlangdoni by the longer ovipositor sheath (approx. 1.3 × longer than body) and largely dark head and mesosoma.
Holotype ♀
Guanacaste, Sector Del Oro, Quebrada Serrano, 11.00025, -85.45614, 585 meters, caterpillar collection date: 19/vii/2007, wasp eclosion date: 10/viii/2007. Depository:
Host data. tortricid 07-SRNP-22617 (Tortricidae) feeding on Hampea appendiculata (Malvaceae).
Caterpillar and holotype voucher codes. 07-SRNP-22618, DHJPAR0021643.
Paratypes
None.
Hymenochaonia keithlangdoni , sp. nov.
Diagnostics
BOLD:AAC3252. Consensus barcode. TATATTATATTTTTTATTTGGAATATGATCGGGAATAATTGGTTTATCAATAAGTATAATTATTCGTTTAGAATTAGGTCAATCAGGAACTTTAATTAATAATGATCAAATTTATAATAGATTTGTAACTTCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGGTTTGGAAATTGATTAATTCCATTAATATTAGGAAGAGTTGATATGGCTTTCCCTCGAATAAATAATATAAGATTTTGATTGTTAATTCCTTCATTAATAATATTAATTTTAAGAGGATTTATAAATATTGGAGTTGGTACAGGATGAACAGTTTATCCACCTTTATCTTTAAATGTAAGTCATATAGGAATTTCAGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGAATTTCATCTATTATGGGAGCTATTAATTTTATTGTTACTATTTTAAATATACGAAATTATGGGGTGTTAATAGATAAAATTAGTTTATTATGTTGATCAATTTTAATTACAGCTATTTTATTATTATTATCATTACCAGTTTTAGCTGGAGCTATTACTATATTATTAACTGATCGAAATTTAAATACATCTTTTTTTGATCCTTCAGGGGGGGGGGATCCAATTTTGTATCAACATTTATTT. Both H. keithlangdoni and H. kazumifukunagae have the ovipositor sheath and antenna of ♀ medially white or ivory combined with a strongly widened vein 1Cua of forewing which is shorter than curved vein cu-a. Differs from H. kazumifukunagae by the shorter ovipositor sheath (approx. as long as body) and largely ivory head, mesopleuron and metapleuron.
Holotype ♀
Guanacaste, Sector Cacao, Sendero Segundo, 10.92679, -85.45332, 1180 meters, caterpillar collection date: 30/vii/2007, wasp eclosion date: 1/ix/2007. Depository:
Host data. gelJanzen01 Janzen356 (Depressariidae) feeding on Hampea appendiculata (Malvaceae)
Caterpillar and holotype voucher codes. 07-SRNP-36311, DHJPAR0021647.
Paratypes
Hosts = Conchylodes salamisalis (Crambidae) Dichomeris Janzen169 (Gelechiidae). DHJPAR0040592, DHJPAR0040597. Depository:
Hymenochaonia keithwillmotti , sp. nov.
Diagnostics
BOLD:AAP9457. Consensus barcode. TATATTATATTTTATATTTGGAATATGGTCAGGGATAATTGGTTTATCAATAAGAATAATTATTCGTTTAGAAYTAGGTCAATCTGGTACTTTAATTAATAATGATCAAATTTATAATAGATTTGTTACTTYTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGGTTTGGTAATTGATTGATTCCTTTAATATTAGGTAGGGTAGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGGTTATTAATTCCATCTTTAATAATATTAATTTTAAGGGGATTTATAAATATTGGTGTAGGAACTGGTTGGACTGTTTATCCTCCTTTATCTTTAAATGYAAGTCATATAGGAATTTCAGTAGATATAGCAATTTTTTCATTACATTTAGCGGGWATTTYTTCTATTTTAGGTGCCATTAATTTTATTATTACTATTATAAATATACGAAATTATGGAGTATTAATAGATAAAATTAGATTATTATCTTGATCAATTTTAATTACAGCAATTTTATTATTATTATCATTACCAGTTTTAGCAGGTGCTATTACTATATTATTAACTGATCGGAATTTAAATACATCTTTTTTTGATCCTTCAGGGGGGGGGGRTCCTATTTTATATCAACATTTATTT. This species and H. kalevikulli sp. nov. share a long and widened vein 1Cua of forewing, antenna (except blackish base) pale yellowish or whitish and distinctly curved vein cu-a of forewing. Hymenochaonia keithwillmotti differs by having vein 1Cua of forewing approx. as long as vein cu-a, tarsal claws with ventral lamella, hind tibia brownish yellow, subbasal cell of forewing setose medially and apical half of metasoma yellowish.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Rio Francia Arriba, 10.89666, -85.29003, 400 meters, caterpillar collection date: 13/ix/2011, wasp eclosion date: 2/x/2011. Depository:
Host data. gelJanzen01 Janzen407 (Gelechiidae) feeding on Clibadium pittieri (Asteraceae).
Caterpillar and holotype voucher codes. 11-SRNP-44212, DHJPAR0048081.
Paratypes
Host = gelJanzen01 Janzen461 (Gelechiidae), elachJanzen01 Janzen272 (Depressaridae), Dichomeris Janzen703 (Gelechiidae), gelJanzen01 Janzen407 (Gelechiidae): DHJPAR0040613, DHJPAR0040570, DHJPAR0040601, DHJPAR0040603, DHJPAR0040606, DHJPAR0040607, DHJPAR0041403, DHJPAR0042758, DHJPAR0043246, DHJPAR0048083, DHJPAR0048086, DHJPAR0048087. Depository:
Hymenochaonia kenjinishidai , sp. nov.
Diagnostics
BOLD:ACG8190. Consensus barcode. TATATTATATTTTATATTTGGGATATGATCAGGGTTAATTGGTTTATCAATAAGATTAATTATTCGAATAGAGTTAGGTCAATCTGGTTTATTTATTGGTAATGATCAAATTTATAATAGTATAGTTACTGCACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGTTTTGGTAATTGATTAATTCCTTTAATATTAGGAAGTGTGGATATGGCTTTCCCTCGGATAAATAATATAAGATTTTGGTTATTATTCCCTTCAATTATAATATTAATTTTTAGAGGTTTTATAAATATTGGTGTTGGTACTGGTTGAACTGTTTATCCTCCTTTATCTATAAATATTAGACATATAGGTATATCAGTTGATATAGCTATTTTTTCTCTTCATTTAGCTGGGATTTCTTCAATTATAGGTTCTATTAATTTTATTGTTACAATTATAAATATACGAAATTATATAGTGTTAATAGATAAAATTAGTTTATTTTGTTGATCAGTATTAATTACAGTTATTTTATTGTTATTATCTTTGCCTGTTTTAGCTGGTGCAATTACAATATTATTAACTGATCGAAATTTAAATACTTCTTTTTTTGATCCTGCTGGAGGGGGGGATCCAGTTTTATATCAACATTTATTT. Very similar to H. pallida (Cresson, 1865) because of the pale colored body and stigma, slender first metasomal tergite and simple tarsal claws. The new species differs by having the first subdiscal cell of forewing approx. 1.5 × longer than wide (approx. 2.5 × in H. pallida), vein cu-a of forewing curved (straight) and mesosoma with dark brown pattern (entirely brownish yellow).
Holotype ♂
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 4/vii/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG06141-G06.
Paratypes
None.
Hymenochaonia kimberleysheldonae , sp. nov.
Diagnostics
BOLD:AAB4677. Consensus barcode. AATATTATATTTTTTATTTGGGTTATGATCTGGAATAATTGGGTTATCAATTAGATTAATTATTCGTATAGAACTAAGACAATCAGGGGTTTTATTAAATAATGATCAAATTTATAATAGGTTAGTTACTATGCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGGTTAATTCCTTTAATGTTGGGTAGAGTGGATATGGCTTTTCCTCGTATAAATAATATAAGATTTTGGTTATTAATTCCTTCATTAATATTATTAATTTTAAGAGGTTTTATAAATATTGGGGTTGGAACTGGATGAACAGTGTATCCTCCTCTTTCATTAAATGTTAGACATATAGGTATTTCTGTAGATATAGCTATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATAGGAGCTATTAATTTTATTAGAACAATTTTAAATATACGTAATTATGGGGTTTTAATAGATAAAATTAGATTAATATGTTGATCTATTTTAATTACTGCTATTTTATTATTATTATCTTTACCTGTATTGGCAGGAGCTATCACAATATTATTAACAGATCGTAATTTAAATACTTCTTTTTTTGATCCTYCGGGAGGGGGGGACCCTATTTTATATCAACATTTATTT. The new species shares with H. maculicoxa (Enderlein, 1920) the elongate and largely transversely striate first metasomal tergite. Hymenochaonia kimberleysheldonae differs because it has vein cu-a of forewing longer and slightly narrower (shorter and slightly wider in H. maculicoxa), hind femur and coxa yellowish (mainly blackish) and first tergite slenderer, especially in front of spiracles (less slender).
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Naciente, 10.98705, -85.42816, 700 meters, caterpillar collection date: 25/i/2014, wasp eclosion date: 24/ii/2014. Depository:
Host data. Desmia BioLep06 (Crambidae) feeding on Hoffmannia liesneriana (Rubiaceae).
Caterpillar and holotype voucher codes. 14-SRNP-30295, DHJPAR0055155.
Paratypes
Hosts = Syngamia florella, Desmia ploralis, Desmia Janzen09, Desmia BioLep06, Desmia Janzen14, Desmia Solis19 (all Crambidae). DHJPAR0036730, DHJPAR0036724, DHJPAR0028982, DHJPAR0037872, DHJPAR0037874, DHJPAR0039388, DHJPAR0040233, DHJPAR0050353, DHJPAR0050914, DHJPAR0055154, DHJPAR0056290. Depository:
Etymology
Hymenochaonia kimberlysheldonae is named to honor Dr. Kimberly Sheldon of the University of Tennessee for her intense and effective efforts to translate why “Mountain Passes are higher in the Tropics” to today’s concern for climate change impacts on biodiversity.
Hymenochaonia krisnorvigae , sp. nov.
Diagnostics
BOLD:ACR5126. Consensus barcode. ATATTATATTTTTTATTCGGTATATGATCTGGTGTAATTGGTTTATCAATAAGATTAATTATTCGAATAGAATTAGGTCAATCTGGGTCTTTGATTGGTAATGATCAAATTTATAATAGTTTTGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGGTTTGGTAATTGATTAATTCCTTTAATATTAGGAAGAGTTGATATAGCTTTCCCTCGGATAAATAATATAAGATTTTGATTATTAATTCCTTCAATTATATTATTAATTATAAGAGGATTTATAAATATTGGTGTAGGAACTGGGTGAACAGTTTATCCTCCTTTATCTTTAAATATTAGACATATAGGGATTTCAGTTGATATATCAATTTTTTCATTACATTTAGCAGGGGTATCCTCAATTATAGGTGCTATTAATTTTATTATTACAATTTTAAATATGCGAAATTATGGTGTATTAATAGATAAGATTAGATTATTATGTTGATCTGTATTAATTACAGCTATTTTATTATTATTATCATTACCAGTATTAGCAGGTGCTATTACTATATTATTAACAGAT------------------------------------------. Very similar to the Nearctic and Neotropical H. delicata (Cresson, 1872) because of its size, narrow temples, robust first metasomal tergite and pale yellow stigma, but differs by having simple tarsal claws (with small ventral lamella in H. delicata), vein cu-a of forewing slightly inclivous and straight (subvertical and slightly curved in H. delicata), vein 1Cua of forewing wider than vein 2-Cu1 (similar in H. delicata), T1–2 largely smooth and shiny (aciculate and less shiny in H. delicata) and malar space ca. as long as basal width of mandible (shorter in H. delicata).
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 16/iv/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG17470-D11.
Paratypes
None.
Hymenochaonia lilianamadrigalae , sp. nov.
Diagnostics
BOLD:AAA7058. Consensus barcode. GATATTGTATTTTTTATTTGGAATATGATCAGGAATAGTAGGTTTATCAAWAAGAATAATTATTCGAATGGAATTAGGTCAAGTGGGAGATTTAATTGGAAATGATCAAATTTATAATAGATTTGTTACTGCTCATGCATTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGATTTGGTAATTGRTTAATTCCTTTAATGTTAGGAAGAGTCGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGGTTATTAATTCCMTCAATTATATTATTAATTTTTAGTGGATTTATGAATATTGGGGCGGGGACTGGTTGAACAGTTTATCCACCTTTATCATTAAATATTAGACATATAGGAGTTTCRGTTGATATGGCAATTTTTTCTTTACATTTAGCTGGAATTTCYTCAATTATGGGTTCAGTTAATTTTATTATTACTATTATAAATATACGAAATTAYGGGGTATTAATAGAAAAAATTAGATTATTATGTTGATCAATTTTAATTACTGTAATTTTATTATTATTATCATTACCYGTATTAGCAGGTGCTATTACTATAYTATTAACTGATCGAAATTTAAATACTTCTTTCTTTGATCCTGCAGGTGGAGGTGATCCAATTTTATATCAGCATTTATTT. Strong sexual dimorphism, subbasal cell of forewing without brownish patch and sparsely setose, tarsal claws with ventral lamella, vein 1Cua of the forewing short and hardly widened, and vertex darkened this species is closely related to H. nupera (Cresson, 1872). Hymenochaonia lilianamadrigalae differs by the yellowish brown mesoscutal lobes (more or less darkened) and POL approx. equal to diameter of posterior ocellus (approx. 1.6 × diameter of posterior ocellus).
Holotype ♀
Guanacaste, Sector Santa Rosa, Sendero Natural, 10.83575, -85.61253, 290 meters, caterpillar collection date: 8/vi/2001, wasp eclosion date: 27/v/2002. Depository:
Host data. Syllepte belialis (Crambidae) feeding on Casearia corymbosa (Salicaceae).
Caterpillar and holotype voucher codes. 01-SRNP-13023, DHJPAR0029366.
Paratypes
Hosts = Syllepte belialis. DHJPAR0029363, DHJPAR0029364, DHJPAR0029361, DHJPAR0029362, DHJPAR0035255, DHJPAR0035290, DHJPAR0055768, DHJPAR0055920.
Etymology
Hymenochaonia lilianamadrigalae is named to honor Mrs. Liliana Madrigal of Costa Rica for her seminal and founding role in all that is ACG since November 2005. Depository:
Hymenochaonia lizlangleyae , sp. nov.
Diagnostics
BOLD:ACK7939. Consensus barcode. AATATTATATTTTWTWTTTGGAATATGATCAGGAATAGTAGGTYTATCAATGAGAATAATTATTCGAATGGAATTAGGTCAAGTAGGGGATTTAATTGGAAATGATCAAATTTATAATAGATTTGTTACTGCTCATGCATTTGTAATAATTTTTTTTATGGTTATACCAATTATAATTGGGGGATTTGGTAATTGATTAATTCCATTAATATTAGGAAGAGTTGATATAGCTTTYCCTCGTATAAATAATATAAGGTTTTGATTATTAATTCCATCAATTATATTATTAATTTTTAGAGGATTYATAAATATTGGTGTAGGKACTGGATGAACAGTTTATCCACCTTTATCATTAAATATTAGACATATAGGAGTTTCAGTTGATATAGCAATTTTTTCTTTACATTTAGCTGGGRTTTCTTCAATTATAGGTTCAGTTAATTTTATTATTACTATTATAAATATACGAAATTAYGGAGTATTAATAGAAAAAATTAGRTTATTATGTTGATCAATTTTAATTACTGTGATTTTATTATTATTATCATTACCTGTATTAGCAGGTGCTATTACTATATTATTAACTGATCGAAATTTAAATACATCTTTTTTTGATCCTGCAGGTGGRGGGGATCCAATTTTATATCAACATTTATTT. Similar to H. lilianamadrigalae sp. nov. and H. nupera (Cresson, 1872) because of sharing the lack of a brownish patch in the subbasal cell of forewing, tarsal claws with ventral lamella, vein 1Cua of the forewing short and hardly widened, and darkened vertex. Hymenochaonia lizlangleyae differs by having the subbasal cell of forewing largely glabrous (sparsely setose in H. nupera), mesoscutal lobes yellowish brown (more or less darkened in H. nupera) and POL approx. 0.8 × diameter of posterior ocellus (approx. 1.6 × diameter of posterior ocellus in H. nupera).
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.84389, -85.61384, 300 meters, caterpillar collection date: 13/vii/2005, wasp eclosion date: 2/viii/2005. Depository:
Host data. Eulepte Solis15 (Crambidae) feeding on Handroanthus ochraceus (Bignoniaceae).
Caterpillar and holotype voucher codes. 05-SRNP-18593, DHJPAR0009306.
Paratypes
Hosts = Eulepte Solis15, Syllepte aechmisalis (Crambidae). DHJPAR0009305, DHJPAR0009309, DHJPAR0009324, DHJPAR0009325, DHJPAR0009326, DHJPAR0009307, DHJPAR0009308, DHJPAR0029409, DHJPAR0028991, DHJPAR0036306, DHJPAR0041541, BIOUG06141-D03. Depository:
Macrocentrus Curtis, 1833
Macrocentrus is worldwide in distribution with 190 described species, a few of which have been widely used in biological control. There are three species from Mexico described by
Key to the species of Macrocentrus in BIN BOLD: ACK7466
| 1 | A. Hind femur partly to entirely black | 2 |
| – | B. Hind femur entirely yellow to reddish brown | 4 |
| 2(1) | A. Mesopleuron black below precoxal groove | M. gustavogutierrezi |
| – | B. Mesopleuron pale yellow to orange-brown below precoxal groove | 3 |
| 3(2) | A. Face and gena predominantly black | M. geoffbarnardi |
| – | B. Face and gena predominantly yellow. | M. gregburtoni |
| 4(1) | A. Mesopleuron black below precoxal groove | M. gretchendailyae |
| – | B. Mesopleuron pale yellow to light brown below precoxal groove. | 5 |
| 5(4) | A. Propodeum and first metasomal tergum black | M. lucianocapelli |
| – | B. Propodeum and first metasomal tergum yellowish brown | 6 |
| 6(5) | A. Hind femur reddish brown. | M. grettelvegae |
| – | B. Hind femur yellow | M. fredsingeri |
Macrocentrus fredsingeri , sp. nov.
Diagnostics
BOLD:ACK7466. Consensus barcode. YATATTRTATTTTTTRTTTGGTATRTGRTCTGGGGWRWTAGGTTTATCAYTAAGTTTAATTATTCGTATAGAATTRGGTCAAATTGGTTYATTTATTGGAAATGATCAAATTTAYAATAGTATTGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATRCCTATTATAATTGGGGGKTTTGGHAATTGRTTRATTCCYTTAATATTAGGRAGTGTTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTARTTCCTTCTTTAATATTATTAATTTTAAGWGGRTTTATAAATATTGGTGTAGGTACAGGATGAACAGTWTAYCCYCCTTTATCAYTAAATGTTAGTCATATAGGRATTTCTGTMGATATAGYTATTTTTTCAYTACATTTGGCTGGWATTTCTTCAATTATAGGKGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATYATGATCAATTTTAATTACRGCTATTTTATTATTRTTATCTTTACCTGTGTTAGCTGGTGCWATTACAATATTRTTAACTGAYCGTAATTTAAATACATCYTTTTTTGAYCCTGCTGGAGGAGGRGRYCCTATTTTATAYCAACATTTATTT. Recognizable by the combinations of largely setose subbasal cell of forewing, vein 1Cua of forewing ca. as long as vein cu-a and moderately widened, POL 0.7 × diameter of posterior ocellus, dorsal carinae of first tergite surpassing middle of tergite, first tergite with oblique striae latero-posteriorly, pterostigma mainly dark brown and contrasting with pale R1b vein and antenna largely pale yellowish.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Conguera, 10.91589, -85.26631, 420 meters, caterpillar collection date: 17/i/2011, wasp eclosion date: 10/ii/2011. Depository:
Host data. Neurophyseta clymenalisDHJ03 (Crambidae) feeding on Cyathea trichiata (Cyatheaceae).
Caterpillar and holotype voucher codes. 11-SRNP-40293, DHJPAR0042876.
Paratypes
Hosts = Neurophyseta clymenalisDHJ03. DHJPAR0042874, DHJPAR0042869, DHJPAR0042865, DHJPAR0041508, DHJPAR0042878, DHJPAR0043228, DHJPAR0041474, DHJPAR0041458, DHJPAR0041522, DHJPAR0041528, DHJPAR0042866, DHJPAR0042868, DHJPAR0042871, DHJPAR0041476, DHJPAR0041477, DHJPAR0041479, DHJPAR0041480, DHJPAR0041481, DHJPAR0041483, DHJPAR0041484, DHJPAR0041486, DHJPAR0041489, DHJPAR0041501, DHJPAR0041505, DHJPAR0051407. Depository:
Macrocentrus geoffbarnardi , sp. nov.
Diagnostics
BOLD:ACK7466. Consensus barcode. YATATTRTATTTTTTRTTTGGTATRTGRTCTGGGGWRWTAGGTTTATCAYTAAGTTTAATTATTCGTATAGAATTRGGTCAAATTGGTTYATTTATTGGAAATGATCAAATTTAYAATAGTATTGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATRCCTATTATAATTGGGGGKTTTGGHAATTGRTTRATTCCYTTAATATTAGGRAGTGTTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTARTTCCTTCTTTAATATTATTAATTTTAAGWGGRTTTATAAATATTGGTGTAGGTACAGGATGAACAGTWTAYCCYCCTTTATCAYTAAATGTTAGTCATATAGGRATTTCTGTMGATATAGYTATTTTTTCAYTACATTTGGCTGGWATTTCTTCAATTATAGGKGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATYATGATCAATTTTAATTACRGCTATTTTATTATTRTTATCTTTACCTGTGTTAGCTGGTGCWATTACAATATTRTTAACTGAYCGTAATTTAAATACATCYTTTTTTGAYCCTGCTGGAGGAGGRGRYCCTATTTTATAYCAACATTTATTT. Belongs to the group with vein M+Cu of forewing distinctly widened apically. Ventral half of mesopleuron and metapleuron brownish yellow; vein 1A of forewing narrow subapically. It differs from M. gregburtoni and M. grettelvegae by the strongly widened vein 1Cua of forewing (vs. approx. 5 × wider than vein 1Cub; approx. 3 × wider in both other species), and largely dark brown hind tibia, head, and first metasomal tergite (vs. mainly yellowish or brownish in both other species).
Holotype ♀
Guanacaste, Sector Pitilla, Bullas, 10.98670, -85.38503, 440 meters, caterpillar collection date: 24/i/2013, wasp eclosion data: 19/ii/2013. Depository:
Host data. Syllepis marialis (Crambidae) feeding on Allophylus psilospermus (Sapindaceae).
Caterpillar and holotype voucher codes. 13-SRNP-70155, DHJPAR0050916.
Paratypes
Host = Syllepis marialis: DHJPAR0035268, DHJPAR0038828, DHJPAR0028992, DHJPAR0028995, DHJPAR0039162, DHJPAR0048712, DHJPAR0028994, DHJPAR0038891, DHJPAR0040081, DHJPAR0049246. Depository:
Macrocentrus gregburtoni , sp. nov.
Diagnostics
BOLD:ACK7466. Consensus barcode. YATATTRTATTTTTTRTTTGGTATRTGRTCTGGGGWRWTAGGTTTATCAYTAAGTTTAATTATTCGTATAGAATTRGGTCAAATTGGTTYATTTATTGGAAATGATCAAATTTAYAATAGTATTGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATRCCTATTATAATTGGGGGKTTTGGHAATTGRTTRATTCCYTTAATATTAGGRAGTGTTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTARTTCCTTCTTTAATATTATTAATTTTAAGWGGRTTTATAAATATTGGTGTAGGTACAGGATGAACAGTWTAYCCYCCTTTATCAYTAAATGTTAGTCATATAGGRATTTCTGTMGATATAGYTATTTTTTCAYTACATTTGGCTGGWATTTCTTCAATTATAGGKGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATYATGATCAATTTTAATTACRGCTATTTTATTATTRTTATCTTTACCTGTGTTAGCTGGTGCWATTACAATATTRTTAACTGAYCGTAATTTAAATACATCYTTTTTTGAYCCTGCTGGAGGAGGRGRYCCTATTTTATAYCAACATTTATTT. Belongs to the group with vein M+Cu of forewing distinctly widened apically. Vein 1Cua of forewing approx. 3 × wider than vein 1Cub, vein 1A of forewing narrow subapically and of metapleuron brownish yellow. Shares with M. grettelvegae the pale (yellowish brown or brownish yellow) hind tibia and pale yellow head (except for black stemmaticum); it differs by the largely dark brown pterostigma (yellowish brown in M. grettelvegae), rather robust second metasomal tergite (slenderer in M. grettelvegae), largely dark brown first tergite (pale brown in M. grettelvegae), and largely blackish dorsal face of mesosoma (mainly yellowish brown in M. grettelvegae).
Holotype ♀
Guanacaste, Sector Pitilla, Estación Pitilla, 10.98931, -85.42581, 675 meters, caterpillar collection date: 5/iv/2010, wasp eclosion date: 21/iv/2010. Depository:
Host data. Herpetogramma phaeopteralis (Crambidae) feeding on Paspalum nutans (Poaceae).
Caterpillar and holotype voucher codes. 10-SRNP-30965, DHJPAR0039541.
Paratypes
None.
Macrocentrus gretchendailyae , sp. nov.
Diagnostics
BOLD:ACK7466. Consensus barcode. YATATTRTATTTTTTRTTTGGTATRTGRTCTGGGGWRWTAGGTTTATCAYTAAGTTTAATTATTCGTATAGAATTRGGTCAAATTGGTTYATTTATTGGAAATGATCAAATTTAYAATAGTATTGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATRCCTATTATAATTGGGGGKTTTGGHAATTGRTTRATTCCYTTAATATTAGGRAGTGTTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTARTTCCTTCTTTAATATTATTAATTTTAAGWGGRTTTATAAATATTGGTGTAGGTACAGGATGAACAGTWTAYCCYCCTTTATCAYTAAATGTTAGTCATATAGGRATTTCTGTMGATATAGYTATTTTTTCAYTACATTTGGCTGGWATTTCTTCAATTATAGGKGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATYATGATCAATTTTAATTACRGCTATTTTATTATTRTTATCTTTACCTGTGTTAGCTGGTGCWATTACAATATTRTTAACTGAYCGTAATTTAAATACATCYTTTTTTGAYCCTGCTGGAGGAGGRGRYCCTATTTTATAYCAACATTTATTT. Belongs to the group with vein M+Cu of forewing distinctly widened apically. Ventral half of mesopleuron and part of metapleuron largely black and first metasomal tergite subparallel-sided. Differs from M. jesseausubeli, M. gustavogutierrezi, and M. jennyphillipsae by having the hind femur pale brown (black in other species) and the first metasomal tergite yellowish brown.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, San Lucas, 10.91846, -85.30338, 320 meters, caterpillar collection date: 8/i/2014, wasp eclosion date: 3/ii/2014. Depository:
Host data. Piletosoma thialis (Crambidae) feeding on Doliocarpus multiflorus (Dilleniaceae).
Caterpillar and holotype voucher codes. 14-SRNP-40250, DHJPAR0054411.
Paratypes
Host = Piletosoma thialis: DHJPAR0017227, DHJPAR0035152, DHJPAR0035306, DHJPAR0035305, DHJPAR0040082, DHJPAR0035262, DHJPAR0035264, DHJPAR0035269, DHJPAR0035151, DHJPAR0038018, DHJPAR0063008, DHJPAR0063009, DHJPAR0063035. Depository:
Macrocentrus grettelvegae , sp. nov.
Diagnostics
BOLD:ACK7466. Consensus barcode. YATATTRTATTTTTTRTTTGGTATRTGRTCTGGGGWRWTAGGTTTATCAYTAAGTTTAATTATTCGTATAGAATTRGGTCAAATTGGTTYATTTATTGGAAATGATCAAATTTAYAATAGTATTGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATRCCTATTATAATTGGGGGKTTTGGHAATTGRTTRATTCCYTTAATATTAGGRAGTGTTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTARTTCCTTCTTTAATATTATTAATTTTAAGWGGRTTTATAAATATTGGTGTAGGTACAGGATGAACAGTWTAYCCYCCTTTATCAYTAAATGTTAGTCATATAGGRATTTCTGTMGATATAGYTATTTTTTCAYTACATTTGGCTGGWATTTCTTCAATTATAGGKGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATYATGATCAATTTTAATTACRGCTATTTTATTATTRTTATCTTTACCTGTGTTAGCTGGTGCWATTACAATATTRTTAACTGAYCGTAATTTAAATACATCYTTTTTTGAYCCTGCTGGAGGAGGRGRYCCTATTTTATAYCAACATTTATTT. Belongs to the group with vein M+Cu of forewing distinctly widened apically. Vein 1Cua of forewing approx. 3 × wider than vein 1Cub. Ventral half of mesopleuron and metapleuron brownish yellow. Vein 1A of forewing narrow subapically and of metapleuron brownish yellow. Shares with M. gregburtoni the pale hind tibia and pale yellow head (except for blackish stemmaticum and its surroundings); it differs by the yellowish brown pterostigma (largely dark brown in M. gregburtoni), comparatively slender second metasomal tergite (rather robust in M. gregburtoni), pale brown first tergite (largely dark brown in M. gregburtoni), and mainly yellowish brown dorsal face of mesosoma (largely blackish in M. gregburtoni).
Holotype ♀
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.86472, -85.41531, 540 meters, caterpillar collection date: 11/vi/2011, wasp eclosion date: 3/vii/2011. Depository:
Host data. Desmia ploralis (Crambidae) feeding on Hamelia patens (Rubiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-2291, DHJPAR0045379.
Paratypes
Host = Desmia ploralisDHJ10 and Desmia benealisDHJ03: DHJPAR0040350, DHJPAR0045377. Depository:
Macrocentrus gustavogutierrezi , sp. nov.
Diagnostics
BOLD:ACK7466. Consensus barcode. YATATTRTATTTTTTRTTTGGTATRTGRTCTGGGGWRWTAGGTTTATCAYTAAGTTTAATTATTCGTATAGAATTRGGTCAAATTGGTTYATTTATTGGAAATGATCAAATTTAYAATAGTATTGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATRCCTATTATAATTGGGGGKTTTGGHAATTGRTTRATTCCYTTAATATTAGGRAGTGTTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTARTTCCTTCTTTAATATTATTAATTTTAAGWGGRTTTATAAATATTGGTGTAGGTACAGGATGAACAGTWTAYCCYCCTTTATCAYTAAATGTTAGTCATATAGGRATTTCTGTMGATATAGYTATTTTTTCAYTACATTTGGCTGGWATTTCTTCAATTATAGGKGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATYATGATCAATTTTAATTACRGCTATTTTATTATTRTTATCTTTACCTGTGTTAGCTGGTGCWATTACAATATTRTTAACTGAYCGTAATTTAAATACATCYTTTTTTGAYCCTGCTGGAGGAGGRGRYCCTATTTTATAYCAACATTTATTT. Belongs to the group with vein M+Cu of forewing distinctly widened apically. Ventral halves of mesopleuron and metapleuron largely black; first metasomal tergite subparallel-sided. Most similar to M. jennyphillipsae because of the largely black hind femur and first metasomal tergite but differs by having large ocelli (POL approx. 0.8 × diameter of posterior ocellus; approx. 1.5 × in M. jennyphillipsae) and face pale yellow medio-dorsally (partly dark brown in M. jennyphillipsae).
Holotype ♀
Guanacaste, Sector Del Oro, Bosque Aguirre, 11.00060, -85.43800, 620 meters, caterpillar collection date: 3/ii/2010, wasp eclosion date: 17/ii/2010. Depository:
Host data. Asturodes fimbriauralisDHJ02 (Crambidae) feeding on Colubrina spinosa (Rhamnaceae).
Caterpillar and holotype voucher codes. 10-SRNP-20281, DHJPAR0038832.
Paratypes
Hosts = Asturodes fimbriauralisDHJ02 and Antaeotricha spurca (Depressariidae). DHJPAR0029383, DHJPAR0050856, DHJPAR0055165, DHJPAR0028986, DHJPAR0029341, DHJPAR0036261, DHJPAR0029386, DHJPAR0029389, DHJPAR0029360, DHJPAR0053557, DHJPAR0050803, DHJPAR0038831, DHJPAR0037707, DHJPAR0028998, DHJPAR0028997, DHJPAR0029393, DHJPAR0029392, DHJPAR0029391, DHJPAR0029390, DHJPAR0029388, DHJPAR0028981, DHJPAR0029076, DHJPAR0029349, DHJPAR0029379, DHJPAR0029384, DHJPAR0029339, DHJPAR0029340, DHJPAR0029338, DHJPAR0036731, DHJPAR0029380. Depository:
Macrocentrus hannahjamesae , sp. nov.
Diagnostics
BOLD:ABA9321. Consensus barcode. TGTATTGTATTTTTTATTTGGTATATGATCCGGGGTATTGGGCTTGTCATTAAGTTTAATTATTCGTATAGAATTAGGTCAAATTGGTTCATTTATTGGAAATGATCAAATTTATAATAGTATTGTTACTTCACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATCGGGGGATTTGGAAATTGATTAATTCCTTTAATATTAGGGAGTGTTGATATAGCTTTCCCTCGAATAAATAACATAAGATTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTAAGAGGATTTATAAATATTGGTGTAGGTACAGGATGAACAGTTTACCCCCCTTTATCATTGAATATTAGTCATATAGGAATTTCTGTAGATATAGCTATTTTTTCATTACATTTGGCTGGGGTTTCTTCAATTATAGGTGCTATTAATTTTATCATTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATCATGATCAATTTTAATTACAGCTATTTTATTATTATTATCTTTACCTGTGTTAGCTGGTGCTATTACAATATTGTTAACTGATCGTAATTTAAATACATCTTTTTTTGATCCTGCTGGAGGGGGGGATCCTATTTTATATCAACATTTATTT. Belongs to the group with vein M+Cu of forewing distinctly widened apically, and mesosternum, ventral half of mesopleuron and metapleuron brownish yellow. It shares with M. lucianocapelli the somewhat widened subapical part of vein 1A of forewing, but differs by the posteriorly parallel-sided first tergite (distinctly widened in M. lucianocapelli), face with dark brown heart-shaped patch (entirely yellowish brown in M. lucianocapelli), and hind tibia largely dark brown (brownish yellow in M. lucianocapelli).
Holotype ♀
Alajuela, Sector San Cristobal, Finca San Gabriel, 10.87766, -85.39343, 645 meters, caterpillar collection date: 14/vii/2011, wasp eclosion date: 7/viii/2011. Depository:
Host data. Syllepis marialis (Crambidae) feeding on Allophylus psilospermus (Sapindaceae).
Caterpillar and holotype voucher codes. 11-SRNP-2806, DHJPAR0045376.
Paratypes
Host = Syllepis marialis: DHJPAR0028984, DHJPAR0036307, DHJPAR0036308, DHJPAR0045386. Depository:
Macrocentrus harisridhari , sp. nov.
Diagnostics
BOLD:ACK7467. Consensus barcode. TATATTGTATTTTTTWWTTGGYATRTGRTCRGGAGTATTAGGTTTATCATTAAGTTTAATTATTCGTATAGAATTAGGTCAAATTGGKTCATTTATTGGAAATGATCAGATTTATAATAGTATYGTTACTTCTCATGCTTTYATTATAATTTTTTTTATAGTTATACCTATTATAATTGGKGGATTTGGMAATTGATTAATTCCTTTRATRTTRGGAAGTGTWGATATRGCYTTYCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTAAGAGGWTTTATAAATATTGGTGTAGGTACAGGATGAACAGTTTATCCYCCYTTATCATTAAATATTAGTCATATAGGAATTTCTGTAGATATAGCTATTTTTTCATTACATTTGGCGGGTATTTCTTCAATTATAGGTGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATCATGATCAATTTTAATCACAGCTATTTTATTATTATTATCTTTACCTGTRTTAGCTGGTGCWATTACAATRTTGTTAACTGATCGTAATTTAAATACATCTTTYTTTGACCCYGCYGGAGGGGGTGACCCTRTTTTATAYCAACATTTATTT. There are two species in this BIN (M. harisridhari and M. hiroshikidonoi) that differ by almost 2%, and both share having the metasoma dark brown medio-dorsally, vein 1Cua of forewing distinctly longer than vein cu-a, R1b paler than pterostigma, and vein cu-a curved and posteriorly widened. Their barcodes form two groups with no intermediates. The two species also differ in color: M. harisridhari is darker than M. hiroshikidonoi i.e., M. harisridhari is dark brown where M. hiroshikidonoi is light reddish brown, and brownish yellow where M. hiroshikidonoi is light yellow. Morphologically, both are very similar, but M. harisridhari differs by having the first metasomal tergite slightly widened posteriorly and slightly shorter POL (0.7 × diameter of posterior ocellus vs. 0.9 × in M. hiroshikidonoi.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Flecha, 10.94741, -85.31501, 491 meters, caterpillar collection date: 21/i/2013, wasp eclosion date: 6/ii/2013. Depository:
Host data. Undulambia Solis02 (Crambidae) feeding on Cyathea multiflora (Cyatheaceae).
Caterpillar and holotype voucher codes. 13-SRNP-69137, DHJPAR0051314.
Paratypes
Hosts = Undulambia Solis02, Neurophyseta camptogrammalisDHJ01 (Pyralidae). DHJPAR0045384, DHJPAR0043243, DHJPAR0041469, DHJPAR0050021, DHJPAR0051386. Depository:
Macrocentrus hillaryrosnerae , sp. nov.
Diagnostics
BOLD:ABA7284. Consensus barcode. TATATTGTATTTTTTATTTGGTATATGATCGGGAGTATTAGGTTTATCATTAAGTTTAATTATTCGTATAGAATTAGGCCAAATTGGTTCATTTATTGGAAATGATCAGATTTATAATAGTATTGTTACTTCTCATGCTTTCATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGAGGATTTGGAAATTGATTGATTCCTTTAATGTTAGGAAGTGTTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTAAGAGGGTTTGTAAATATTGGTGTAGGTACAGGATGAACGGTCTATCCTCCTTTATCATTAAATATTAGTCATATAGGAATTTCTGTAGATATCGCTATTTTTTCATTACATTTGGCGGGTATTTCTTCAATTATAGGTGCTATTAACTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATCATGATCAATTTTAATTACAGCTATTTTATTATTATTATCTTTACCTGTGTTAGCTGGTGCTATTACAATATTGTTAACTGATCGTAATTTAAACACATCTTTTTTTGATCCTGCCGGAGGGGGTGATCCTATTTTATATCAGCATTTATTT. Recognizable by the wide elliptical bicolored pterostigma and elongate (approx. twice as long as vein cu-a) moderately widened vein 1Cua.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Camino Porvenir, 10.90383, -85.25964, 383 meters, caterpillar collection date: 12/iii/2011, wasp eclosion date: 30/iii/2011. Depository:
Host data. Undulambia Solis02 (Crambidae) feeding on Alsophila firma (Cyatheaceae).
Caterpillar and holotype voucher codes. 11-SRNP-41204, DHJPAR0043202.
Paratypes
None.
Macrocentrus hiroshikidonoi , sp. nov.
Diagnostics
BOLD:ACK7467. Consensus barcode. TATATTGTATTTTTTWWTTGGYATRTGRTCRGGAGTATTAGGTTTATCATTAAGTTTAATTATTCGTATAGAATTAGGTCAAATTGGKTCATTTATTGGAAATGATCAGATTTATAATAGTATYGTTACTTCTCATGCTTTYATTATAATTTTTTTTATAGTTATACCTATTATAATTGGKGGATTTGGMAATTGATTAATTCCTTTRATRTTRGGAAGTGTWGATATRGCYTTYCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTAAGAGGWTTTATAAATATTGGTGTAGGTACAGGATGAACAGTTTATCCYCCYTTATCATTAAATATTAGTCATATAGGAATTTCTGTAGATATAGCTATTTTTTCATTACATTTGGCGGGTATTTCTTCAATTATAGGTGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATCATGATCAATTTTAATCACAGCTATTTTATTATTATTATCTTTACCTGTRTTAGCTGGTGCWATTACAATRTTGTTAACTGATCGTAATTTAAATACATCTTTYTTTGACCCYGCYGGAGGGGGTGACCCTRTTTTATAYCAACATTTATTT. There are two species in this BIN (M. harisridhari and M. hiroshikidonoi) that differ by almost 2%; both share having the metasomal dark brown medio-dorsally, vein 1Cua of forewing distinctly longer than vein cu-a, and vein cu-a curved and posteriorly widened. Their barcodes form two groups with no intermediates (Suppl. material
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Quebrada Guarumo, 10.90445, -85.28412, 400 meters, caterpillar collection date: 22/xi/2012, wasp eclosion date: 14/xii/2012. Depository:
Host data. Undulambia Solis02 (Crambidae) feeding on Alsophila firma (Cyatheaceae).
Caterpillar and holotype voucher codes. 12-SRNP-86847, DHJPAR0051422.
Paratypes
Hosts = Neurophyseta clymenalisDHJ03, Neurophyseta Janzen229, Diacme BioLep02, Neurophyseta Janzen222, Undulambia Solis02, Neurophyseta completalis, Neurophyseta clymenalisDHJ03, Neurophyseta camptogrammalisDHJ01 (all Crambidae), Paramorbia Brown001 (Tortricidae). DHJPAR0050022, DHJPAR0051418, DHJPAR0051318, DHJPAR0051320, DHJPAR0051322, DHJPAR0052193, DHJPAR0052327, DHJPAR0052380, DHJPAR0053148, DHJPAR0053667, DHJPAR0053668, DHJPAR0053669, DHJPAR0054926, DHJPAR0055590, DHJPAR0028990, DHJPAR0023710, DHJPAR0038403, DHJPAR0038466, DHJPAR0038021, DHJPAR0041399, DHJPAR0041406, DHJPAR0041469, DHJPAR0041490, DHJPAR0041495, DHJPAR0041498, DHJPAR0041499, DHJPAR0041504, DHJPAR0041509, DHJPAR0041512, DHJPAR0041519, DHJPAR0041544, DHJPAR0042870, DHJPAR0043239, DHJPAR0043243, DHJPAR0043250, DHJPAR0045381, DHJPAR0045382, DHJPAR0045384, DHJPAR0045503, DHJPAR0048239, DHJPAR0050011, DHJPAR0050012, DHJPAR0050013, DHJPAR0050014, DHJPAR0050017, DHJPAR0050016, DHJPAR0050017, DHJPAR0050018, DHJPAR0050020, DHJPAR0028989, DHJPAR0041459, DHJPAR0041462, DHJPAR0041465, DHJPAR0041397. Depository:
Etymology
Macrocentrus hiroshikidonoi is named to honor Dr. Hiroshi Kidono of Japan, formerly of JICA, for his very direct and enthusiastic support for ACG’s Parataxonomist Program and INBio, and personally to DHJ and WH in the germination and growth of ACG and INBio.
Macrocentrus iangauldi , sp. nov.
Diagnostics
BOLD:ABY7812. Consensus barcode. TATATTGTATTTTTTATTTGGTGTATGATCTGGGGTATTAGGTTTGTCACTAAGTTTAATTATTCGTATAGAATTAGGTCAAATTGGTTCATTTATTGGAAATGATCAAATTTATAATAGTATTGTTACTTYTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGGTTTGGAAATTGATTAATTCCTTTAATATTAGGAAGTGTTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTAAGAGGATTTATAAATATTGGTGTAGGTACAGGATGAACAGTTTATCCTCCTTTATCATTAAATATTAGTCATATAGGAATTTCTGTAGATATATCTATTTTTTCATTACACTTGGCTGGTATTTCTTCAATTATAGGTGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATCATGATCKATTTTAATTACAGCTATTTTATTATTATTATCTTTACCTGTTTTAGCTGGTGCTATTACAATATTGTTAACTGATCGTAATTTAAATACATCTTTTTTTGATCCTGCCGGAGGGGGGGATCCTATTTTATACCAACATTTATTT. Easily recognizable by the pale yellowish R1 vein contrasting with mainly dark brown veins of the infuscated middle third of the forewing. In addition, is the first metasomal tergite robust, approx. 2.3 × longer than its apical width and vein 1Cua slightly widened and slightly longer than vein cu-a.
Holotype ♀
Alajuela, Brasilia, Piedrona, 11.01618, -85.35902, 340 meters, caterpillar collection date: 26/x/2011, wasp eclosion date: 10/xi/2011. Depository:
Host data. Antaeotricha Janzen290 (Depressariidae) feeding on Inga oerstediana (Fabaceae).
Caterpillar and holotype voucher codes. 11-SRNP-66121, DHJPAR0046828.
Paratypes
Hosts = Janzen433, Antaeotricha marmorea, Antaeotricha Janzen31, Antaeotricha Janzen77 (Depressariidae). DHJPAR0045869, DHJPAR0045871, DHJPAR0045874, DHJPAR0046846, DHJPAR0048262, DHJPAR0048274. Depository:
Etymology
Macrocentrus iangauldi is named to honor Dr. Ian Gauld (RIP) for his decades of highly enthusiastic support for all things taxonomic and bioinventory of Costa Rica in general and ACG specifically, with special emphasis on wasps in the family Ichneumonidae.
Macrocentrus jennyphillipsae , sp. nov.
Diagnostics
BOLD:ADB4234. Consensus barcode. ATATTGTATTTTTTATTTGGTATATGATCTGGTATATTAGGCTTATCATTAAGTTTAATTATTCGAATAGAATTAGGTCAAATTGGTTCATTTATTGGAAATGATCAAATTTATAATAGTATCGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGGTTTGGGAATTGATTAATCCCCCTAATATTAGGAAGTGTTGATATGGCTTTTCCTCGGATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTAAGAAGATTTATAAATATTGGTGTAGGGACAGGATGGACAGTTTATCCACCTTTATCAATAAATATTAGTCATATAGGTATTTCTGTAGATATGGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATGGGGGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGTGTATTAATAGATAAAATTAGATTATTATCATGATCAATTTTAATTACAGCTATTTTATTGTTATTATCTTTACCTGTATTAGCTGGTGCTATTACAATATTATTGACTGAT------------------------------------------------. Belongs to the group with vein M+Cu of forewing distinctly widened apically, mesosternum, ventral half of mesopleuron and of metapleuron largely black and first metasomal tergite subparallel-sided. Most similar to M. gustavogutierrezi because of the largely black hind femur and first metasomal tergite but differs by having comparatively small ocelli (POL approx. 1.5 × diameter of posterior ocellus; approx. 0.8 × in M. gustavogutierrezi) and face partly dark brown medio-dorsally (pale yellow in M. gustavogutierrezi).
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 19/x/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29288-B09.
Paratypes
None.
Etymology
Macrocentrus jennyphillipsae is named to honor Dr. Jenny Phillips of BioAlfa for her decades of enthusiastic taxonomic and administrative support of INBio, parataxonomists, and all matters biodiversity of Costa Rica coupled with intensely developing BioAlfa of Costa Rica.
Macrocentrus jesseausubeli , sp. nov.
Diagnostics
BOLD:ACK7403. Consensus barcode. TATCTTATATTTTATATTTGGGCTATGGTCTGGGGTATTAGGTTTATCAATAAGGTTAATTATTCGTATAGAGTTAGGTCAAATTGGTTCATTTATTGGAAATGATCAAATTTATAATAGTATTGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGGTTAATTCCTTTAATATTAGGTAGAGTTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTGATTCCTTCTTTAACTTTATTAATTTTGAGAGGATTTATAAATATTGGGGTAGGYACAGGATGAACAGTTTATCCTCCTTTATCATTAAATATTAGACATATAGGARTTTCTGTAGATATGGCTATTTTTTCTTTACATTTGGCGGGTGTTTCTTCAATTATAGGTGCTATTAATTTTATTGTTACTATTATAAATATRCGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATTATGATCAGTTTTAATTACAGCTATTTTATTGTTATTATCTTTACCTGTATTAGCTGGTGCCATTACTATATTATTAACTGATCGTAATTTAAATACATCTTTTTTTGATCCTGCTGGTGGGGGGGACCCTATTTTGTACCAACATTTATTT. Belongs to the group with vein M+Cu of forewing distinctly widened apically, mesosternum, ventral half of mesopleuron and metapleuron largely black and first metasomal tergite subparallel-sided. Differs from M. gretchendailyae, M. gustavogutierrezi, and M. jennyphillipsae by having vein 1A of forewing slender subapically and vein cu-a of forewing widened posteriorly.
Holotype ♀
Alajuela, Sector San Cristobal, Río Blanco Abajo, 10.90037, -85.37254, 500 meters, caterpillar collection date: 15/x/2005, wasp eclosion date: 4/xi/2005. Depository:
Host data. spiloBioLep01 BioLep382 (Crambidae) feeding on Aphelandra aurantiaca (Acanthaceae).
Caterpillar and holotype voucher codes. 05-SRNP-6429, DHJPAR0028985.
Paratypes
Host = Herpetogramma Solis10 (Crambidae): DHJPAR0028983, DHJPAR0028988, DHJPAR0041515, DHJPAR0041543, DHJPAR0048254. Depository:
Macrocentrus jessemaysharkae , sp. nov.
Diagnostics
BOLD:AAW4582. Consensus barcode. TATATTATATTTTTTATTTGGAATATGATCTGGTATATTGGGGTTATCAATAAGTTTAATTATTCGAATAGAATTAGGTCAAATTGGTTCATTTATTGGTAATGATCAAATTTATAATAGAATTGTCACATCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGAAATTGATTAATTCCTTTAATATTGGGTAGTGTTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGGTTATTAATTCCATCTTTAATATTACTAATTTTAAGGGGATTTTTAAATATTGGTGTAGGTACAGGTTGAACAGTTTATCCTCCTTTATCTATAAATGTTAGTCATATAGGAATTTCTGTTGATATAGCTATTTTTTCTTTGCATTTGGCTGGTATTTCTTCAATTATAGGTGCTATTAATTTTATTGTTACTATTTTAAATATACGAAATTATGGAGTATTAATAGATAAAATTAGATTATTATCATGATCAATTTTAATTACAGCTATTTTATTATTATTATCATTACCAGTATTAGCTGGTGCTATTACTATATTATTAACTGATCGTAATTTGAATACATCATTTTTTGATCCTGCAGGAGGGGGGGATCCTATTTTATATCAACATTTATTT. Recognizable by the combination of the robust (subparallel-sided and approx. 2.3 × as long as wide posteriorly) and dark brown first tergite, with dorsal carinae only basally distinct, vein cu-a of forewing curved but not widened posteriorly, and the antenna largely pale yellowish.
Holotype ♀
Guanacaste, Sector Cacao, Sendero a Maritza, 1 km NW Estación Cacao, 10.92691, -85.46822, 1150 meters, caterpillar collection date: 1/ix/2010, wasp eclosion date: 20/ix/2010. Depository:
Host data. cram 10-SRNP-36093 (Crambidae) feeding on Ficus pertusa (Moraceae)
Caterpillar and holotype voucher codes. 10-SRNP-36093, DHJPAR0042077.
Paratypes
None.
Etymology
Macrocentrus jessemaysharkae is named in honor of science writer Ms. Jesse Mayshark in recognition of her 2011 stellar reporting on the All Taxa Biodiversity Inventory of the Great Smoky Mountains National Park, an intellectual inspiration from the same project once anticipated as the ATBI of ACG.
Macrocentrus jimwhitfieldi , sp. nov.
Diagnostics
BOLD:ABX5241. Consensus barcode. TATATTATATTTTTTATTTGGTTTATGATCTGGTGTGTTAGGATTATCTATAAGGCTAATTATTCGTATTGAATTAGGYCAAATTGGTTCTTTTATCGGTAATGATCAGATTTATAATAGAATTGTTACATCACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGATTTGGAAATTGATTAATTCCTTTAATATTAGGAAGAGTTGATATAGCTTTYCCTCGTATAAATAATATAAGATTTTGGTTATTAGTTCCTTCATTAATATTACTAATTTTAAGGGGATTTATAAATATTGGRGTAGGTACTGGTTGAACAGTCTATCCCCCTTTATCTTTAAATATTAGGCATATAGGAGTTTCTGTTGATATAGCTATTTTTTCATTACATTTAGCAGGAATTTCATCAATTATAGGTGCTATTAATTTTATTATTACTATTATGAATATACGAAATTATGGAGTATTGATAGATAAAATTAGATTATTATCTTGATCTATTTTAATTACAGCAATTTTRTTATTATTATCTTTACCTGTTTTAGCAGGGGCTATTACAATATTATTAACTGATCGAAATTTAAATACATCTTTTTTTGAYCCTGCTGGAGGAGGGGATCCTATTTTGTATCAACATTTATTT. Easy to recognize because of the mostly black hind femur tibia (except base and apex) combined with the posteriorly widened vein cu-a of forewing; metasoma dorsally black; vein 1A of forewing slender.
Holotype ♀
Guanacaste, Sector Pitilla, Leonel, 10.99637, -85.40195, 510 meters, caterpillar collection date: 27/i/2010, wasp eclosion date: 27/ii/2010. Depository:
Host data. Chlamydastis Janzen04 (Depressariidae) feeding on Pouteria campechiana (Sapotaceae).
Caterpillar and holotype voucher codes. 10-SRNP-70517, DHJPAR0039160.
Paratypes
Hosts = Stenoma Janzen199, Chlamydastis Janzen04, and Antaeotricha fascicularis (all Depressariidae). DHJPAR0035285, DHJPAR0039156, DHJPAR0039161, DHJPAR0041521, DHJPAR0041523, DHJPAR0041525, DHJPAR0045356, DHJPAR0045361, DHJPAR0049997, DHJPAR0050004, DHJPAR0052194. Depository:
Etymology
Macrocentrus jimwhitfieldi is named to honor Dr. Jim Whitfield of the University of Illinois for his decades of mentorship to DHJ about Braconidae taxonomy and stimulus for their collection and rearing from ACG, while carrying out many braconid taxonomic projects for ACG specimens.
Macrocentrus johnbrowni , sp. nov.
Diagnostics
BOLD:ADJ5301. Consensus barcode. TATATTATATTTTTTATTTGGTATATGATCTGGGATATTAGGGTTATCATTAAGTTTAATTATTCGAATAGAATTAGGTCAAATTGGGGCATTTATTGGGAATGATCAAATTTATAATAGTATTGTTACATCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCGATTATAATTGGGGGATTTGGTAATTGATTAATTCCTTTAATATTGGGAAGAGTTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCGTCTTTAATATTATTAATTTTAAGGGGATTTGTAAATATTGGGGCAGGTACAGGGTGAACAGTTTACCCCCCTTTATCTTTAAATATTAGACATATAGGAGTTTCTGTTGATATAGTTATTTTTTCATTACATTTAGCTGGAATTTCATCAATTATAGGGGCTATTAATTTTATTGTTACTATTTTAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGGTTATTATCATGATCTATTTTAATTACAGCTATTTTATTGTTATTATCTTTACCTGTATTAGCTGGTGCTATTACAATATTATTAACTGATCGTAATTTAAATACATCTTTTTTTGATCCTTCAGGAGGGGGGGATCCTATTTTATATCAACATTTATTT. Shares with M. iangauldi the pale yellowish stigma contrasting with mainly dark brown veins of middle third of forewing and the robust first tergite (approx. 2.3 × longer than its apical width). Differs because of the slightly wider vein 1Cua of forewing, which is approx. as long as vein cu-a (slightly narrower and ca. 1.2 × in M. iangauldi), basal half of vein C+SC+R of forewing dark brown (yellow in M. iangauldi) and head pale yellow (pale brown in M. iangauldi).
Holotype ♀
Guanacaste, Sector Santa Maria, Crater Bosque Sendero Adentro, 10.80348, -85.32729, 1594 meters, light trapped, 7/iv/2016. Depository:
Host data. None.
Caterpillar and holotype voucher codes. 17-SRNP-11371, DHJPAR0061182.
Paratypes
None.
Macrocentrus johnburnsi , sp. nov.
Diagnostics
BOLD:ADB2238. Consensus barcode. ATATTATATTTTTTATTTGGAATATGATCAGGTATTTTAGGTTTATCTTTAAGGTTAATTATTCGGATAGAATTAGGTCAAGTTGGTTCATTTATTGGGAATGATCAAATTTATAATAGGATTGTTACATCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGGGGGTTTGGTAATTGATTTATTCCTTTGATATTAGGTAGGGTTGATATAGCATTCCCTCGAATAAATAATATAAGGTTTTGATTATTGATTCCATCATTGTTTTTATTAATTTTAAGGGGGTTCTTAAATATTGGAGTAGGTACTGGTTGGACTGTTTATCCTCCATTATCATTAAATATTAGTCATATAGGGATTTCTGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGGGTTTCTTCTATTATAGGGTCTATTAATTTTATTGTTACTATTCTTAATATACGAAATTATGGTGTTTTAATAGATAAAATTAGTTTGTTATCTTGATCAATTTTGATTACAGTTATTTTATTGTTATTATCATTACCTGTTTTAGCTGGGGCTATTACAATATTGTTAACTGATCGTAATTTAAATACT---------------------------. Similar to M. jonathanfranzeni sp. nov. because of the largely black hind femur and tibia and largely blackish head, but differs by the longer vein 1Cua of forewing (approx. as long as vein cu-a; much shorter than vein cu-a in M. jonathanfranzeni), widened vein 1A of forewing, first tergite more widened posteriorly (ca. 2.3 × longer than wide apically; ca. 2.8 × in M. jonathanfranzeni) and partly yellowish brown.
Holotype ♀
Guanacaste, Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, 5/x/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28712-G07.
Paratypes
None.
Etymology
Macrocentrus johnburnsi is named to honor Dr. John Burns of the Smithsonian Institution for his decades of taxonomic projects and mentorship of DHJ and WH about the Hesperiidae of ACG, as well as happily accepting their DNA barcoding as part of their taxonomic discovery.
Macrocentrus jonathanfranzeni , sp. nov.
Diagnostics
BOLD:AAX3465. Consensus barcode. ATATTATATTTTTTATTTGGAATATGATCAGGTATATTAGGTTTATCTATAAGTTTAATTATTCGAATAGAATTAGGTCAAGTAGGGTCATTAATTGGAAATGATCAAATTTATAATAGATTTGTAACTGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATTATGATTGGGGGGTTTGGTAATTGATTAATTCCATTAATATTAGGTAGAGTTGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGATTGTTAATTCCTTCATTAATATTATTAATTTTAAGGGGATTTATAAATATTGGTGTAGGGACAGGATGAACAGTTTATCCTCCTTTATCTTTAAATATTAGACATATAGGTATTTCTGTAGATATAGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATAGGGGCTATAAATTTTATTATTACTATTATTAATATACGTAATAATGGAGTATTAATAGATAAAATTAGATTATTATGTTGATCTATTTTAATTACAGCTTTTTTATTATTATTATCTTTACCTGTATTAGCAGGAGCAATTACTATATTATTAACTGATCGTAATTTAAATACTTCTTTTTTTGATCCGGCCGGTGGGGGAGATCCTATTTTATATCAACATTTATTT. Characterized by the combination of short vein 1Cua of forewing (distinctly shorter than vein cu-a), metasoma dark brown dorsally, white face, and elongate maxillary palp.
Holotype ♂
Guanacaste, Sector Cacao, Sendero Derrumbe, 10.92918, -85.46426, 1220 meters, caterpillar collection date: 21/iii/2007, wasp eclosion date: 15/iv/2007. Depository:
Host data. Pantographa suffusalis (Crambidae) feeding on Hampea appendiculata (Malvaceae).
Caterpillar and holotype voucher codes. 07-SRNP-35658, DHJPAR0028189.
Paratypes
None.
Macrocentrus jonathanrosenbergi , sp. nov.
Diagnostics
BOLD:AAT9449. Consensus barcode. TATATTATATTTTTTATTTGGAATATGGTCTGGTATAGTGGGTTTATCAATAAGTTTAATTATTCGGATGGAATTAGGTTGTGTAGGATCATTAATTGGAAATGATCAAATTTATAATAGATTTGTTACAGCTCATGCTTTTGTTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGTAGTGTGGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCAATTATATTATTAATAATAAGGGGATTTATAAATATTGGAGTAGGAACTGGATGAACTGTTTATCCTCCTTTATCATTAAATATTAGTCATATAGGTATTTCTGTTGATATAGCTATTTTTTCATTACATTTAGCTGGTATTTCTTCAATTATAGGGGCTATCAATTTTATTGTTACTATTTTAAATATACGAAATTATGGTGTTTTAATGGATAAAATTAGTTTATTATCTTGATCTATTTTGATTACAGCTATTTTATTATTATTATCTTTACCTGTATTAGCTGGTGCAATTACTATATTATTAACGGATCGTAATTTAAATACTTCTTTTTTTGATCCAGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT. Easily recognizable species because of the large ventral lamella (slender basal lobe) of the tarsal claws, anterior ocellus close to level of antennal sockets, metapleuron distinctly punctate, outer side of middle femur with numerous pegs, and elongate maxillary palp.
Holotype ♀
Guanacaste, Sector Mundo Nuevo, Estación La Perla, 10.76737, -85.43313, 325 meters, caterpillar collection date: 31/v/2010, wasp eclosion date: 30/vii/2010. Depository:
Host data. Hahncappsia BioLep471 (Crambidae) feeding on Iresine calea (Amaranthaceae).
Caterpillar and holotype voucher codes. 10-SRNP-55504, DHJPAR0040239.
Paratypes
None.
Macrocentrus jorgebaltodanoi , sp. nov.
Diagnostics
BOLD:ADB3971. Consensus barcode. ATATTATATTTTTTATTTGGTATATGATCAGGTATATTGGGTTTATCAATAAGTTTAATTATTCGAATAGAATTAGGTCAAGTAGGTTCATTAATTGGTAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATYGGGGGATTTGGAAATTGATTAATTCCTTTAATATTAGGAAGTGTGGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGGTTATTAATTCCTTCATTAATATTATTAATTTTAAGGGGATTTTTAAATATTGGGGTCGGAACAGGTTGAACAGTTTATCCTCCTTTATCATTAAATATTAGTCATATAGGTATTTCTGTGGATATAGCTATTTTTTCATTACATTTAGCTGGTGTCTCATCTATTATAGGAGCTATTAATTTTATTGTTACAATTTTAAATATACGAAATTATGGGGTTTTAATAGATAAAATTAGATTATTATCTTGATCAATTTTAATTACAGCTATTTTATTATTATTATCTTTACCTGTTTTAGCAGGAGCTATTACTATATTATTAACAGAT. Easily recognizable because of the slender and long vein 1Cua of forewing (much longer than vein cu-a) and largely setose subbasal cell of the forewing.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 6/iii/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29687-D11.
Paratypes
All Malaise-trapped: BIOUG29620-B02, BIOUG30041-H09, BIOUG30934-A02. Depository:
Etymology
Macrocentrus jorgebaltodanoi is named to honor Sr. Jorge Baltodano (RIP) for his many years as a neighboring collaborator with ACG and for accepting the challenge of being the first President of the Comité Local for ACG; part of his ranch is now a permanent part of ACG thanks to his foresight and spirit for conservation.
Macrocentrus lucianocapelli , sp. nov.
Diagnostics
BOLD:ACK7466. Consensus barcode. YATATTRTATTTTTTRTTTGGTATRTGRTCTGGGGWRWTAGGTTTATCAYTAAGTTTAATTATTCGTATAGAATTRGGTCAAATTGGTTYATTTATTGGAAATGATCAAATTTAYAATAGTATTGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATRCCTATTATAATTGGGGGKTTTGGHAATTGRTTRATTCCYTTAATATTAGGRAGTGTTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTARTTCCTTCTTTAATATTATTAATTTTAAGWGGRTTTATAAATATTGGTGTAGGTACAGGATGAACAGTWTAYCCYCCTTTATCAYTAAATGTTAGTCATATAGGRATTTCTGTMGATATAGYTATTTTTTCAYTACATTTGGCTGGWATTTCTTCAATTATAGGKGCTATTAATTTTATTGTTACTATTATAAATATACGAAATTATGGGGTATTAATAGATAAAATTAGATTATTATYATGATCAATTTTAATTACRGCTATTTTATTATTRTTATCTTTACCTGTGTTAGCTGGTGCWATTACAATATTRTTAACTGAYCGTAATTTAAATACATCYTTTTTTGAYCCTGCTGGAGGAGGRGRYCCTATTTTATAYCAACATTTATTT. Belongs to the group with vein M+Cu of forewing distinctly widened apically. Ventral halves of mesopleuron and metapleuron brownish yellow. It shares with M. hannahjamesae the somewhat widened subapical part of vein 1A of forewing, but differs by the comparatively robust and posteriorly distinctly widened first tergite (parallel-sided in M. hannahjamesae), face entirely yellowish brown (with dark brown heart-shaped patch in M. hannahjamesae), and hind tibia brownish yellow (largely dark brown in M. hannahjamesae).
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Palomo, 10.962, -85.28, 96 meters, caterpillar collection date: 15/x/2013, wasp eclosion date: 2/xi/2013. Depository:
Host data. Apogeshna stenialisDHJ03 (Crambidae) feeding on Bravaisia integerrima (Acanthaceae).
Caterpillar and holotype voucher codes. 13-SRNP-79776, DHJPAR0054348.
Paratypes
None.
Chapter 9: Orgilinae
Members of all genera of Orgilinae are koinobiont endoparasitoids of caterpillars from a wide range of families. All species with previously known life-histories are solitary, but here we describe a gregarious species, Orgilus ebbenielsoni Sharkey, sp. nov. (BOLD:ABA8537). Eleven species were recorded from Costa Rica and ca. 20 from the entire neotropics prior to this publication. The genera of New World Orgilinae may be identified with the following key. For the Orgilinae NJ tree, see Suppl. material
Key to the New World genera of the subfamily Orgilinae
Orgilus Haliday, 1833
Orgilus have been recorded from a wide range of Lepidoptera larvae (
Orgilus amyrossmanae , sp. nov.
Diagnostics
BOLD:ADY7227. Consensus barcode. AATTTTATATTTTTTATTTGGAATTTGATCAGGTATTTTAGGTATATCAATAAGTTTAATTATTCGTTTGGAATTAGGTATACCTGGTAGATTGATTGGTAATGATCAAATTTATAATAGGATTGTGACAGCACATGCTTTTGTAATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCTATAATATTAGGATGTCCAGATATGGCATTTCCTCGTATAAATAATATGAGATTTTGATTATTAATTCCTTCAATAATTTTTTTGATTTTTTCAAGAATTTTAAATATTGGTGTAGGAACTGGGTGAACAGTGTATCCTCCTTTATCATTAAGATTAGGACATGGTGGAATTTCAGTGGATATAGCTATTTTTTCTTTACATTTAGCTGGGGCTTCTTCAATTATAGGAGCTATTAATTTTATTACAACAATTTTTAATATACGATCAAGTATGATTACTATGGATAAAATTTCTTTATTAAGATGGTCAGTGTTAATTACTGCTATTTTATTATTATTATCATTACCTGTTTTAGCTGGAGCTATTACAATATTATTAAGTGATCGAAATTTAAATACTTCTTTTTTTGATCCTGCAGGGGGGGGCGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Rio Francia Arriba, 10.89666, -85.29003, 400 meters, caterpillar collection date: 9/ix/2010, wasp eclosion date: 1/x/2010. Depository:
Host data. Rectiostoma eowilsoni (Depressariidae) feeding on Turpinia occidentalis (Staphyleaceae).
Caterpillar and holotype voucher codes. 10-SRNP-43220, DHJPAR0041408.
Paratype
DHJPAR0064113. Depository:
Etymology
Orgilus amyrossmanae is named in honor of Amy Rossman for her participation in the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus carrolyoonae , sp. nov.
Diagnostics
BOLD:ADB0533. Consensus barcode. ATTTTATATTTTATATTTGGTATTTGATCAGGTATTTTAGGAATATCTATAAGATTAATTATTCGAATGGAATTAGGTATACCTGGTAGATTGATTGGAAATGATCAAATTTATAATAGAATTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATGCCAATTATAATTGGTGGTTTTGGAAATTGATTAATTCCTATAATACTAGGATGTCCAGATATAGCATTTCCTCGAATAAATAATATAAGGTTTTGGTTATTAATTCCTTCAATAATTTTTTTAATTTTTAGAAGAATTTTAAATGTTGGGGTTGGTACTGGTTGAACAGTGTATCCACCTTTATCTTTAATAATTGGTCATGGGGGTTTATCTGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATGGGTGCAATTAATTTTATTACTACTATTTTAAATATACGATCTGATAAAGTTTTAATGGATAAAATTTCATTATTAAGATGATCTGTTTTAATTACAGCTATTTTATTATTATTATCATTACCTGTATTGGCAGGG------------------------------------------------------------------------.
Holotype ♂
Guanacaste, Sector Pailas Dos, PL12-1, 10.7642, -85.335, 828 meters, 12/vi/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29134-B08.
Paratypes
None.
Orgilus christhompsoni , sp. nov.
Diagnostics
BOLD:ADW2886. Consensus barcode. TTTATATTTTTTATTTGGAATTTGAGCTGGTATTTTAGGTATATCAATAAGATTAATTATTCGAATAGAATTAGGTATACCAGGAAGATTAATTGGAAATGATCAGATTTATAATAGAATTGTTACAGCTCATGCATTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGRGGATTTGGTAATTGATTAATTCCTATAATATTAGGATGTCCTGATATAGCTTTCCCACGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCACTTGTATTTTTAATTTTTAGAAGGGTATTAAATGTAGGGGTAGGTACTGGTTGAACTGTTTATCCTCCTTTATCTTTATTAATTGGTCATGGAGGTTTGTCAGTTGATATAGCTATTTTTTCTTTACATTTGGCAGGTATTTCTTCAATTATAGGTGCAGTTAATTTTATTACAACAATTTTAAATATGCGATCTGATTATKTTTATATAGATAAAATTTCTTTATTATGTTGATCAGTTTTAATTACAGCTATTTTATTATTATTATCTTTACCAGTATTAGCTGGAGCAATTACTATATTATTAACAGATCGGAATTTAAATACTTCATTTTTTGACCCTTCTGGAGGTGGAGATCCAATTTTATATCAACATTTATT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 2/iv/2015, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG44707-C12.
Paratype
BIOUG44846-A07. Depository:
Etymology
Orgilus christhompsoni is named in honor of Chris Thompson attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus christinemcmahonae , sp. nov.
Diagnostics
BOLD:AAU1670. Consensus barcode. AATTTTATATTTTTTATTTGGTATTTGATCTGGAATTTTAGGAATATCTATAAGTTTATTAATTCGTATAGAATTAGGTATACCTGGGAGATTAATTGGTAATGATCAAATTTATAATAGTATTGTAACAGCACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCTATAATATTAGGATGTCCAGATATAGCTTTTCCTCGTATAAATAATATAAGGTTTTGATTATTGATTCCTTCTTTAATTTTTTTAATTTTTAGTAGTGTTTTAAATGTTGGAGTTGGTACAGGATGAACAGTTTATCCTCCTTTATCTTTAACTATTGGTCATGGGGGATTATCAGTTGATATGGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTATAGGGGCTATTAATTTTATTACAACTATTTTGAATATACGTTCTGATAAGGTTTTAATAGATAAAATTTCTTTATTAAGATGATCTGTTTTAATTACTGCTATTTTATTATTATTATCTTTACCAGTTTTAGCTGGAGCAATTACAATATTATTAACAGATCGTAATTTAAATACATCTTTTTTTGATCCTTCTGGTGGGGGAGATCCTATTTTATATCAGCATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Finca San Gabriel, 10.87766, -85.39343, 645 meters, caterpillar collection date: 26/vi/2013, wasp eclosion date: 20/vii/2013. Depository:
Host data. Antaeotricha Janzen24 (Depressariidae) feeding on Coccoloba peltata (Polygonaceae).
Caterpillar and holotype voucher codes. 13-SRNP-3328, DHJPAR0053050.
Paratypes
DHJPAR0040025, DHJPAR0040027, DHJPAR0052982, DHJPAR0053050. Depository:
Etymology
Orgilus christinemcmahonae is named in honor of Christine McMahon attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus dianalipscombae , sp. nov.
Diagnostics
BOLD:AAM1105. Consensus barcode. TATTTTATATTTTATATTTGGTATTTGATCAGGTATTTTAGGAATATCAATAAGTTTACTTATTCGAATAGAACTTGGAATACCTGGTAGTTTAATTGGTAATGATCAAATTTATAATAGAATTGTAACAGCTCATGCATTTATTATAATTTTTTTTATGGTTATACCAATTATAATTGGTGGATTTGGAAATTGATTGATTCCTATAATATTAGGATGTCCAGATATAGCATTTCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCAATAATTTTTTTAATTTTTAGCAGAGTTTTAAATGTAGGGGTTGGTACTGGATGAACAGTTTATCCACCTTTATCTTTAACAATTGGGCATGGGGGATTATCTGTTGATATAGCTATTTTTTCTTTACATTTGGCGGGTATTTCTTCTATTATAGGAGCAATTAATTTTATTACTACAATTTTAAATATACGATCTGATAAAGTTTTAATAGATAAAATTTCATTGTTAAGATGATCTGTTTTAATTACAGCAATTTTATTATTATTATCATTACCTGTATTGGCAGGTGCTATTACAATATTATTAACTGATCGAAATTTAAATACTTCTTTTTTTGATCCTTCAGGTGGTGGAGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Mundo Nuevo, Vado Miramonte, 10.77175, -85.43400, 305 meters, caterpillar collection date: 17/ii/2010, wasp eclosion date: 4/iii/2010. Depository:
Host data. elachJanzen01 Janzen786 (Depressariidae) feeding on Inga vera (Fabaceae).
Caterpillar and holotype voucher codes. 10-SRNP-55161, DHJPAR0039036.
Paratypes
None.
Etymology
Orgilus dianalipscombae is named in honor of Diana Lipscomb attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus ebbenielsoni , sp. nov.
Diagnostics
BOLD:ABA8537. Consensus barcode. AATTTTATATTTTTTATTTGGAATTTGATCAGGAATTTTAGGAATATCAATAAGTTTAATTATTCGTATAGAATTGGGGATACCAGGGAGATTAATTGGTAATGATCAAATTTATAATAGTGTTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGGTTTGGAAATTGATTGATTCCTATAATATTAGGATGTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGGTTTTGATTGTTGATTCCATCTATAATTTTTTTAATTTTTAGAAGAATTTTAAATGTTGGTGTTGGYACAGGGTGAACTGTATATCCACCTTTATCTTTAACAATTGGTCATGGGGGGGTATCTGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCTATTATAGGAGCAATTAATTTTATTACTACAATTTTAAATATACGTTCTGATAAAGTTTTAATAGATAAAATTTCTTTATTAAGTTGATCTGTATTAATTACAGCAATTTTATTATTATTATCATTACCTGTATTAGCAGGGGCAATTACAATATTATTAACAGATCGTAATTTAAATACATCTTTTTTTGATCCTTCAGGAGGGGGGGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Medrano, 11.01602, -85.38053, 380 meters, caterpillar collection date: 17/iii/2012, wasp eclosion date: 3/iv/2012. Depository:
Host data. Gregarious parasitoid on Megalota crassana (Tortricidae) feeding on Croton billbergianus (Euphorbiaceae). Seven specimens emerged from the host caterpillar.
Caterpillar and holotype voucher codes. 12-SRNP-70637, DHJPAR0049088.
Paratypes
Hosts = Megalota crassana, Megalota Janzen03, Megalota ochreoapex. DHJPAR0038979, DHJPAR0043266, DHJPAR0049086. Depository:
Etymology
Orgilus ebbenielsoni is named in honor of Ebbe Nielson (RIP) attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus elizabethpennisiae , sp. nov.
Diagnostics
BOLD:AAW8981. Consensus barcode. TATTTTGTATTTTGTATTTGGTATTTGATCAGGTATTTTGGGAATATCAATAAGGTTAATTATTCGAATAGAATTAGGGATACCAGGTAGAATAATYGGAAATGATCAGATTTATAATAGAATTGTTACAGCACATGCATTTATTATAATTTTTTTTATAGTTATGCCAATTATAATTGGTGGGTTTGGAAATTGACTTATTCCAATAATATTAGGATGTCCAGATATAGCGTTTCCACGAATAAATAATATAAGATTTTGATTGTTAATTCCTTCAATAATTTTATTGATTTTTAGAAGAATTTTAAATGTGGGTGTTGGGACAGGTTGGACAGTTTATCCACCTTTATCTTTAATAATTGGTCATGGGGGGGTATCTATTGATATAGCTATTTTTTCTTTACATTTAGCAGGAATTTCTTCTATTATGGGGTCAATTAATTTTATTACTACAATTTTAAATATGCGTTCTGATAAAATTTTAATAGATAAAATTTCATTATTAAGATGATCTATTTTAATTACAGTAATCTTATTGTTGTTATCGTTACCTGTATTAGCAGGTGCTATTACAATGTTATTGACTGATCGTAATTTAAATACATCTTTTTTTGATCCTTCAGGTGGGGGGGAYCCTATTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93, -85.372, 527 meters, 18/iv/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. DHJPAR0049293.
Paratypes
Both Malaise – trapped. DHJPAR0051816, DHJPAR0049469. Depository:
Orgilus evertlindquisti , sp. nov.
Diagnostics
BOLD:AAI2109. Consensus barcode. TTTTTATTTGGAATTTGATCGGGAATTTTGGGTATATCTATAAGATTAATTATTCGTATAGAGTTAGGTATACCTGGAAGATTGATTGGGAATGATCAAATTTATAATAGAATTGTTACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGTTTTGGAAATTGATTAATTCCAATAATATTAGGATGTCCGGATATAGCTTTTCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCAATAACTTTTTTAATTTTTAGRGGGGTTTTAAATGTTGGAGTGGGAACAGGTTGAACAGTTTATCCTCCATTATCATTAATTATTGGTCATGGTGGGTTATCTGTTGATATAGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATAGGGGCAATTAATTTTATTACTACAGTTTTAAATATACGATCTGATAAAGTTTATATAGATAAAATTTCTTTATTGAGTTGATCTATTTTGATTACAGCTATTTTGTTATTATTATCTTTACCGGTTTTAGCTGGGGCTATTACTATATTATTGACTGATCGTAATTTGAATACATCATTTTTTGATCCTTCAGGAGGGGGAGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Potrero Argentina, 10.89021, -85.38803, 520 meters, 24/i/2008, Malaise trap. Depository:
Host data. None.
Holotype voucher code. DHJPAR0027553.
Paratypes
Both Malaise-trapped. DHJPAR0027581, BIOUG28185-A09. Depository:
Etymology
Orgilus evertlindquisti is named in honor of Evert Lindquist attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus genestoermeri , sp. nov.
Diagnostics
BOLD:ADH3907. Consensus barcode. TATTTTTGGAATTTGGTCAGGGATTTTAGGAATATCAATAAGATTACTTATTCGATTAGAATTAGGTATGCCAGGTAGATTAATTGGTAATGATCAAATTTATAATAGAGTCGTAACAGCTCATGCATTTTTAATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTATAATATTAGGGTGCCCAGATATAGCATTTCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATTTTGTTAATTTTTAGAGGAATTTTAAATATTGGAGTAGGGACTGGGTGAACAGTTTATCCTCCTTTATCATTAAATATTGGTCATGGGGGTATATCAGTAGATATAGCAATTTTTTCTTTACATTTGGCAGGAATTTCATCAATTATAGGAGCAATTAATTTTATTACAACGATTTTAAATATACGATCTAAAGGAGTAAGAATAGATAAAATTTCATTATTTAGATGGTCAGTGTTAATTACTGCTGTATTATTATTATTATCTTTACCAGTT.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, 2/iv/2015, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG33118-F04.
Paratypes
None.
Etymology
Orgilus genestoermeri is named in honor of Gene Stoermer attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus jamesriegeri , sp. nov.
Diagnostics
BOLD:ACM9544. Consensus barcode. AATTTTATATTTTTTATTTGGTATTTGATCTGGAATTTTAGGAATATCTATAAGTTTATTAATTCGTGTAGAATTAGGTATACCTGGAAGATTAATTGGTAATGATCAAATTTATAATAGTATTGTAACAGCACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCTATAATATTGGGGTGTCCAGATATAGCTTTCCCTCGTATAAATAATATAAGTTTTTGATTATTGATTCCTTCTTTAATTTTTTTAATTTTTAGTAGTGTTTTAAATGTTGGAGTAGGAACTGGATGAACAGTTTATCCTCCTTTATCTTTAACTATTGGTCATGGGGGATTATCAGTTGATATGGCTATTTTTTCTTTACATTTGGCAGGTATTTCTTCAATTATAGGGGCTATTAATTTTATTACAACTATTTTGAATATACGTTCTGATAAAGTTTTAATAGATAAAATTTCTTTATTAAGATGATCTATTTTAATTACTGCTATTTTATTATTATTATCTTTACCAGTTTTAGCTGGAGCAATTACAATATTATTAACAGATCGTAATTTAAATACATCTTTTTTTGATCCTTCTGGTGGAGGGGATCCAATTTTATATCAGCATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Comejen, 10.95669, -85.28933, 213 meters, caterpillar collection date: 7/ii/2014, wasp eclosion date: 19/ii/2014. Depository:
Host data. Brenthia Janzen13 (Choreutidae) feeding on Heliconia irrasa (Heliconiaceae).
Caterpillar and holotype voucher codes. 14-SRNP-45281, DHJPAR0055301.
Paratypes
None.
Etymology
Orgilus jamesriegeri is named in honor of James Rieger attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus jeanmillerae , sp. nov.
Diagnostics
BOLD:ADA6087. Consensus barcode. GTTTTATATTTTATATTTGGAATTTGATCAGGAATTTTAGGTATATCTATAAGTTTAATTATTCGTTTAGAATTAGGTATATTAGGTAGATTGATTGGTAATGATCAAATTTATAATAGAGTAGTTACAGCTCATGCGTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGTTTTGGAAATTGATTAATTCCAATGATATTAGGATGTCCAGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAGTTCCTTCTTTATTTTTTTTAATTTTTAGTAGAATTTTAAATATTGGTGTTGGGACTGGATGAACAGTTTATCCACCTTTATCTTTAAATATTGGTCATGGAGGATTATCAGTTGATATATCAATTTTTTCTTTACATTTAGCTGGTATTTCATCTATTATAGGAGCTATTAATTTTATTACTACAATTATTAATATACGATCAAGAATAATTTATATAGATAAAATTTCTTTATTAAGATGATCTATTTTGATTACAGCAATTTTATTATTATTATCTTTGCCTGTTTTGGCTGGTGCAATTACTATATTATTGACTGATCGT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 26/vi/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29912-B11.
Paratypes
BIOUG29685-A09, BIOUG28777-A07, BIOUG28807-F04, BIOUG44866-F09. Depository:
Orgilus jeffmilleri , sp. nov.
Diagnostics
BOLD:ACT7418. Consensus barcode. TTTATTTTTGGTATTTGATCAGGAATTTTAGGTATGTCAATAAGATTGATTATTCGATTAGAATTAGGTATATCTGGAAGATTAATTGGTAATGATCAAATTTATAATAGAATTGTTACTGCACATGCATTTTTAATAATTTTTTTTATAGTAATGCCTATTATAATTGGAGGGTTTGGTAATTGATTAATTCCTATAATATTAGGATGTCCAGATATAGCATTCCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTGATTTTTTTAATATTTAGTGGTATTTTAAATATTGGAGTAGGAACAGGTTGAACAGTTTATCCTCCTTTATCTTTAAGAATTGGTCATGGTGGGATATCTGTAGATATAGCAATTTTTTCTTTACATTTAGCTGGGATTTCATCAATTATAGGAGCAGTTAATTTTATTACAACAATTTTAAATATACGATCTGAAGGAGTTACTATAGATAAAATTTCATTATTAAGATGATCAGTATTAATTACAGCTGTATTATTATTATTATCTTTACCAGTTTTAGCTGGAGCTATTACAATATTATTAACTGAT------------------------------------------------------------.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.88, -85.389, 575 meters, 26/xiii/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG19395-H01.
Paratypes
None.
Orgilus jerrypowelli , sp. nov.
Diagnostics
BOLD:ABZ9478. Consensus barcode. AATTTTATATTTTTTGTTTGGAATTTGATCAGGAATTTTAGGTATATCAATAAGATTAATTATTCGAATGGARTTAGGGATACCGGGYAGRTTAATYGGTAATGATCAAATTTATAATAGAATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCTATAATATTAGGRTGTCCTGATATAGCTTTYCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCRATAATTTTTTTAATTTTTAGGAGAGTGTTAAATGTTGGTGTTGGAACGGGATGAACTGTTTATCCTCCTYTATCTTTAAYTATTGGTCATGGAGGTTTATCTGTTGATATAGCTATTTTTTCTTTACATTTRGCTGGAATTTCATCTATTATAGGAGCAATTAATTTTATTACAACAATTTTAAATATACGTTCTGAYAAAGTTTTTATAGATAAAATTTCTTTATTAAGTTGATCTGTTTTWATTACAGCTATTTTATTATTATTATCTTTACCAGTATTAGCTGGGGCAATTACAATATTATTAACTGATCGAAATTTAAATACTTCMTTTTTTGATCCGTCAGGTGGAGGTGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.86472, -85.41531, 540 meters, caterpillar collection date: 2/ix/2011, wasp eclosion date: 21/ix/2011. Depository:
Host data. Antaeotricha Janzen412 (Depressariidae) feeding on Semialarium mexicanum (Celastraceae).
Caterpillar and holotype voucher codes. 11-SRNP-3427, DHJPAR0050158.
Paratypes
Hosts = Antaeotricha marmorea, Antaeotricha Janzen88, Antaeotricha Janzen412. DHJPAR0048258, DHJPAR0048266, DHJPAR0048271, DHJPAR0048277, DHJPAR0051415, DHJPAR0048251, DHJPAR0048267, DHJPAR0048268, DHJPAR0055593, DHJPAR0053161. Depository:
Etymology
Orgilus jerrypowelli is named in honor of Jerry Powell participating in the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus jimtiedjei , sp. nov.
Diagnostics
BOLD:ADX8196. Consensus barcode. TTTATATTTTATGTTTGGTATTTGATCAGGTATTTTAGGGATATCAATAAGTTTAATTATTCGAATAGAATTAGGTATACCAGGAAGTTTGATTGGTAATGATCAAATTTATAACAGAATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATGCCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTATAATATTAGGATGTCCGGATATGGCATTTCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTATAATTTTTTTAATTTTTAGAAGAATTTTAAATGTTGGAGTTGGGACTGGATGAACAGTTTATCCACCTTTATCTTTAACTATTGGTCATGGAGGTTTATCTGTTGATATAGTTATTTTTTCTTTACATTTAGCAGGAATTTCTTCTATTATAGGGGCAATTAATTTTATTACAACAATTTTAAATATACGATCTGATAAAGTTTTAATAGATAAAATTTCATTATTAAGGTGATCTGTTTTAATTACAGCAATTTTGTTATTGTTATCATTACCTGTATTAGCAGGTGCTATTACAATATTATTGACTGATCGTAATTTAAATACATCTTTTTTTGATCCTTCAGGTGGAGGTGACCCAATTTTATACCAACATTTATT------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 23/iv/2015, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG44165-G01.
Paratypes
None.
Etymology
Orgilus jimtiedjei is named in honor of Jim Tiedje attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus johnlundbergi , sp. nov.
Diagnostics
BOLD:AAT9015. Consensus barcode. TATTTTATATTTTTTATTTGGAATTTGATCTGGRATTTTAGGAATATCTATAAGATTAATTATTCGATTAGAATTAGGTATACCTGGGAGTTTAATTGGTAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGATTTGGAAATTGATTAATTCCAATAATATTAGGATGTCCTGATATGGCTTTCCCTCGGATAAATAATATAAGTTTTTGATTGTTAATTCCTTCAATAATTTTTTTAATTTTTAGAAGGGTTTTAAATGTTGGTGTAGGAACTGGTTGAACTGTRTATCCTCCTTTATCATTAATTATTGGTCATGGGGGGGTATCTGTTGATATARCTATTTTTTCTTTACATTTAGCAGGAATTTCTTCAATTATAGGAGCAATTAATTTTATTACAACAATTTTAAATATACGATCTGATAAAGTTTTAATGGATAAAATTTCTTTATTAAGATGATCAGTTTTAATTACAGCAATTTTATTATTATTATCTTTACCTGTTTTAGCAGGAGCTATTACTATATTATTAACAGATCGTAATTTAAATACATCTTTTTTTGAYCCTTCAGGAGGAGGTGATCCTATTTTATATCAGCATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Rio Francia Arriba, 10.89666, -85.29003, 400 meters, caterpillar collection date: 14/v/2011, wasp eclosion date: 29/v/2011. Depository:
Host data. elachBioLep01 BioLep81 (Depressariidae) feeding on Conceveiba pleiostemona (Euphorbiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-42377, DHJPAR0042873.
Paratypes
Hosts = elachBioLep01 BioLep81, elachBioLep01 Janzen48, elachJanzen01 Janzen698 (Depressaridae). DHJPAR0061511, DHJPAR0040026, DHJPAR0040028, DHJPAR0040029, DHJPAR0049918, DHJPAR0050141, DHJPAR0050150, DHJPAR0049671, DHJPAR0049679, DHJPAR0054529, DHJPAR0040553. Depository:
Etymology
Orgilus johnlundbergi is named in honor of John Lundberg attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus johnpipolyi , sp. nov.
Diagnostics
BOLD:ABU7457. Consensus barcode. TATTTTGTATTTTATATTTGGTATTTGATCTGGGGTATTAGGTATATCAATGAGATTTGTAGTTCGGATAGAATTAGGTATACCTGGGAGTTTAATTGGTAACGATCAAATTTATAATAGAATTGTTACTGCTCATGCTTTTTTAATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGGAATTGATTAATTCCTGTAATATTAGGATGTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGTTTTTGGTTATTAATTCCTTCAATTTTATTTTTAATTTTTAGGGGAATTTTAAATATTGGTGTAGGGACTGGGTGAACCGTTTATCCTCCTTTATCTTTGAATATTGGTCATGGAGGGTTATCAGTTGATATAGCTATTTTTTCTTTACATTTAGCGGGGGCGTCATCTATTATGGGAGCTATTAATTTTATTACGACAATTATAAATATACGATCTAGAATGGTTTTTATAGATAAAATTTCCTTGTTATGTTGATCAGTATTAATTACTGCTGTTTTATTATTATTATCTTTACCAGTATTAGCTGGGGCTATTACTATATTATTAACTGATCGTAATATAAATACTTCTTTTTTTGATCCTTCCGGGGGTGGTGATCCAATTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Sendero Anonas, 10.90528, -85.27882, 405 meters, caterpillar collection date: 9/iii/2012, wasp eclosion date: 10/iv/2012. Depository:
Host data. Anadasmus Janzen25 (Depressariidae) feeding on Ocotea atirrensis (Lauraceae).
Caterpillar and holotype voucher codes. 12-SRNP-41056, DHJPAR0048772.
Paratypes
None.
Etymology
Orgilus johnpipolyi is named in honor of John Pipoly attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus jorgellorentei , sp. nov.
Diagnostics
BOLD:AAN2501. Consensus barcode. AATTTTATATTTTTTATTTGGAATTTGATCAGGTATTTTAGGAATATCGATAAGATTAATTATTCGAATAGAATTAGGTATACCAGGAAGATTAATTGGTAATGATCAAATTTATAATAGAATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGAAATTGGTTAATTCCTATAATATTAGGATGTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGTTTTTGGTTATTAATTCCTTCAATAATTTTTTTAATTTTTAGAAGAGTATTAAATGTTGGAGTAGGAACAGGATGAACTGTTTATCCTCCTTTATCATTAGGTATTGGTCATGGAGGATTATCTGTTGATATAGCAATTTTTTCTTTACATTTAGCAGGAATTTCTTCAATTATAGGAGCAATTAATTTTATTACAACAATTTTAAATATACGATCTGATAAAGTTTTAATAGATAAAATTTCTTTATTAAGGTGATCGGTTTTAATTACAGCAATTTTATTATTATTATCTTTACCTGTTTTAGCTGGGGCAATTACTATATTATTAACTGATCGTAATTTAAATACATCATTTTTTGATCCCTCAGGAGGTGGAGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Puente Palma, 10.9163, -85.37869, 460 meters, caterpillar collection date: 21/v/2014, wasp eclosion date: 13/vi/2014. Depository:
Host data. Antaeotricha radicalis (Depressariidae) feeding on Vochysia guatemalensis (Vochysiaceae).
Caterpillar and holotype voucher codes. 14-SRNP-2556, DHJPAR0055612.
Paratype
Host = Stenoma Janzen148 (Depressariidae): DHJPAR0039489. Depository:
Etymology
Orgilus jorgellorentei is named in honor of Jorge Llorente attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus larryspearsi , sp. nov.
Diagnostics
BOLD:ACL4839. Consensus barcode. TATTTTATATTTTATGTTTGGTATTTGATCAGGTATTTTAGGAATATCAATAAGTTTAATTATTCGAATAGAATTAGGTATACCAGGAAGTTTAATTGGTAATGATCAAATTTATAACAGAATTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTAATGCCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTATAATATTAGGATGTCCGGATATGGCATTCCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTATAATTTTTTTAATTTTTAGAAGAATTTTAAATGTTGGGGTTGGAACTGGGTGAACAGTTTATCCACCTTTATCTTTAACTATTGGTCAYGGAGGGTTATCTGTTGATATAGCTATTTTTTCTTTACATCTAGCGGGAATTTCTTCTATTATAGGAGCAATTAATTTTATTACAACAATTTTAAATATACGATCTGATAAAGTTTTAATAGATAAAATTTCATTATTAAGGTGATCTGTTTTAATTACAGCAATTTTATTATTATTATCATTACCTGTATTAGCAGGTGCTATTACAATATTATTGACTGATCGTAATTTAAATACATCTTTTTTTGATCCTTCAGGTGGAGGTGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 6/ii/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29625-H11.
Paratypes
All Malaise-trapped. BIOUG09739-H07, BIOUG17496-G11, BIOUG18248-D12, BIOUG29423-E02, BIOUG29479-G11, BIOUG29660-B09, BIOUG29724-F11. Depository:
Etymology
Orgilus larryspearsi is named in honor of Larry Spears attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus marlinricei , sp. nov.
Diagnostics
BOLD:ACR9910. Consensus barcode. ATTTTATATTTTATATTTGGTATTTGATCAGGTATTTTAGGAATATCAATAAGTTTAATTATTCGAATAGAATTAGGTATACCTGGAAGTTTAATTGGAAATGATCAAATTTATAATAGAATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCTATAATATTAGGATGCCCAGATATAGCATTTCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCAATAACTTTTTTAATTTTTAGAAGAATTTTGAATGTTGGTGTTGGAACTGGATGAACAGTATATCCACCTTTATCTTTAACAATTGGTCACGGAGGTTTATCTGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATAGGGGCAATTAATTTTATTACTACAATTTTAAATATACGATCTGATAAAGTTTTAATAGATAAAATCTCATTATTAAGATGATCTGTATTAATTACAGCAATTTTATTATTATTATCATTACCAGTATTAGCAGGTGCTATTACAATATTATTAACTGAT---------------------.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 28/v/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG18748-D07.
Paratypes
None.
Orgilus mellissaespinozae , sp. nov.
Diagnostics
BOLD:ADG8044. Consensus barcode. TTTTTATTTGGTATTTGATCAGGAATTTTAGGAATATCAATAAGTTTAATTATTCGATTAGAATTAGGTATGCCAGGCAGATTAATTGGTAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGGAATTGATTAGTACCTATAATATTAGGATGTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTAGTACCATCATTAATTTTTTTAATTTTTAGAGGAATTTTAAATATTGGAGTTGGGACAGGATGAACAGTTTATCCCCCTTTATCTTTAAGAATTGGACATGGGGGGGTATCAGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGTATCTCTTCAATTATAGGAGCTATTAATTTTATTACTACTATTTTAAATATACGTTCAAGAATAGTTTATATAGATAAAATTCCTTTATTTGTTTGATCAGTATTAATTACTGCAATTTTATTATTATTATCTTTACCAGTATTAGCTGGTGCTATTACTATA-------------------------------------------------------.
Holotype ♂
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, 26/ii/2015, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG32860-D06.
Paratypes
None.
Orgilus mikesmithi , sp. nov.
Diagnostics
BOLD:ADF7540. Consensus barcode. ATATTATATTTTTTATTTGGGATTTGGGCAGGCATTTTAGGGATATCAATAAGGTTAATTATTCGACTAGAATTGGGAATACCTGGGGGATTAATTGGTAACGATCAAATTTATAATAGAGTTGTTACTGCTCATGCTTTTATTATGATTTTTTTTATGGTTATACCAATTATAATTGGAGGGTTTGGTAATTGGTTGATTCCTTTAATATTAAGATGTCCTGATATAGCTTTCCCTCGAATGAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATATTTTTAATTTTTAGTAGAATCTTAAATATTGGGGTAGGCACAGGGTGAACTGTTTATCCCCCTTTATCTTTATCTATTGGTCATGGAGGGCTTTCTGTTGATATGGCTATTTTTTCTTTACATTTAGCGGGTATTTCTTCAATTATGGGGGCTATTAATTTTATTACTACTATTATAAATATACGATTAAATATAATTTATATAGATAAAGTTTCTTTATTTATTTGGTCAGTTTTAATTACAGCAATTTTATTATTGTTATCTTTACCTGTATTAGCGGGG-------------------------------------------------------------------.
Holotype ♂
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, 18/xii/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG31641-F04.
Paratypes
None.
Etymology
Orgilus mikesmithi is named in honor of Mike Smith attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus normplatnicki , sp. nov.
Diagnostics
BOLD:ABX5811. Consensus barcode. TGTTTTATATTTTATATTTGGTATTTGATCAGGTATTTTAGGAATATCAATAAGGTTAATTATTCGTATAGAATTGGGCATACCAGGTAGGTTAATTGGTAATGATCAAATTTATAATAGAATTGTTACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATCATAATTGGTGGGTTTGGAAATTGATTAATTCCTATAATATTAGGATGTCCTGATATAGCATTCCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCAATAATTTTTTTAATTTTTAGAAGAATTTTAAATGTTGGAGTGGGAACTGGTTGGACAGTATATCCTCCATTATCTTTAACAATTGGTCATGGAGGATTATCTGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATTATAGGAGCAATTAATTTTATTACTACAATTTTAAATATACGGTCTGAAAAAGTTTTTATAGATAAAATTTCATTATTAAGATGATCAATTTTAATTACAGCAATTTTATTGTTATTATCATTACCTGTTTTAGCAGGTGCTATTACAATATTATTAACTGATCGTAATTTAAATACATCCTTTTTTGATCCTTCAGGTGGCGGCGATCCAATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.86472, -85.41531, 540 meters, caterpillar collection date: 2/ix/2011, wasp eclosion date: 23/ix/2011. Depository:
Host data. pyrBioLep01 BioLep761 (Pyralidae) feeding on Semialarium mexicanum (Celastraceae).
Caterpillar and holotype voucher codes. 11-SRNP-3414, DHJPAR0048166.
Paratypes
Host = phyBioLep01 BioLep761: DHJPAR0045298, DHJPAR0048187. Depository:
Etymology
Orgilus normplatnicki is named in honor of Norm Platnick (RIP) attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus peterrauchi , sp. nov.
Diagnostics
BOLD:ACJ4626. Consensus barcode. AATTTTATATTTTTTGTTTGGAATTTGATCAGGAATTTTAGGTATGTCAATAAGATTGATTATTCGAATGGAATTAGGTATACCAGGTAGATTAATTGGTAATGATCAAATTTATAATAGGATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCTATAATATTAGGATGTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGRTTTTGATTATTAATTCCTTCAATAATTTTTTTAATTTTTAGGAGAGTATTAAATGTTGGTGTTGGAACAGGATGAACTGTTTATCCTCCCYTATCTTTAACTATTGGTCATGGGGGTTTATCTGTTGATATGGCTATTTTTTCTTTACATTTAGCTGGAATTTCATCAATTATAGGGGCAATTAATTTTATTACAACAATTTTAAATATACGTTCTGATAAAGTTTTTATAGATAAAATTTCTTTATTAAGTTGATCTGTTTTAATTACAGCTATTTTATTATTATTATCTTTACCTGTATTAGCTGGGGCAATTACAATATTATTAACTGATCGAAATTTAAATACTTCATTTTTTGATCCGTCAGGTGGGGGTGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Perdido, 10.87940, -85.38607, 620 meters, caterpillar collection date: 7/i/2013, wasp eclosion date: 31/i/2013. Depository:
Host data. Antaeotricha Janzen146 (Depressariidae) feeding on Lonchocarpus oliganthus (Fabaceae).
Caterpillar and holotype voucher codes. 13-SRNP-68, DHJPAR0051319.
Paratype
Host = Antaeotricha Janzen401: DHJPAR0055603. Depository:
Etymology
Orgilus peterrauchi is named in honor of Peter Rauch attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus richardprimacki , sp. nov.
Diagnostics
BOLD:ADB9574. Consensus barcode. ATTTTATATTTTTTATTTGGAGTTTGATCTGGTATTTTAGGTATATCAATAAGTTTTATTATTCGTTTAGAATTAGGTATACCTGGCAGACTTATTGGGAATGATCAAATTTATAATAGAATTGTTACGGCTCACGCTTTTATTATAATTTTTTTTATGGTTATACCAATTATAATTGGGGGGTTTGGTAATTGATTAGTTCCTATAATATTGGGGTGTCCTGATATAGCTTTCCCTCGTATAAATAATATGAGATTTTGATTATTAGTTCCTTCAATAATTTTTTTAATTTTTAGAGGGGTATTAAATATTGGGGTAGGTACTGGGTGAACTGTTTATCCTCCTTTATCTTTAATAATTGGACATGGTGGAGTTTCAGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCCTCTATTATAGGTGCAATTAATTTTATTACTACTATTTTAAATATACGTTCTAATATAATTTATATAGATAAAATTTCTTTATTTATTTGGTCAGTATTGCTTACTGCTATTTTATTATTATTATCTTTACCTGTATTAGCAGGTGCTATTACTATATTATTAAGTGATCGA------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 18/ix/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG30111-A11.
Paratypes
None.
Orgilus sandraberriosae , sp. nov.
Diagnostics
BOLD:ADB0231. Consensus barcode. GTTTTATATTTTTTATTTGGTATTTGAGCTGGAATTTTAGGTATGTCAATAAGATTAATTATTCGAATGGAATTAGGAATACCAGGAAGATTAATTGGTAATGATCAAATTTATAATAGGATTGTTACAGCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGTTTTGGGAATTGATTAATTCCTATAATATTAGGTTGTCCTGATATAGCTTTTCCACGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCTCTTGTGTTTTTAATTTTTAGGAGAGTTTTAAATGTTGGGGTAGGAACTGGTTGAACTGTTTATCCTCCTTTATCTTTATTAATTGGACATGGTGGATTATCAGTTGATATAGCTATTTTTTCGTTACATTTAGCTGGTATTTCTTCAATTATAGGTGCAATTAATTTTATTACAACAATTTTGAATATACGGTCTGATAATGTTTATATAGATAAAATTTCTTTATTATGTTGATCAGTTTTAATTACAGCAATTTTATTATTATTATCTTTACCAGTTTTAGCTGGGGCAATTACTATATTATTAACAGATCGAAATTTAAATACT------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 27/ii/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28825-B03.
Paratypes
None.
Orgilus sarahmirandae , sp. nov.
Diagnostics
BOLD:ADB0825. Consensus barcode. ATTTTGTATTTTTTATTTGGAATTTGATCAGGAATTTTAGGAATATCTATGAGTTTAATTATTCGAATGGAATTAGGGATACCTGGGAGATTAATTGGTAATGATCAAATTTATAATAGAATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGGTTTGGGAATTGATTAATTCCTATAATATTAGGATGTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCAATAATTTTTTTAATTTTTAGAAGAATTTTAAATGTTGGGGTGGGAACTGGTTGAACTGTTTATCCTCCTTTATCTTTAATAATTGGTCATGGTGGATTATCAGTTGATATAGCTATTTTTTCTTTACATTTAGCAGGAATTTCATCAATTATAGGGGCAATTAATTTTATTACTACTATTTTAAATATACGTTCTGATAAAGTTTTAATAGATAAAATTTCTTTATTAAGTTGATCAGTATTAATTACTGCTATTTTATTATTATTATCATTGCCTGTATTAGCAGGAGCAATTACAATATTATTAACGGATCGT.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, 23/i/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28804-H03.
Paratype
Malaise-trapped. BIOUG28680-C02. Depository:
Orgilus scottmilleri , sp. nov.
Diagnostics
BOLD:AAW8985. Consensus barcode. TATTTTATATTTTTTATTTGGTATTTGATCTGGTATTTTAGGTATATCAATAAGTTTAATTATTCGTATAGAATTAGGTATACCTGGGACATTAATTGGAAATGATCAAATTTATAATAGAGTTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATGCCAATTATAATTGGGGGGTTTGGTAATTGATTAATTCCAATAATATTAGGATGTCCTGATATGGCTTTTCCTCGTATAAATAATATAAGTTTTTGATTATTAATTCCCTCATTAATTTTTTTAATTTTTAGTGGGGTTTTAAATGTTGGAGTTGGTACAGGGTGAACTGTTTATCCTCCCTTATCTTTAGGTATTGGTCATGGTGGATTTTCAGTGGATATAGCTATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATGGGGGCAATTAATTTTATTACTACTATTTTAAATATACGTTCTGATAAAGTTTTAATAGATAAAATTTCTTTATTAAGATGATCGGTATTAATTACAGCTATTTTGTTGTTATTATCTTTACCGGTATTAGCAGGGGCAATTACAATATTATTAACAGATCGTAATTTAAATACATCTTTTTTTGATCCTTCTGGAGGTGGGGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Quebrada Cementerio, 10.87124, -85.38749, 700 meters, caterpillar collection date: 6/vii/2009, wasp eclosion date: 21/vii/2009. Depository:
Host data. Anadasmus Janzen11 (Depressariidae) feeding on Nectandra hihua (Lauraceae).
Caterpillar and holotype voucher codes. 09-SRNP-3511, DHJPAR0040031.
Paratypes
None.
Etymology
Orgilus scottmilleri is named in honor of Scott Miller attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Orgilus scottmorii , sp. nov.
Diagnostics
BOLD:ABA8481. Consensus barcode. TATTTTATATTTTTTATTTGGTATTTGATCTGGTATTTTAGGTATATCAATAAGTTTAATTATTCGTATAGAATTAGGTATACCTGGGACATTAATTGGAAATGATCAAATTTATAATAGAGTTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATGCCAATTATAATTGGGGGGTTTGGTAATTGATTAATTCCAATAATATTAGGATGTCCTGATATGGCTTTTCCTCGTATAAATAATATAAGTTTTTGATTATTAATTCCCTCATTAATTTTTTTAATTTTTAGTGGGGTTTTAAATGTTGGAGTTGGTACAGGGTGAACTGTTTATCCTCCCTTATCTTTAGGTATTGGTCATGGTGGATTTTCAGTGGATATAGCTATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTATGGGGGCAATTAATTTTATTACTACTATTTTAAATATACGTTCTGATAAAGTTTTAATAGATAAAATTTCTTTATTAAGATGATCGGTATTAATTACAGCTATTTTGTTGTTATTATCTTTACCGGTATTAGCAGGGGCAATTACAATATTATTAACAGATCGTAATTTAAATACATCTTTTTTTGATCCTTCTGGAGGTGGGGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.86472, -85.41531, 540 meters, caterpillar collection date: 13/v/2011, wasp eclosion date: 27/v/2011. Depository:
Host data. Antaeotricha Janzen233 (Depressariidae) feeding on Tapirira brenesii (Anacardiaceae).
Caterpillar and holotype voucher codes. 11-SRNP-1951, DHJPAR0043217.
Paratypes
Hosts = elachBioLep01 BioLep754 (Depressariidae), Antaeotricha Janzen233. DHJPAR0049310, DHJPAR0043221, DHJPAR0043234, DHJPAR0043237, DHJPAR0048269, DHJPAR0048244, DHJPAR0043209, DHJPAR0043210, DHJPAR0043242, DHJPAR0043249. Depository:
Etymology
Orgilus scottmorii is named in honor of Scott Mori attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Stantonia Ashmead, 1904
There are eight species of Stantonia recorded from Costa Rica (Braet and Quicke 2004), but only three with the type specimen from Costa Rica, i.e., S. osa, S rossa, and S. sirena (all Braet & Quicke, 2004.) The holotypes of the others are from Brazil or the Caribbean. Based on the descriptions, none of the eight species is conspecific with those treated below. The following families have been recorded as hosts: Pyralidae, Crambidae, Noctuidae, and Tortricidae. Our data concur with those of Braet and Quick (2004) in that most hosts are in the family Crambidae. Here we report two new host families for Stantonia, Oecophoridae and Immidae.
Stantonia billalleni , sp. nov.
Diagnostics
BOLD:AAM1059. Consensus barcode. TATTTTATATTTAAAATTTGGAATTTGAGCTGGTATTTTAGGTATGTCTATAAGATTAATTGTACGATTAGAATTAGGTATACCTGGTAGTATAATTGGTAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCTTTTGTAATAATTTTTTTTATGGTTATACCAATTATAATTGGGGGGTTTGGTAATTGATTAATTCCAATAATATTGGGATGTCCAGATATAGCTTTCCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATATTATTAATTTTTAGAAGAGTTTTAAATATTGGGGTAGGTACTGGATGAACAGTATATCCACCATTATCYTTATTGATTGGCCATGGTGGATTATCAGTTGATATAGCAATTTTTTCTTTGCATTTGGCTGGTATTTCTTCTATTATAGGAGCAATTAATTTTATTACTACAATTTTAAATATGCGAGTTGATAAAGTTTATATAGATAAAATTTCTTTATTATCTTGATCAGTTTTTATTACTGCAATTTTATTATTATTATCTTTACCTGTATTAGCTGGTGCTATTACTATATTATTAACTGATCGTAATTTAAATACTTCTTTTTTTGATCCTTCAGGTGGRGGTGATCCAATTTTATATCAACATTTGTTT.
Holotype ♀
Guanacaste, Sector Mundo Nuevo, Estación La Perla, 10.76737, -85.43313, 325 meters, caterpillar collection date: 16/x/2012, wasp eclosion date: 2/xi/2012. Depository:
Host data. Hyalorista exuvialis (Crambidae) feeding on Hyptis verticillata (Lamiaceae).
Caterpillar and holotype voucher codes. 12-SRNP-56280, DHJPAR0051309.
Paratypes
Hosts = Hyalorista exuvialis and Hyalorista exuvialisDHJ02. DHJPAR0038188, DHJPAR0051311. Depository:
Stantonia brookejarvisae , sp. nov.
Diagnostics
BOLD:AAK5505. Consensus barcode. AATTTTGTATTTAAAATTTGGRATTTGGGCTGGGATTTTAGGTATATCATTAAGATTAATTATTCGATTAGAGTTAGGAACTCCTGGGAGATTAATTGGTAATGATCAAATTTATAATAGTGTGGTAACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGTTTTGGAAATTGGCTAATTCCTATAATATTGGGGTGTCCTGATATAGCATTTCCTCGTATAAATAATATAAGATTTTGGTTGTTAATTCCTTCTTTAATAATATTAATTTTTAGAGGTATTTTAAATATTGGAGTTGGTACAGGTTGAACTGTTTATCCTCCTTTATCTTTAAATATTGGTCATGGTGGAGTTTCAGTTGATATATCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCCATYATAGGYGCCGTAAATTTTATTACTACAGTTTTAAATATACGAATTAAATTAATTTTAATGGATAAAATTTCTTTATTAATTTGATCAGTTTTTATYACAGCTATTTTATTGTTATTATCTTTACCTGTTTTAGCTGGGGCTATTACTATATTGTTAACAGATCGTAATTTAAATACTTCTTTTTTTGATCCWTCTGGGGGGGGTGATCCAGTTTTGTATCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Rio Francia Arriba, 10.89666, -85.29003, 400 meters, caterpillar collection date: 28/vi/2010, wasp eclosion date: 12/vii/2010. Depository:
Host data. Pilocrocis purpurascens (Crambidae) feeding on Pentagonia donnell-smithii (Rubiaceae).
Caterpillar and holotype voucher codes. 10-SRNP-42400, DHJPAR0040550.
Paratypes
Hosts = Desmia benealisDHJ02, Phostria latiapicalis, Pilocrocis purpurascens (all Crambidae). DHJPAR0050354, DHJPAR0050911, DHJPAR0050920, DHJPAR0057266, DHJPAR0036321, DHJPAR0052099, DHJPAR0053534, DHJPAR0053538. Depository:
Stantonia donwilsoni , sp. nov.
Diagnostics
BOLD:AAG3930. Consensus barcode. TATTTTATATTTTAAATTTGGTATTTGATCTGGTATTATRGGAATATCTTTAAGATTAATTGTACGAATAGAGTTAGGTATACCTGGTAGTTTAATTGGTAATGATCAAATTTATAATAGGGTAGTAACTGCTCATGCATTTGTAATAATTTTTTTTATAGTTATACCTATTATAATTGRGGGATTTGGCAATTGATTAATTCCTTTAATATTAGGTTGTCCAGATATAGCTTTCCCTCGTATAAATAATATAAGGTTTTGATTATTAATYCCTTCTTTAATATTATTGATTTTTAGAAATGTTCTGAATATTGGGGTTGGTACAGGTTGAACTGTTTATCCTCCTTTATCTTTATTAATTGGTCATGGTGGTTTATCAGTTGATATAGCAATTTTTTCTTTACATTTAGCAGGTATTTCTTCTATTATAGGGGCTGTTAATTTTATTACTACAATTTTAAATATGCGTGTTGAAAAAATTTATATGGATAAAATTTCTTTATTATCTTGATCTATTTTTATTACTGCTATTTTATTATTATTGTCTTTACCTGTTTTAGCCGGTGCAATTACTATATTATTAACTGATCGTAATATAAATACTTCTTTTTTTGATCCATCTGGTGGTGGAGATCCTATTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Santa Rosa, Area Administrativa, 10.83764, -85.61871, 295 meters, caterpillar collection date: 27/vii/2013, wasp eclosion date: 6/viii/2013. Depository:
Host data. Portentomorpha xanthialis (Crambidae) feeding on Margaritaria nobilis (Phyllanthaceae).
Caterpillar and holotype voucher codes. 13-SRNP-19529, DHJPAR0052706.
Paratypes
Hosts = Omiodes humeralis, Psara obscuralisDHJ02, Eulepte concordalis, Portentomorpha xanthialis, Phostria latiapicalis, Hahncappsia BioLep471 (all Crambidae). DHJPAR0029187, DHJPAR0029429, DHJPAR0041487, DHJPAR0052872, DHJPAR0052875, DHJPAR0063891, DHJPAR0063896. Depository:
Etymology
Stantonia donwilsoni is named in honor of Don Wilson attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Stantonia erikabjorstromae , sp. nov.
Diagnostics
BOLD:ABX0808. Consensus barcode. AATTTTATATTTAAAATTTGGAATTTGGTCAGGTATTTTAGGCATATCTTTAAGTTTAATTGTTCGAATAGAATTAGGGATACCYGGTAATTTAATTGGTAATGATCAAATTTATAATAGAATTGTTACATTTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGGTTTGGAAATTGATTAATTCCTTTAATATTAGGATGTCCTGATATAGCTTTTCCTCGAATGAATAATATAAGATTTTGATTATTAATTCCTTCTTTATTGTTATTAATTTTTAGAAGAATTTTAAATATTGGTGTGGGGACTGGTTGAACAGTTTATCCYCCTTTATCTTTATCTATAGGTCATGGAGGAATTTCTGTAGATATGGCTATTTTTTCTTTACATTTAGCCGGAGCTTCTTCYATTATAGGYGCYGTAAATTTTATTACTACTATTTTAAATATACGAATTAATAAAATTTATTTAGATAAAATTTCTTTATTATCATGATCTGTTTTTATTACTGCTATTTTATTATTATTATCTTTACCTGTTTTAGCYGGAGCAATTACTATATTATTAACTGATCGTAATTTAAATACTTCTTTTTTTGATCCTTCTGGGGGTGGTGATCCTATTTTATACCAGCATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Malaguenya, 10.9555, -85.28381, 221 meters, caterpillar collection date: 16/v/2012, wasp eclosion data: 4/vi/2012. Depository:
Host data. Omiodes Janzen05 (Crambidae) feeding on Entada gigas (Fabaceae).
Caterpillar and holotype voucher codes. 12-SRNP-67833, DHJPAR0048713.
Paratypes
DHJPAR0046874, DHJPAR0046876, DHJPAR0046879, DHJPAR0046880, DHJPAR0049677. Depository:
Stantonia garywolfi , sp. nov.
Diagnostics
BOLD:AAL9316. Consensus barcode. TTTTAGGTATATCTTTAAGTTTATTAATTCGTATAGAATTAGGTACTCCAGGAAGATTAATTGGTAATGATCAAATTTATAATAGAGTTGTTACATCTCATGCTTTTATTATAATTTTCTTTATAGTTATACCTATTATAATTGGTGGTTTTGGAAATTGATTAATTCCAATAATATTAGGGTGTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGGTTATTAATTCCTTCATTGTTAATATTACTTTTTAGTGGAGTTTTAAATATTGGTGTTGGTACTGGATGAACAGTTTATCCTCCTTTATCTTTAAATATTGGTCATGGTGGAATTTCTGTTGATATTGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATAGGTGCAATTAATTTTATTACTACAGTACTAAATATACGAACTGGAAAAATTATAATAGATAAGGTTACTTTATTAATTTGATCAGTTTTTATTACAGCTATCTTATTACTTTTGTCTTTACCAGTTTTAGCTGGTGCTATTACTATATTATTAACTGATCGTAATTTAAATACTTCATTTTTTGATCCTTCAGGTGGTGGTGATCCTGTTTTATATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Pasmompa, 11.01926, -85.40997, 440 meters, caterpillar collection date: 29/xi/2005, wasp eclosion date: 2/i/2006. Depository:
Host data. immidJanzen01 Janzen02 (Immidae) feeding on Securidaca sylvestris (Polygalaceae).
Caterpillar and holotype voucher codes. 05-SRNP-70049, DHJPAR0029189.
Paratype
Host = same as holotype: DHJPAR0029188. Depository:
Stantonia henrikekmani , sp. nov.
Diagnostics
BOLD:AAI2111. Consensus barcode. AATTTTATATTTAAAATTTGGAATTTGGGCTGGGATTTTAGGTATATCATTAAGTTTAATTATTCGGTTAGAATTAGGTACTCCTGGAAGATTAATTGGGAATGATCAGATTTATAATAGTGTAGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGYTTYGGRAATTGATTAATTCCTATAATATTAGGATGTCCTGATATAGCATTTCCTCGAATGAATAATATAAGATTTTGGTTATTAATTCCTTCTTTAATAATATTAATTTTTAGTGGTATTTTAAATATTGGTGTTGGGACAGGTTGAACTGTTTATCCACCTTTATCTTTAAATATTGGTCATGGGGGAATTTCAGTTGATATATCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCYATCATAGGYGCYGTAAATTTTATTACTACGGTTTTAAATATACGAATTAAATTAATTTTAATAGATAAAATTTCTTTATTAATTTGATCAGTTTTTATTACAGCTATTTTATTATTATTATCTTTRCCTGTTTTAGCTGGAGCAATTACTATACTATTAACGGATCGTAATTTAAATACTTCTTTTTTTGATCCATCTGGAGGAGGAGATCCAGTATTATATCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Quebrada Bambu, 10.93009, -85.25204, 109 meters, caterpillar collection date: 5/viii/2014, wasp eclosion date: 28/viii/2014. Depository:
Host data. Ceratocilia sixolalis (Crambidae) feeding on Neea psychotrioides (Nyctaginaceae).
Caterpillar and holotype voucher codes. 14-SRNP-76550, DHJPAR0056233.
Paratypes
Hosts = Pilocrocis Solis20, Desmia ploralisDHJ03, Eulepte Janzen03, Ceratocilia sixolalis, Desmia benealisDHJ02 (all Crambidae). DHJPAR0035220, DHJPAR0035221, DHJPAR0050358, DHJPAR0052825, DHJPAR0054462, DHJPAR0054465, DHJPAR0054466, DHJPAR0056229, DHJPAR0056230, DHJPAR0056231, DHJPAR0056232, DHJPAR0056234. Depository:
Stantonia luismirandai , sp. nov.
Diagnostics
BOLD:AAH9964. Consensus barcode. TATTTTATATTTAAAATTTGGTATTTGATCTGGTATTTTGGGTATATCATTAAGGTTAATTGTRCGATTAGAGTTRGGRATACCAGGTGGTTTAATTGGTAATGATCAAATTTATAATAGAGTTGTTACTGCTCATGCKTTTGTAATAATTTTTTTTATAGTAATGCCTATTATAATTGGGGGGTTTGGAAATTGGYTAATTCCAATAATGTTAGGGTGTCCAGATATGGCTTTYCCTCGAATAAATAATATAAGATTTTGGTTGTTAATTCCTTCATTAATTTTATTAATTTTTAGAAGAATTTTAAATATTGGTGTRGGGACTGGGTGAACTGTTTAYCCTCCATTATCTTTATTAATTGGTCATGGGGGTGTATCGGTTGATATGGCTATTTTTTCTTTACATTTGGCTGGTATTTCTTCAATTATGGGGGCCATTAATTTTATTACAACAATTTTAAATATACGTGTTAATAWAATTTATATAGATAAGATTTCTTTATTATGTTGGTCAGTTTTTATTACTGCTATTTTATTATTGTTATCTTTACCTGTTTTAGCYGGGGCTATTACTATATTGTTAACTGATCGAAATTTAAATACTTCTTTTTTTGATCCTTCTGGKGGAGGRGAYCCAATTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 6/ii/2014, Malaise trap. Depository:
Host data. None for the holotype but see below.
Holotype voucher code. BIOUG29625-H09.
Paratypes
Hosts = Oryctometopia fossulatella and Spoladea recurvalis (both Crambidae). DHJPAR0029190, DHJPAR0029191. Depository:
Stantonia miriamzunzae , sp. nov.
Diagnostics
BOLD:ACB1896. Consensus barcode. AATTTTATATTTAAAATTTGGTATTTGAGCAGGAATTTTAGGTATGTCTATAAGGTTAATTATTCGGTTAGAATTAGGAATGCCCGGAAGTTTATTAGGTAATGATCAAATTTATAATAGAATTGTAACATCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCAATAATATTAGGATGTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATAATATTAATTTTTAGAAGAATTTTAAATATTGGTGTTGGTACAGGTTGAACAGTTTATCCTCCTTTATCATTAAATATTAATCATGGTGGAATTTCTGTTGATATAGCAATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATAGGAGCAATTAATTTTATTACTACAATTTTTAATATACGAATAAAATTAATTTTAATAGATAAAATTTCTTTATTAATTTGATCAGTATTTATTACAGCTATTTTATTATTATTATCTTTACCAGTTTTAGCTGGAGCTATTACAATATTATTAACTGATCGAAATTTAAATACTTCATTTTTTGATCCTTCAGGTGGTGGTGATCCAATTTTATATCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Quebrada Bambu, 10.9301, -85.25205, 109 meters, caterpillar collection date: 18/v/2012, wasp eclosion date: 2/vi/2012. Depository:
Host data. Ategumia lotanalis (Crambidae) feeding on Conostegia xalapensis (Melastomataceae).
Caterpillar and holotype voucher codes. 12-SRNP-75835, DHJPAR0048704.
Paratypes
None.
Stantonia quentinwheeleri , sp. nov.
Diagnostics
BOLD:AAD5731. Consensus barcode. AATCTTATATTTAAAATTTGGAATTTGGGCTGGGATTTTAGGTATGTCATTAAGTTTAATTATTCGATTAGAATTAGGAACTCCAGGGAGATTAATTGGTAATGATCAAATTTATAATAGTGTAGTAACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGGTTTGGAAATTGGTTAATTCCTATAATATTGGGGTGTCCTGATATAGCATTCCCTCGAATAAATAATATAAGATTTTGGTTATTAATTCCTTCTTTAATAATATTAATTTTTAGAGGTATTTTAAATATTGGTGTTGGGACAGGGTGAACTGTTTATCCTCCTTTATCTTTAAATATTGGTCATGGTGGAATTTCAGTTGATATATCTATTTTTTCTTTACATTTGGCTGGTATTTCTTCCATTATAGGAGCCGTAAATTTTATTACAACAGTTTTAAATATACGAATTAAAATAATTTTAATGGATAAAATTTCTTTATTAATTTGATCAGTTTTTATTACAGCTATTTTATTATTATTATCTTTACCTGTTTTAGCTGGGGCAATTACTATATTATTAACTGATCGTAATTTAAATACTACTTTTTTTGATCCGTCTGGAGGTGGGGATCCAGTTTTATATCAACATTTATTT.
Holotype ♂
Guanacaste, Sector San Cristobal, Tajo Angeles, 10.86472, -85.41531, 540 meters, caterpillar collection date: 2/viii/2011, wasp eclosion date: 23/viii/2011. Depository:
Host data. Syllepis marialis (Crambidae) feeding on Allophylus psilospermus (Sapindaceae).
Caterpillar and holotype voucher codes. 11-SRNP-3032, DHJPAR0045054.
Paratypes
Hosts = Syllepis hortalis and Syllepis marialis. DHJPAR0029430, DHJPAR0029431, DHJPAR0029432, DHJPAR0029433, DHJPAR0036719, DHJPAR0029434. Depository:
Etymology
Stantonia quentinwheeleri is named in honor of Quentin Wheeler attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Stantonia robinkazmierae , sp. nov.
Diagnostics
BOLD:ACB2340. Consensus barcode. AATTTTATATTTAAAATTTGGTATTTGAGCTGGTATTTTGGGTATATCTTTAAGATTAATTGTTCGGTTAGAATTGGGTATACCTGGAAGATTAATTGGTAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCTTTTGTTATAATTTTTTTTATGGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCTATAATATTAGGATGCCCTGATATGGCTTTTCCTCGAATAAATAATATGAGATTTTGGTTATTAATTCCTTCTTTGAGATTATTAATTTTTAGAAGAATTTTAAATATTGGTGTCGGTACAGGTTGAACTGTTTATCCTCCTTTATCTTTATTAATTGGCCATGGAGGAGTTTCAGTTGATATAGCAATTTTTTCTTTACATTTAGCTGGGATTTCTTCTATTATAGGGGCTATTAATTTTATTACAACAATTTTAAATATACGAATTAATAAAATTTATATAAATAATATTTCTTTATTGTCTTGATCTGGATTTATTACTGCTATTTTATTATTATTATCTTTGCCTGTTTTAGCTGGTGCTATTACTATATTATTGACTGATCGAAATTTAAATACATCTTTTTTTGATCCTTCTGGTGGAGGGGATCCTGTTTTGTATCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93050, -85.37223, 527 meters, caterpillar collection date: 28/iv/2012, wasp eclosion date: 14/v/2012. Depository:
Host data. Inga Janzen61 (Oecophoridae) feeding on Neea psychotrioides (Nyctaginaceae).
Caterpillar and holotype voucher codes. 12-SRNP-1693, DHJPAR0049288.
Paratypes
Host = Inga Janzen61: DHJPAR0049427. Depository:
Stantonia ruthtifferae , sp. nov.
Diagnostics
BOLD:AAB3996. Consensus barcode. TATTTTATATTTTAAATTTGGTATTTGATCTGGTATTATAGGAATATCTTTAAGATTAATTGTACGAATAGAGTTAGGTATACCTGGTAGTTTAATTGGTAATGATCAAATTTATAATAGGGTAGTAACTGCTCATGCATTTGTAATGATTTTTTTTATAGTTATACCTATTATAATTGGAGGTTTTGGAAATTGATTAATTCCCTTAATATTAGGGTGTCCAGATATAGCTTTTCCTCGTATAAATAATATAAGGTTTTGATTATTAATTCCTTCTTTAATATTATTGATTTTTAGAAATGTTTTAAATATTGGGGTTGGGACAGGTTGAACTGTTTATCCTCCTTTATCTTTATTAATTGGTCATGGTGGTTTATCAGTTGATATAGCAATTTTTTCTTTACATTTAGCAGGTATTTCTTCTATTATAGGAGCTGTTAATTTTATTACTACTATTTTAAATATACGTGTTGATAWAATTTATATAGATAAAATTTCTTTGTTATCTTGATCTATTTTTATTACTGCTATTTTGTTACTATTGTCTTTACCTGTATTAGCTGGTGCAATTACTATATTATTAACTGATCGTAATATAAATACTTCTTTTTTTGATCCATCTGGTGGTGGAGATCCTA.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Estación Llanura, 10.93332, -85.25331, 135 meters, caterpillar collection date: 09/x/2009, wasp eclosion date: 23/x/2009. Depository:
Host data. Microthyris prolongalisDHJ03 (Crambidae) feeding on Ipomoea philomega (Convolvulaceae).
Caterpillar and holotype voucher codes. 09-SRNP-76086, DHJPAR0037144.
Paratype
Host = Stenoma luctificaDHJ02 (Depressariidae): DHJPAR0049372. Depository:
Etymology
Stantonia ruthtifferae is named in honor of Ruth Tiffer attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Chapter 10: Proteropinae
Proteropinae are treated here as a subfamily following
Key to the New World genera of Proteropinae
Hebichneutes Sharkey & Wharton, 1994
Members of the genus are restricted to the Neotropics. The genus was erected and revised by
Hebichneutes tricolor
Diagnostics
BOLD:ACJ8785. Consensus barcode. TATTTTGTATTTTATATTTGGAATATGAAGAGGTATATTAGGTTTATCTATAAGTTTAATTATTCGGTTAGAATTAGGAATACCAGGAAGATTATTATTTAATGATCAAATTTATAATAGGATTGTAACTTCACATGCTTTTGTAATAATYTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATATTATTATTATTAAGAAGTTTAGTTAATGTAGGAGCTGGAACTGGTTGAACAGTTTACCCYCCTTTATCTTTAATTATAGGACATGGAGGAATTTCTGTTGATTTATGTATTTTTTCTTTACATTTAGCTGGTATATCATCTATTATAGGAGCAATTAATTTTATTAGAACTATTTTAAATATACGTGTTTTTGGAATAAAAATAGAAAATATTTCATTATTGAGTTGATCAATTTTTATTACTGCTTTTTTATTATTATTATCATTACCTGTTTTAGCAGGAGCCATTACTATATTATTAACTGATCGTAATTTAAATACAAGATTTTTTGATCCATCTGGAGGAGGTGATCCAATTTTATATCAACATTTATTT.
Illustrated specimen
BIOUG17956-G03. Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, Malaise trap, 11/vi/2012. Depository:
Other specimens
BIOUG18749-D04, BIOUG18822-C03, BIOUG18748-A03, BIOUG08345-F02, BIOUG08345-F03, BIOUG08583-E09, DHJPAR0015420, DHJPAR0015418, DHJPAR0015417, DHJPAR0015419. Depository:
Host data. The DHJPAR specimens were reared from sawfly larvae (family unknown because none reached adulthood) feeding on Xylophragma seemannianum (Bignoniacae).
Proterops Wesmael, 1835
Members of this genus are widespread in the Holarctic and Oriental regions, but not recorded from Africa or Australia. There are 12 described species. Here we describe the first Neotropical species, although there are a dozen or more undescribed. Members are parasitoids of tenthredinid and argid sawfly larvae (
Proterops iangauldi , sp. nov.
Diagnostics
BOLD:ADD0222. Consensus barcode. ATTTTATATTTTATATTTGGTATAGGAGAGGTATAATTGGATTTCTTTAAGAATAATTATTCGATTAGAGTTAGGTTTACCAGGAAAATTTTTAGGTAATGATCAAATTTATAACAGAATTGTTACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAGCATTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTATTTTTATTATTATTAAGAAGATTTATTAATAATGGGGTAGGAACTGGATGAACAGTTTATCCTCCTTTGTCATTATTAATAGGACATAGAGGTATTTCTGTTGATTTGAGAATTTTTTCTTTACATTTAGCTGGTATATCTTCTATTATAGGAGCTATTAATTTTATTACTACTATTTTTAATATACGATTAATTGGAATGAGAATAGATAAAATTTCTTTATTGGTTTGATCAGTTTTAATTACAGCATTTTTATTATTATTATCTTTACCTGTTTTAGCGGGAGCTATTACTATATTATTAACTGATCGTAATTTT------------------------------------------------------------.
Holotype ♀
Guanacaste, Sector Horizontes, Quebrada San Pancho, 10.74769, -85.58577, 90 meters, sawfly larva collection date: 26/v/2000, wasp eclosion date: 12/vi/2000. Depository:
Host data. Sphacophilus edus (Argidae) feeding on Heteropterys laurifolia (Malpighiaceae).
Sawfly larva and holotype voucher codes. 00-SRNP-7495, DHJPAR0015459.
Paratypes
DHJPAR0015457, DHJPAR0015458, DHJPAR0015460, DHJPAR0015461, DHJPAR0015462, DHJPAR0015463, DHJPAR0015464. Depository:
Etymology
Proterops iangauldi is named in honor of Ian Gauld (RIP) attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing his wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Proterops vickifunkae , sp. nov.
Diagnostics
BOLD:AAM1715. Consensus barcode. AATATTATATTTTTTATTTGGGATTTGATCGGGAATAGTAGGTCTTTCATTAAGATTAATTATTCGGTTGGAATTAGGAATCCCTGGAAAATTTTTAGGTAATGATCAAATTTATAATAGTATGGTTACTATACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGAAATTGGTTAATTCCTTTAATATTAGGGGCTCCTGATATAGTATTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAATTTTATTATTATTAAGTGGATTTATAGGTAATGGTGTTGGGACTGGATGAACAGTTTATCCACCATTGTCTTTAATTATAGGTCATTCTAGTATATCTGTAGATATAAGAATTTTTTCTTTACATTTAGCAGGTATATCTTCTATTATAGGAGCTATAAATTTTATTACTACAATTTTTAATATGCGATTATTAGGGATAAGTATGGATAAAATTTCTTTATTAGTATGATCAATTTTTATTACTGCTTTTTTATTATTAATATCTTTACCAGTTTTAGCTGGTGCAATTACTATATTATTAACTGATCGTAATTTAAATACATGTTTTTTTGATCCTTCTGGTGGAGGGGATCCTATTTTGTATCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Bullas, 10.98670, -85.38503, 440 meters, sawfly larva collection date: 8/x/2009, wasp eclosion date: 3/xi/2009. Depository:
Host data. Ptenos leucopoda (Argidae) feeding on Inga oerstediana (Fabaceae).
Sawfly larva and holotype voucher codes. 09-SRNP-73070, DHJPAR0037943.
Paratype
Ptenos leucopoda (Argidae): DHJPAR0052901. Depository:
Etymology
Proterops vickifunkae is named in honor of Vicki Funk (RIP) attending the international NSF-funded planning meeting for the All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems, and contributing her wisdom to the planning that was the founding of Costa Rica’s national BioAlfa today.
Michener , gen. nov.
Type species
Michener charlesi Sharkey, sp. nov.
Diagnosis
Similar to Proterops but differing in the following aspects: 1. Michener with tergum 1 with a pair of prominent, precurrent, longitudinal carinae. Proterops with tergum 1 lacking carinae. 2. Michener with tergum 1 slightly narrowed at midlength and almost 3 × longer than wide at narrowest point. Proterops with tergum 1 not narrowed at midlength, rather narrowest at base or apex; tergum 1 shorter, usually ca. as long as wide but always less than 2 × longer than wide. 3. Michener with tergum 2 short, half the length of tergum 3, measured along midline. Proterops with tergum 2 ca. the same length as tergum 3, measured along midline. 4. Michener with laterotergites of tergum 1 densely setose. Proterops with laterotergites sparsely setose.
Description
Head . Occipital carina absent; antenna with 34 flagellomeres; first flagellomere without patch of basiconic sensillae; maxillary palpus with four segments; labial palpus with three segments; mandible not twisted for scissor-like function; base of mandible not depressed and without microsculptured area set off by acute transverse ridge. Wings. Vein 1M of forewing evenly curved; vein 1RS of forewing present; venation complete; R of hind wing not sharply curved posteriad near apex; three hamuli in one cluster. Mesosoma. Propleuron without longitudinal carina; pronotum with weak dorsolateral pit (subpronope); epicnemial carina absent; precoxal depression absent; notaulus smooth, weakly indicated as a smooth line, posterior scutellar depression absent; propodeum smooth; tarsal claws simple, without basal tooth or lobe. Metasoma. Tergum 1 with a pair of prominent, precurrent, longitudinal carinae; tergum 1 narrowest at midlength and 2.8 × longer than wide at narrowest point; tergum 2 short less than half the length of tergum 3, measured along the midline. Female hypopygium without dense patch of setae apically
Distribution
Known only from the type locality of the type species in Costa Rica.
Etymology
Named with profound admiration for the lifetime achievements of Professor Charles Michener (RIP) of the University of Kansas, Lawrence, and the many professional and personal forms of support offered to Janzen’s entire professional academic career, as well as to many others.
Michener charlesi , sp. nov.
Diagnosis
Length 5.8 mm. Other details are illustrated in Fig.
Holotype ♀
Guanacaste, Área de Conservación Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.84389, -85.61384, 300 meters, 01/vi/1978, D. H. Janzen, Depository:
Host data. None.
Paratype
♀, same data as holotype, except 18/xi/1977.
Etymology
See generic etymology above.
Chapter 11: Rhysipolinae
Rhysipolinae can be differentiated from all other New World braconid subfamilies by having the following suite of characters: oral cavity circular dorsally and labrum concave (cyclostome); occipital carina not meeting hypostomal carina; rather, meeting, or almost meeting, base of mandible; epicnemial carina present; propodeum lacking pair of lateral spines; second submarginal cell of forewing present and 4-sided, i.e., 2RS vein meeting (RS+M)b or junction of (RS+M)a and (RS+M)b. Rhysipolis may be differentiated from the other New World genera of Rhysipolinae using the key that follows.
Little is known about their life histories. They are solitary, koinobiont, ectoparasitoids of late instar leaf-miners and leaf-tiers, and that they move from the host leaves and/or otherwise feed and/or pupate elsewhere (
Key to New World genera of Rhysipolinae
Pseudorhysipolis Scatolini, Penteado-Dias & van Achterberg, 2002
There are ten previously described species, all from the neotropics (
Pseudorhysipolis luisfonsecai , sp. nov.
Diagnostics
BOLD:ACW5686. Consensus barcode. GTATTATATTTTTTATTTGGAATTTGATCTGGTATAGTAGGTTTATCTATGAGTTTAATTATTCGTTTAGAGTTAGGTATACCTGGTAGTTTATTATTTAATGATCAGATTTATAATACTATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTGTAATGATTGGGGGGTTTGGTAATTGATTAGTTCCATTAATATTGGGGGCTCCTGATATAGCTTTCCCTCGTATGAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATTTTATTATTTTTAAGGGGTTTAGTAAATGTTGGGGTAGGTACTGGGTGAACTGTTTATCCTCCTTTATCTTCTTCTATAGGTCATAGAGGTATTTCTGTTGATTTGGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCAATTATAGGAGCTATTAATTTTATTTCAACAATTTTTAATATATGTTTATATTCAATTAATATAGATCAAATTAGTTTATTTGTTTGATCTATTTTGATTACTGCTTTTTTATTATTATTATCTTTACCTGTCTTGGCAGGG------------------------------------------------------------------------. Keys to P. areolaris in
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.88, -85.389, 575 meters, 23/xi/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG22620-D04.
Paratypes
None.
Pseudorhysipolis mailyngonzalezae , sp. nov.
Diagnostics
BOLD:ACL1561. Consensus barcode. TGTTTTGTATTTTATTTTTGGTATATGAGCTGGAATAGTTGGTTTATCTATAAGATTAAT-TATTCGATTAGAATTAGGGGTATCTGGAAGATTATTAGGGAATGATCAAAT------TTATAATACTATTGTTACATCTCATGCTTTTGTAATAATTTTTTTTATAGTTATACCTATTATATTAGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGGGCTCCTGATATAGCATTTCCTCGAATAAATAATATAAGATTTTGATTAT-----------------TGATTCCATCATTAATTTTATTATTTTTAAGTAGA------------TCAATAAATTTAGGAGC-----------------------------------TGGAACGGGGTGAACTATATATCCTCCTTTATCT-------------------TCAAGAATTGGT--------CATAGAGGAATATCTGTTGATTTAACAATTTTT--TCTTTACATTTAGCTGG---TTGTTCTTCTATTATAGGATCAATTAATTTTATTTGTACAATTTTTAATATA--AAAATTAATTTT---TTAAAAATAGAACAATTAAGTTTATTTGTTTGGTCAGTTTTAATTACAACAATTTTATTATTATTATCTTTACCAGTTT--TAGCTGGTGCTATTACTATATTATTAACAGATCGTAATTTAAATACATC------TTTTTTTGATTTTTCAGGTGGTGGTGATCCAATTTTATTTCAACATTTATTT. Keys to P. fuscaoicalis in
Holotype ♀
Guanacaste, Sector Santa Rosa Bosque San Emilio, 10.8438, -85.6138, 300 meters, 10/xii/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG09442-D01.
Paratypes
Both Malaise-trapped. BIOUG09441-H03, BIOUG09740-B12. Depository:
Etymology
Pseudorhysipolis mailyngonzalezae is named to honor Dr. Mailyn Gonzalez of the Humboldt Institute, Bogota, Colombia, for instigating the growing mutualism between Costa Rica’s BioAlfa and Colombia’s interest in biodevelopment of its massive biodiversity.
Rhysipolis Foester, 1862
Little is known about their life histories.
Rhysipolis julioquirosi , sp. nov.
Diagnostics
BOLD:AAT9029. Consensus barcode. TGTTTTGTATTTTTTATTTGGAATATGATCAGGTATAGTTGGATTATCAATAAGTTTGATTATTCGATTAGAATTAGGGATACCTGGAAGTTTAATTGGTAATGATCAAATTTATAATACAATGGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGATTTGGAAATTGATTAGTTCCATTAATATTAGGGGCTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGGTTTTGATTATTAATTCCTTCTTTAATTTTATTATTTTTAAGAAGAATTGTAAATGTTGGGGTTGGTACTGGTTGAACAGTTTATCCGCCCCTATCTTCAATATTAGGTCATAGGGGTATTTCTGTAGATTTGGCTATTTTTTCTTTACACTTGGCTGGTGTTTCTTCTATTATAGGAGCTATTAATTTTATTTCTACAATTTTTAATATATGTATATAYTCAATTAAATTAGATCAACTTAGTTTGTTTGTTTGATCTATTTTAATTACTGCAATTTTATTATTATTATCTTTRCCTGTTTTGGCAGGGGCTATTACTATATTATTGACAGATCGTAATTTGAATACTACTTTTTTTGATTTTTCTGGTGGGGGGGATCCAATTTTATTTCAACATTTATTT. Neither of the two described Neotropical species, R. annulator Achterberg & Pentiado-Dias, 2002, and R. chiapas Spencer, 1999, have deeply infuscated wings.
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Huerta, elevation: 10.93050, -85.37223, 527 meters, caterpillar collection date: 13/vi/2013, wasp eclosion date: 02/vii/2013. Depository:
Host data. Dichomeris Janzen273 (Gelechiidae) feeding on Mikania cordifolia (Asteraceae).
Host caterpillar and holotype wasp voucher codes. 13-SRNP-3016, DHJPAR0053002. The host caterpillar rolls the food plant leaf to make a “nest,” and the wasp larva spins a dull white, thin-walled cocoon in the host’s pupal chamber (Fig.
Paratypes
Hosts = seven rearing records from Dichomeris Janzen273, Anacampsis Janzen301 (both Gelechiidae), elachJanzen01 Janzen397 (Depressariidae). DHJPAR0041996 DHJPAR0041520. DHJPAR0043148, DHJPAR0049862, DHJPAR0054828, DHJPAR0054834. Depository:
Etymology
Chapter 12: Rogadinae
Rogadinae is a large, cosmopolitan subfamily. The New World genera of Rogadinae may be distinguished using the key that follows. A total of 231 species is recorded from the Neotropical realm and 38 from Costa Rica, not including those described below. Members of all genera are koinobiont endoparasitoids of caterpillars from a wide range of families. Some are gregarious but this applies to only two described here, one species of Aleiodes and one of Triraphis. For the Rogadinae NJ tree, see Suppl. materia 10.
Key to the New World genera of Rogadinae
Note: Macrostomion is not included in this key as it was in
Bioalfa , gen. nov.
Type species
Bioalfa pedroleoni Sharkey, sp. nov.
Description
Female: Head. Antenna length approximately 1.5 × forewing length, with ca. 48 flagellomeres. Terminal flagellomere acuminate apically. Median flagellomeres transverse, ca. 2 × as long as wide. Face with oblique and transverse striation. Clypeus shallow, not protruding. Malar suture absent. Eye glabrous, large, and deeply emarginate, occupying entire height of head resulting in very small malar space. Eye strongly emarginated opposite antennal socket. Anterior tentorial pit large, very close to eye. Maxillary palp with six segments, all slender in female. Occipital carina complete dorsally, without dorsolateral posterior bulge, connecting to hypostomal carina ventrally. Mesosoma. Smooth and shiny. Notauli deeply impressed, meeting at the midline and 2/3 length of mesonotum; a single weak medial depression continues to the scuto-scutellar suture. Mesoscutal mid-pit absent. Scutellar sulcus wide with single strong mid-longitudinal carina. Epicnemial carina complete. Mesopleuron smooth with mostly widely spaced minute setiferous punctures; precoxal sulcus wide, almost entirely smooth with a few weak transverse carinae at midlength. Median area of metanotum without mid-longitudinal carina. Propodeum with one precurrent median carina or two weak diverging carinae defining a narrow, elongate medial area, otherwise, finely rugose with or without weak posterolateral depressions set off by weak carinae. Forewing. Crossvein m-cu weakly-curved, forming a slightly obtuse angle with 1Cub. First submarginal cell evenly setose. Second submarginal cell approximately twice as long as wide. 2RS and r-m not parallel. Crossvein 1cu-a postfurcal. First subdiscal cell rectangular. Subbasal cell evenly setose. Hind wing. Vein R short, as long as wide to ca. 2 × as long as wide. Veins M+Cu and 1-M approximately equal length. Vein RS strongly curved. Vein m-cu absent. Base of hind wing evenly setose. Legs shining, with sparse long setae. Legs. Apex of hind tibia with comb of specialized setae medially. Mid and hind tibial spurs strongly curved and glabrous. Claws with squared or slightly acute basal lobe. Metasoma. Tergites 1–3 finely longitudinally striate. First metasomal tergite elongate, strongly and angularly widened in front of sub-basal constriction; with dorsal carinae uniting almost immediately and forming a weak carina along midline almost indistinguishable from longitudinal striae. Second metasomal tergite with midlongitudinal carina arising from small, transverse basal triangular area. Tergites 3–5 with sharp lateral crease. Hypopygium large, ventrally moderately to strongly convex. Ovipositor straight or curved ventrally. All four known species have patterned wings and colorful (for Rogadinae) bodies, suggesting diurnal activity. Male: As in the female but with swollen labial and maxillary palpi.
Biology
Caterpillar hosts are known for two species, which are both external leaf-feeders in the family Uraniidae.
Distribution
Neotropical rainforest, Costa Rica, Amazonian Colombia, and Peru.
Remarks
Morphologically, the new genus displays a mixture of character states associated with the Colastomion-Cystomastax group of genera, i.e., the curved, glabrous tibial spurs, and the enlarged hypopygium. Bioalfa differs from all other genera in this group by the presence of squared to slightly acute basal lobes on the tarsal claws. DNA data (Quicke et al. unpublished) place it deeply within the Colastomion-Cystomastax complex and separate from other New World members.
Etymology
The new genus Bioalfa, from “bioalfabetizacion,” is dedicated to Costa Rica’s brave and optimistic new concept of creating itself to be bioliterate about all of its wild Eukaryota biodiversity, through the combination of national sweat equity, international collaborations, political willpower, and DNA barcoding (c.f. https://news.mongabay.com/2020/04/bold-project-hopes-to-dna-barcode-every-species-in-costa-rica/ and https://ibol.org/barcodebulletin/features/how-a-tropical-country-can-dna-barcode-itself/).
Bioalfa pedroleoni , sp. nov.
Diagnostics
BOLD:AAX3381. Consensus barcode. TGTTTTATATTTTATTTTTGGAATTTGATCTGGTATATTAGGTTTATCTATAAGAATTATTATTCGAATAGAATTAAGTAATCCTGGTGGATTAATTAAAAATGATTATATTTATAATTCAATAGTTACTGCTCATGCTTTTATTATGATTTTTTTTATAGTTATACCAATTATATTAGGGGGATTTGGAAATTGATTAATTCCTATAATATTAGGATCTCCTGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCATTATTTTTATTATTAATAAGTTCATTTGTTAATATTGGTGTTGGTACTGGTTGAACTATATATCCTCCATTATCTTCTTTATTGGGTCATAGTGGATTTTCTGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGGATATCTTCTATTATGGGGTCTATTAATTTTATTTCTACAATTTTTAATATAAAATTATATTCTTTAAATATAGATCAATTAAATTTATTTATTTGGTCAATTTTAATTACTGTAATTTTATTACTTTTATCATTACCTGTTTTAGCAGGTGCTATTACTATATTATTTACTGATCGTAATTTAAATACTACTTTTTTTGATTTTTCTGGTGGGGGGGATCCTATTTTATTTC. Differing from the five other known species (two undescribed and not included here) in many characters including coloration, e.g., it is the only species with one melanic band on the forewing, a terminal band. The others have two melanic bands, one at midlength and one terminal. Body length 5.5 mm. Other details can be gleaned from the images in Fig.
Holotype ♀
Alajuela, Área de Conservación Guanacaste, Sector Rincon Rain Forest, Laureles, 10.933, -85.253, 95 meters, caterpillar collection date: 5/viii/2005, wasp eclosion date: 18/viii/2005. Depository:
Host data. The host caterpillar is Syngria druidaria (Uraniidae) feeding externally on new full-sized leaves of Amphilophium crucigerum, a woody vine in the Bignoniaceae in the dry-rain forest interface.
Host caterpillar and holotype wasp voucher codes. 07-SRNP-42145, DHJPAR0021184.
Paratypes
None.
Etymology
Bioalfa pedroleoni is named in honor of Professor Pedro Leon of the Universidad de Costa Rica for his founding role in the initiation of Área de Conservación Guanacaste in 1986 and his founding role in the initiation of BioAlfa by alerting Costa Rica’s Ambassador to the United States, Dr. Román Macaya, to the potential of DNA barcoding Costa Rica’s biodiversity in 2017.
Bioalfa alvarougaldei , sp. nov.
Diagnosis
Differing from the five other known species (two undescribed and not included here) in many characters including a hypopygium that is much more convex than the others. Body length 7.1 mm. Other details can be gleaned from the images in Fig.
Holotype ♀
Costa Rica, Heredia Prov., La Selva Biological Station, 10.44, -84.01, 100 meters, 1/ii/1994, Malaise trap. Depository:
Host data. None.
Holotype voucher code. None.
Paratypes
4♀♀, all from La Selva Biological Station on the following dates: 1/ix/1993, 1/iv/1985, iv-v/1993, iv/1993. Three specimens deposited in
Etymology
Bioalfa alvarougaldei is named in honor of Sr. Alvaro Ugalde (RIP) in recognition of his major and critical support and role in the founding of Parque Nacional Santa Rosa and its subsequent development as Área de Conservación Guanacaste, and eventual stimulus for BioAlfa, the concept of facilitating Costa Rica to become bioliterate.
Bioalfa rodrigogamezi , sp. nov.
Diagnosis
Differing from all other species of Bioalfa in its lack of color and lack of banding on the forewing (Fig.
Holotype ♂
Costa Rica, Alajuela, Área de Conservación Guanacaste, Sector Rincon Rain Forest, Sendero Parcelas, 10.9077, -85.29137, 375 meters, caterpillar collection date: 10/i/2005, wasp eclosion date: 13/i/2005. Depository:
Host data. The host caterpillar is Hypena Poole17 (Erebidae) feeding on Urera caracasana (Urticaceae).
Host caterpillar and holotype wasp voucher codes. 05-SRNP-40098, DHJPAR0009401.
Paratypes
None.
Note
This is the only known male specimen of Bioalfa. As in related rogadine genera, it has swollen labial and maxillary palpi.
Etymology
Bioalfa rodrigogamezi is named in honor of Professor Rodrigo Gámez, co-founder and executor of the Instituto Nacional de Costa Rica (INBio) in 1989, and co-founder of Área de Conservación Guanacaste in 1985.
Hermosomastax , gen. nov.
Type species
Hermosomastax clavifemorus Quicke, sp. nov.
Description
Female: Head. Antenna approximately as long as forewing, with ca. 40 flagellomeres. Terminal flagellomere acuminate. Median flagellomeres very elongate, > 3 × longer than wide. Face protruding. Clypeus shallow, not protruding. Malar suture absent. Eyes glabrous, distinctly emarginated opposite antennal sockets. Anterior tentorial pits large, very close to eye. Maxillary palp with six segments. Occipital carina complete dorsally and ventrally, with dorsolateral posterior angulation and bulge, connecting to hypostomal carina ventrally. Mesosoma. smooth and shiny. Pronotum wide and truncate anteriorly in dorsal view. Notauli deep, crenulate anteriorly. Middle lobe of mesoscutum anteriorly steep, nearly vertical. Mesoscutum longitudinally striate posteriorly. Scutellar sulcus strongly reniform with single strong midlongitudinal carina. Scutellum narrow. Epicnemial carina complete. Mesopleuron shiny; precoxal sulcus deep and strongly crenulate. Median area of metanotum with midlongitudinal carina and protruding posterodorsally into a point. Propodeum and metapleuron foveate rugose, with weak but distinct angulate apophyses. Forewing. Second submarginal cell approximately twice as long as wide. Second submarginal cell very long (ca. 3 × longer than wide). Vein m-cu nearly straight; vein m-cu interstitial with 2RS. Vein 1cu-a postfurcal. First subdiscal cell rectangular. Subbasal cell evenly setose. Hind wing. Vein R short longitudinal. Vein RS straight. Veins M+Cu and 1-M approximately equal length. Vein RS strongly curved. Vein m-cu absent. Base of hind wing evenly setose. Legs. smooth, with sparse long setae. Hind femur clavate. Apex of hind tibia with comb of specialized setae medially. Mid and hind tibial spurs strongly curved and glabrous. Claws simple without basal lobe. Metasoma. All tergites foveolate-punctate. First metasomal tergite not strongly narrowing subbasally; dorsope deep; with dorsal carinae uniting to form a weak, thin longitudinal carina. Second metasomal tergite with thin mid-longitudinal carina arising from small medio-basal triangular area. Tergites 3–5 with sharp lateral crease. Hypopygium large, ventrally strongly convex. Ovipositor curved ventrally. Male: Unknown.
Biology
Unknown.
Distribution
Only known from type specimen of the type species from Ecuador.
Remarks
Morphologically, the new genus displays a mixture of character states associated with different genus groups. The curved glabrous tibial spurs suggest an affinity with the Colastomion-Cystomastax group of genera; however, it lacks the curved forewing vein m-cu and the elongate and basally narrowed first metasomal tergite that is characteristic of other members of that group. Nevertheless, DNA data (barcode and 28S; Quicke et al. unpublished) place it deeply derived within the Colastomion-Cystomastax group, and therefore the stouter first tergite and straight vein m-cu are presumed to be character state reversals.
Etymology
From Spanish hermoso meaning beautiful, and the suffix mastax from the name of the closely related genus Cystomastax. The genus is masculine.
Hermosomastax clavifemorus , sp. nov.
Diagnostics
No BIN is assigned, presumably because the barcode is only 411 bp long. Consensus barcode. ATTTGATCAGGTATANTAGGTTTATCTATAAGTATTATTATTCGAATAGAACTTAGTAATCCTGGAGGTTTAATTAAAAATGATTATATTTATAATTCAATAGTTACAGCTCATGCTTTTGTAATGATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCTTTGATATTAGGGGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTGTTAATTCCATCTTTATCAATATTATTAATTAGTTCATTTACTAATATTGGGGTAGGTACTGGATGAACAATATATCCTCCTTTATCTTCTTTAATTGGTCATAGTGGTGTTTCTGTAGATTTAGCAATTTTTTCTTTACATTTAGCAGGAATTTCTTCTATTATGGGG. Unique within Rogadinae, this species possesses a strongly petiolate (clavate) hind femur. The patchily patterned wings are also diagnostic within the Colastomion-Cystomastax group.
Description
Length of body 5.4 mm, of forewing 4.0 mm and of antenna 6.8 mm. First flagellomere 1.25 and 1.35 × longer than the 2nd and third respectively, the latter 4 × longer than wide. Width of head: width of face: height of eye = 2.6: 1.0: 1.5. Inter-tentorial distance 3.5 × shortest distance between tentorial pit and eye. Shortest distance between posterior ocelli: transverse diameter of posterior ocellus: shortest distance between posterior ocellus and eye = 1.0: 1.7: 1.7. Occiput smooth and shiny. Length of head behind eye 0.35 × length of eye in dorsal view. Lengths of forewing veins r-rs: 3RSa: 3RSb = 1.0: 5.0: 6.5: Forewing vein 1CUb 7.5 × 1CUa. Hind wing vein 1-M 1.15 × M+CU. Hind coxa deeply punctured basodorsally. Lengths of fore femur (excluding trochantellus): tibia: tarsus = 1.0: 1.2: 1.25. Hind tibia 1.3 × hind femur (excluding trochantellus). First tergite 1.6 × longer than maximally wide. Second tergite as long as wide. Third tergite 1.3 × wider than long.
Coloration. Body, scape and pedicel, and hind coxa largely orange brown becoming darker ventrally. Flagellomeres white each with narrow darker apex. Lower part of face, labial and maxillary palps, fore and mid legs white. Hind leg white except swollen part of femur which is dark brown. Forewing grey with clear subapical transverse band and darker apical margin.
Holotype ♀
Ecuador, Sucumbios, Sacha Lodge, 0.8, -76.1, 270 meters, 24/vi/1994, coll. Peter Hibbs. Depository:
Host data. None.
Holotype voucher code. BOLD process Id: ASQSQ128-09.
Paratypes
None
Aleiodes Wesmael, 1838
Aleiodes are found worldwide. There are records of approximately ten Costa Rican species scattered in publications. There is not a comprehensive generic treatment, and it is remotely possible that a few of the species below are junior synonyms of previously described species. When the holotypes are barcoded, this will become clear. Members are parasitoids of various macrolepidopterans, especially Noctuoidea, Geometroidea, and Bombycoidea (Shaw 1993).
Aleiodes adrianaradulovae , sp. nov.
Diagnostics
BOLD:AAG1413. Consensus barcode. AGTTTTATATTTTTTATTTGGAATATGATCAGGAATAGTAGGTTTATCAATAAGAATAATTATTCGATTAGAATTAAGAATTAGCGGAAGAATTTTGAAAAATGATCAAATTTATAATGGAATAGTTACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCTTTAATATTAAGAGTYCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTATTTTTTTTTTATTATTTAGAGGATTAATTAATTCAGGAGTAGGTACAGGATGAACTATATACCCTCCTTTATCTTCATTAATTGGTCATAGAGGGATATCTGTAGATATATCAATTTTTTCTTTACATTTAGCAGGTATATCTTCTATTATAGGTTCAGTTAATTTTATTACAACAATTTTTAATATTAATTTAAAAAGTATAAAAATAGATCAAATTTCATTATTTACTTGATCAGTAATAATTACTACAATTTTATTACTTTTATCATTACCTGTTTTAGCAGGAGCAATTACAATATTATTGACTGATCGAAATTTAAATTCAAGATTTTTTGATTTT---------------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, 16/i/2014, forest, Malaise trap. Depository:
Holotype voucher code. BIOUG28804-F01.
Paratypes
Both Malaise-trapped. BIOUG28817-F07, BIOUG10014-B04. Depository:
Other material
Specimens in the same BIN are from Belize (BMNHE897719), Chiapas Mexico (07TAPACH-01734), and Kentucky, USA (BCLDQ01649). The specimen from Chiapas has an image on BOLD that appears to be similar to the Costa Rican specimens and therefore it is likely the same species. The Belizean specimens do not have images but are presumably conspecific with the Costa Rican specimens. The Kentucky specimen does not have an image, and its identity is uncertain.
Aleiodes adrianforsythi , sp. nov.
Diagnostics
BOLD:ADJ1086. Consensus barcode. AATTTTATATTTTTTATTTGGTATATGAGCAGGAATAATTGGTATATCAATAAGATTAATTATTCGTCTAGAATTAAGAACGGGAGGAAGAATTTTAAAAAATGATCAAATCTACAATGGAATAGTAACATTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGGTTTGGTAATTGATTAATTCCTTTAATACTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATGAGATTTTGATTATTAATTCCTTCTCTAATACTTTTATTAACTAGAGGTATTATTAATACAGGTGTAGGAACAGGATGAACAATATATCCCCCCTTATCATCATTAATTGGTCATAATGGTATTTCAGTAGATATATCTATTTTTTCATTACATCTTGCTGGAACGTCTTCAATTATAGGAGCAATTAATTTTATTTCTACTATTTTTAATATAAATCTTTTAAGAATTAAGATAGACCAAATTATATTATTAATTTGATCTATCTTAATTACCACAATTTTATTATTATTATCTCTTCCAGTTCTTGCAGGAGCAATTACAATATTATTAACAGATCGAAATTTAAATACTAGATTTTTTGATTTTTCTGGAGGAGGGGACCCAATTTTATTCCAACATCTTTTT.
Holotype ♂
Guanacaste, Sector Pitilla, Bullas, 10.98670, -85.38503, 440 meters, caterpillar collection date: 06/v/2017, wasp eclosion date: 21/v/2017. Depository:
Host data. Eupithecia Janzen774 (Geometridae) feeding on Entada gigas (Fabaceae)
Host caterpillar and holotype wasp voucher codes. 17-SRNP-71403, DHJPAR0061499.
Paratypes
None.
Aleiodes agnespeelleae , sp. nov.
Diagnostics
BOLD:AAH8705. Consensus barcode. GTTTTATATTTTATATTTGGGATATGGGCTGGTATAATTGGTTTATCGATAAGATTAATTATTCGATTGGAGTTAAGAGTTAGAGGAAGAATTTTAAAAAATGATCAAATTTATAATGGTATGGTTACATTACATGCTTTTGTAATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGAAATTGGTTGGTACCATTAATATTAAGGGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTGTTAATTCCATCTTTTTTTTTATTATTAATTAGAGGTATTATTAATGCAGGAGTTGGAACTGGATGAACAATATACCCTCCCTTATCTTTATTAATTGGTCATAATGGAATTTCAGTTGATATATCTATTTTTTCTTTACATTTAGCTGGTGCTTCTTCAATTATAGGGTCAATTAATTTTATTACTACAATTTTTAATATAAAATTAAAAGAAATTAAGTTAGATCAAATTTCTTTATTAGTTTGATCTATTTTAATTACTACAATTTTATTATTACTTTCATTACCTGTTTTAGCAGGTGCTATTACTATATTATTAACT------------------------------------------------------------------------.
Holotype ♂
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 02/i/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29423-E01.
Paratypes
None.
Other material
Specimens from, Bolivia, Belize, and French Guiana are in the same BIN. There are no images of the specimens from Bolivia and Belize, though the specimens from Belize are likely conspecific with the Costa Rican specimens. The specimens from French Guiana are darker and may not be conspecific with the Costa Rican specimen.
Aleiodes alaneaglei , sp. nov.
Diagnostics
BOLD:AAM5650. Consensus barcode. ATTCTTTACTTTTTATTTGGTATATGAGCAGGTATAATTGGTCTATCAATAAGATTAATTATTCGATTAGAATTAAGGACAAGAGGAAGAATCTTAAAAAATGATCAAATTTACAACGGAATAGTAACTTTACACGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGACTCATTCCTTTAATATTAGGTGCTCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATACTTTTATTAATTAGAGGAATAATTAATACAGGAGTAGGTACAGGATGAACAATATACCCTCCATTATCTTCATTAATTGGTCATAATGGATTTTCAGTAGATATATCTATTTTTTCTTTACATCTTGCTGGTGCTTCATCAATTATAGGAGCCATTAATTTTATTTCAACTATTTTTAATATAAATCTATTAAAAATTAAAATAGATCAAATTATATTATTAATTTGATCTATTTTAATTACTACAATTTTATTATTATTATCTTTACCAGTTCTTGCTGGTGCTATTACTATATTATTAACAGAT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, dry forest, 17/xii/2012, forest, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG17566-G02.
Paratypes
None.
Other material
Two specimens from Belize are in the same BIN (BMNHE897725, BMNHE897745) but were not examined.
Aleiodes alanflemingi , sp. nov.
Diagnostics
BOLD:AAM5670. Consensus barcode. ATTCTATATTTTTTATTTGGGRTATGAGCAGGTATGATTGGTTTATCTATAAGGTTAATTATTCGTATAGAATTAAGAATAAGAGGGAGTATTTTAAAAAATGATCAGATTTATAATGGGATAGTGACATTACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGGTTTGGTAATTGATTAATTCCTTTRATATTAGGAGCCCCTGATATGGCTTTYCCTCGAATAAATAATATAAGGTTTTGATYGTTGATCCCTTCATTATTTTTACTTTTAAATAGTGGAATTATTAATTTAGGAGTTGGTACAGGGTGAACTATATATCCTCCTTTATCTTCTTTAATTGGGCATAATGGTATATCTGTAGATATATCAATTTTTTCTTTACAYTTAGCTGGGGCTTCTTCAATTATAGGAGCTATTAATTTTATTTCTACAATTTTTAATATAAAAATAATAAAAATTAAAATAGATCAAATTAATTTATTTATTTGATCTRTTTTAATTACAGTAATTTTATTATTACTTTCATTGCCTGTTTTAGCGGGAGCTATTACTATATTATTAACTGATCGAAATTTAAATACAAGATTTTTTGATTTTTCTGGAGGAGGAGACCCTATTTTATTTCAACATCTT.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, rain forest/laguna, 12/v/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG27868-C03.
Paratypes
None.
Other material
One specimen from Belize (BMNHE897785) is in the same BIN but was not examined.
Aleiodes alanhalevii , sp. nov.
Diagnostics
BOLD:AAM5683. Consensus barcode. GTGTTATATTTTTTATTTGGTATATGAGCGGGTATRTTAGGTTTATCTATAAGTTTAATTATTCGATTGGAATTGAGAGTTGCTGGAAGATTGTTAAAGAATGATCAAATTTATAATGGTATAGTGACATTACATGCTTTTGTAATAATTTTTTTTATAGTTATACCTATTATGATTGGGGGGTTTGGTAATTGGTTAATTCCTTTGATATTAGGAGCTCCTGATATAGCTTTCCCTCGAATAAATAATATRAGATTTTGATTATTAATTCCTTCTTTTTTGTTATTGTTAATTAGAGGGGTAATTAAYTCAGGAGTAGGGACTGGTTGAACTATGTATCCTCCTCTTTCTTTATTAATTGGTCATAGTGGTTTTTCTGTGGATATATCTATTTTTTCATTACATTTAGCTGGGGCTTCTTCAATTATAGGATCTATTAATTTTATTTCAACTATTTTTAATATAAATTTATTWAAAATTAAATTAGATCARATTTCTTTATTTGTTTGATCAGTATTAATTACTACTGTATTGTTATTATTATCTTTACCTGTATTGGCAGGGGCTATTACTATATTGTTGACTGATCGTAATTTAAATACTAGATTTTTTGATTTTTCTGGGGGGGGTGATCCAATTTTATTTCAGCACTTA.
Holotype ♀
Guanacaste, Pailas Dos, PL12-7, 10.7612, -85.3353, 791 meters, 09/i/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28817-F08.
Paratype
BIOUG29646-B02.
Other material
One specimen from Belize (BMNHE897841) is in the same BIN but was not examined.
Aleiodes alejandromasisi , sp. nov.
Diagnostics
BOLD:ACG6844. Consensus barcode. AATTTTATATTTTTTATTTGGATTATGATCAGGAATAATTGGTATATCAATAAGCTTAATTATTCGATTAGAACTAAGGACAGGCGGAAGAATTTTAAAAAATGATCAAATTTATAATGGAATAGTAACTTTACATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGTTTTGGAAATTGATTAATTCCTATCATACTAGGAGCTCCTGATATAGCTTTCCCTCGAATAAATAACATAAGATTTTGATTATTAATCCCATCTTTAATATTTTTATTAGTAAGAGGAATTATTAATATAGGGGTAGGGACAGGGTGAACTATATATCCCCCATTATCCTCATTAATTGGTCATAATAGAATATCAGTTGATATATCAATTTTCTCTTTACATATAGCAGGAGCATCTTCAATTATAGGATCAATTAACTTCATTTCAACAATCTTCAATATAAATTTAATAAAAATTAAACTAGACCAAATTATATTATTAGTTTGATCAGTATTAATTACAACTATTTTATTATTATTATCATTACCTGTATTAGCAGGAGCTATTACAATATTATTAACTGATCGTAATTTAAATACAAGTTTTTTTGATTTTTCAGGAGGAGGGGATCCAATTTTATTCCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 30/vii/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG05466-G08.
Paratypes
None.
Aleiodes alessandracallejae , sp. nov.
Diagnostics
BOLD:ACJ2417. Consensus barcode. AGTTTTATATTTCCTTTTTGGCATATGAGCAGGAATAATTGGAATATCTATAAGCTTAATTATTCGATTAGAATTAAGAATGAGTGGAAGAATTTTAAAAAATGATCAAATTTATAACGGTATAGTAACTTTACATGCTTTCATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGRTTAGTTCCTCTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATACTTTTATTAATTAGTGGGGTAATTAATACAGGAGTAGGAACTGGTTGAACAATATACCCTCCATTATCATCATTAATTGGTCATAATGGAATTTCTGTAGATATATCTATTTTTTCATTACATCTAGCTGGAGCTTCATCAATTATAGGAGCCATTAACTTTATCTCAACTATTTTTAATATAAATTTAATAACAATTAAAATAGACCAAATCATACTATTAATTTGATCTATTCTAATTACCACAATTTTATTACTCTTATCTTTACCTGTTCTAGCTGGAGCTATTACTATATTATTAACAGATCGAAATTTAAACACAAGATTTTTTGACTTCACAGGAGGAGGGGATCCAATTTTATTCCAACATCTTTTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 29/v/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29863-G04.
Paratypes
BIOUG07616-F05, BIOUG09740-D04, BIOUG18397-D04, BIOUG29728-B07, BIOUG29725-E09, BIOUG29836-H03, BIOUG29862-A04, BIOUG29883-H11, BIOUG29879-F03, BIOUG30963-C06. Depository:
Aleiodes alexsmithi , sp. nov.
Diagnostics
BOLD:AAV7512. Consensus barcode. AATTTTATACTTTTTATTTGGYATTTGAGCTGGAATAATTGGAATATCTATAAGTCTAATTATTCGAATAGAATTAAGAACAAATAAAAGAATTCTAAATAATGAACAAATCTATAATGGAATAGTAACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCCATTATAATTGGAGGATTTGGTAATTGATTAATTCCTTTAATGCTAGGAGCCCCTGATATAGCTTTCCCACGAATAAATAAYATAAGATTTTGRTTATTAATTCCATCTTTATTATTTTTATTAACAAGAGGAATTATTAATTCAGGGGTAGGAACAGGATGAACAATATAYCCACCTTTATCATCATTAATTGGTCATAATGGAATATCAGTAGATATATCAATTTTTTCTTTACATTTAGCAGGAGCTTCTTCAATTATAGGAGCTATTAATTTCATCTCCACTATTTTCAATATAAATTTAAAAATAATTAAAATAGATCAATTACCTCTTCTTATTTGATCTATTTTAATTACAACAATTTTATTACTTTTATCTTTACCAGTYTTAGCAGGAGCTATTACCATATTATTAACAGATCGAAATCTAAATACTAGATTTTTTGATTTTTCAGGAGGAGGAGACCCCATTCTTTTCCAACATCTTTTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Cuesta Canyon Tigre, 10.81703, -85.64366, 270 meters, caterpillar collection date: 20/v/2006, wasp eclosion date: 21/v/2007. Depository:
Host data. Holochroa ochra (Geometridae) feeding on Combretum farinosum (Combretaceae).
Host caterpillar and holotype wasp voucher codes. 06-SRNP-14583, DHJPAR0062173.
Paratypes
None.
Other material
A specimen (BCLDQ01401) collected in Chamela, Jalisco, Mexico is in the same BIN. It was not examined.
Aleiodes alfonsopescadori , sp. nov.
Diagnostics
BOLD:ACR4858. Consensus barcode. AATTTTATATTTTTTATTTGGCATATGAGCAGGAATGGTTGGAGTATCAATAAGATTAATTATTCGTATAGAATTAAGAATAAGAGGAAGTATTTTAAAAAATGATCAAATTTATAATGGYATAGTTACATTACATGCTTTTATTATAATTTTTTTTATAGTTATRCCTATTATAATTGGGGGATTTGGRAATTGATTAGTTCCTTTAATATTGGGGGCTCCTGATATAGCTTTCCCYCGAATAAATAATATAAGATTTTGATTGTTAATYCCTTCATTATTTTTACTTTTAAATAGTGGAATTATTAATTTAGGTGTTGGYACAGGATGAACTATATAYCCTCCTTTATCTTCTTTAATTGGRCATAATGGTATATCTGTAGATATATCAATTTTYTCTTTACATTTAGCCGGAGCTTCTTCAATTATAGGAGCTGTTAATTTTATTTCTACAATTTTTAATATAAAAATAATAAAAATTAAAATAGATCAGATTAATTTATTTATTTGATCTATTTTAATTACAGTAATTTTATTATTACTTTCATTACCTGTTTTAGCGGGGGCTATTACTATATTATTAACTGATCGAAATTTAAATACAAGATTTTTTGATTTTTCTGGAGGGGGGG.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 11/ii/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG18398-F08.
Paratypes
BIOUG17569-F04, BIOUG18290-A11, BIOUG18499-F03. Depository:
Aleiodes alisundermieri , sp. nov.
Diagnostics
BOLD:ADB3278. Consensus barcode. ATTTTATATTTTTTATTTGGTATATGAGCAGGAATAATTGGRATATCTATAAGTCTAATTATTCGTATAGAATTAAGAATAAATGGTAGAATTTTAAAAAATGATCAAATTTATAATGGTATAGTAACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAGTAGGAGGGTTTGGAAATTGACTAATTCCTTTAATATTAGGRAGGCCTGAYATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATACTTTTATTAATTAGAGGAATAATCAATACAGGAGTAGGRACRGGTTGAACAATATACCCCCCATTATCATCATTAATTGGTCACAATGGAATTTCAGTAGATATATCTATTTTCTCACTTCATTTAGCTGGAGCTTCATCAATYATAGGATCTATTAATTTTATTTCTACTATTTTTAATATAAATCTTATAGGAATTAAAATAGATCAAATTATACTATTTATTTGATCTATTTTAATTACTACAATTTTACTATTATTATCKTTACCAGTTCTTGCAGGAGCAATTACTATATTATTAACAGATCGAAATTTAAATACA.
Holotype ♀
Guanacaste Pailas Dos PL12-3, 10.7631, -85.3344, 820 meters, 23/i/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29481-G05.
Paratypes
BIOUG29288-B08, BIOUG29288-A10.
Aleiodes almasolisae , sp. nov.
Diagnostics
BOLD:ADF6889. Consensus barcode. ATCTTATATTTTTTATTTGGAATATGATCAGGTATAATCGGGTTAGCAATAAGGTTAATTATTCGAATAGAATTAGGAATTAGAGGGAGAGTTTTAAAAAATGATCAAATTTATAATGGTGTAGTAACATTGCATGCTTTTATTATAATTTTTTTTATGGTTATACCTATTATAATTGGAGGGTTTGGAAATTGATTAATTCCTTTAATATTAGGGGCCCCAGATATAGCATTCCCTCGTATAAATAATATAAGGTTTTGGTTATTAATTCCTTCATTATTTTTACTTTTAATAAGAGGAATTATTAACTTAGGAGTAGGAACGGGGTGAACAATATATCCTCCTTTATCTTCTTTAATTGGCCATAATGGGATATCTGTTGATATGTCTATTTTTTCTTTACATTTAGCAGGAATTTCTTCAATTATAGGAGCCATTAATTTTATTTCTACAATTTTTAATATAAATTTAATAAAAATTAAAATAGATCAAATTATATTATTAGTATGATCAATTTTAATTACAGTAATTTTATTATTACTTTCTTTGCCAGTTTTAGCGGGAGCAATTACTATACTATTAACTGACCGCAAT-------------------------.
Holotype ♀
Guanacaste, Sector Cacao, Derrumbe, 10.9292, -85.4643, 1220 meters, 04/xii/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG31583-C08.
Paratypes
None.
Aleiodes alvarougaldei , sp. nov.
Diagnostics
BOLD:ABW3270. Consensus barcode. AGTTTTATATTTCCTTTTTGGTATATGAGCAGGAATAATTGGGATATCTATAAGTTTAATTATTCGGTTAGAATTAAGAATGAGTGGAAGAATTTTAAAAAATGATCAAATTTATAACGGTATAGTAACTTTACATGCTTTCATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGCTTTGGRAATTGATTARTTCCTCTAATRTTAGGAGCTCCTGAYATAGCTTTCCCACGAATAAACAATATAAGATTTTGATTATTAATCCCTTCTTTAATACTTTTATTAATTAGYGGAATAATTAATACAGGTGTAGGAACTGGMTGAACAATATACCCYCCATTATCATCATTAATTGGTCATAATGGAATTTCTGTAGATATATCTATTTTTTCATTACAYCTAGCTGGAGCCTCATCAATTATAGGAGCCATTAATTTTATTTCAACTATTTTTAATATAAATTTAATAACAATTAAAATAGACCAAATCATATTATTAATTTGGKCTATTTTAATTACCACAATTTTATTACTCTTATCTTTACCTGTYCTAGCTGGAGCCATTACTATATTACTAACAGATCGAAATTTAAACACAAGATTTTTTGATTTCTCAGGAGGAGGTGAYCCAATTTTATTCCAACATCTTTTT.
Specimen ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 05/ii/2015, Malaise trap. Depository:
Host data. None.
Specimen voucher code. BIOUG44435-B10.
Paratypes
None.
Etymology
Aleiodes alvarougaldei is named in honor of Alvaro Ugalde’s (RIP) long-appreciated contributions to publicity for ACG and GDFCF.
Note
A second specimen in the same BIN on BOLD (BIOUG01036-F12) was collected in Ontario, Canada and identified as A. wyomingensis Shaw & Marsh, 2006, however the holotype of A. wyomingensis has a bicolored stigma and differs morphologically in other ways. Although placed in the same BIN the two specimens are not likely to be conspecific. The Canadian specimen has a similar color but is far more robust in the body and appendages than A. alvarougaldei.
Aleiodes alvaroumanai , sp. nov.
Diagnostics
BOLD:AAM1443. Consensus barcode. AATTTTATATTTTTTATTTGGAATATGAGCTGGTATAATTGGGATATCAATAAGACTTATTATTCGATTAGAATTAAGAACTAACGGAAGAATTTTAAAAAATGATCAAATTTATAATAGAATAGTAACTTTACACGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAACTGATTAATTCCTTTAATATTAGGAGCCCCTGATATAGCTTTCCCACGTATAAATAATATAAGATTTTGATTATTAGTCCCTTCTTTAATACTATTATTAATTAGAGGTATTATCAACACAGGGGTAGGTACTGGATGAACAATATATCCACCATTATCATCCTTAATTGGCCACACCGGTATATCTGTAGATATATCTATTTTCTCACTACATTTAGCAGGAGCTTCATCAATTATAGGAGCAATTAATTTTATTTCCACTATCTTTAATATAAACTTAATAAAAATTAAAATAGATCAAATCATACTTTTAATTTGATCTATTTTAATTACTACTATTTTATTACTATTATCTTTACCTGTATTAGCAGGTGCAATTACTATATTATTAACTGATCGAAACTTAAATACAAGATTTTTTGATTTTTCAGGAGGAGGAGACCCAATTTTATTCCAGCATCTTTTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, 17/iv/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29092-H10.
Paratypes
Host = Syngamia florella (Crambidae): DHJPAR0038017, DHJPAR0038019.
Aleiodes angelsolisi , sp. nov.
Diagnostics
BOLD:AAM1722. Consensus barcode. GGTTTTATATTTTTTATTTGGAATTTGGGCAGGAATAGTAGGTTTATCAATAAGGTTAATTATTCGGTTAGAATTAAGGGTTTGTGGAAGAATTTTGAAAAATGATCAAATTTATAATGGTATAGTTACTTTACATGCTTTTGTGATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGGTTTGGAAATTGGTTAATTCCTTTAATATTAGGGGCCCCAGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTGGTGCCATCTTTTTTTTTATTATTAATAAGAGGGTTAATTAATTCAGGAGTGGGGACGGGTTGAACCATATACCCCCCTTTATCTTTATTAATTGGTCATAGTGGTATTTCAGTTGATATATCTATTTTTTCTTTACATTTAGCGGGGGCTTCTTCAATTATAGGGGCTATTAATTTTATTTCAACTATTTTTAATATAAATTTAATAAAAATTAAATTAGATCAAATTTCTTTATTTATTTGGTCAGTTTTAATTACTACTATTTTATTACTTTTATCTTTACCGGTATTAGCGGGGGCTATTACTATGTTATTGACTGATCGTAATTTAAATACAAGATTTTTTGATTTTTCAGGAGGGGGGGATCCAATTTTGTTTCAGCATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Charia, 10.99339, -85.40271, 530 meters, caterpillar collection date: 20/xi/2009, wasp eclosion date: 10/xii/2009. Depository:
Host data. Nicetas Poole11 (Erebidae) feeding on algae.
Host caterpillar and holotype wasp voucher codes. 09-SRNP-73747, DHJPAR0038456.
Paratypes
None.
Aleiodes annhowdenae , sp. nov.
Diagnostics
BOLD:AAM5664. Consensus barcode. AATTTTATATTTTTTATTTGGAATATGAKCAGGRATAATCGGTATATCGATAAGATTAATTATTCGCATGGAATTAAGAACTAGAGGAAGAATTTTAAAAAATGATCAAATTTATAATAGAGTAGTAACTTTACATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGTAATTGATTAATTCCACTAATATTAGGGGCTCCAGATATAGCTTTCCCRCGAATAAATAATATAAGATTTTGACTTTTAATCCCATCTTTATTATTACTTCTAAACAGGGGTATTATTAACTCAGGGGTGGGAACAGGGTGAACAATATACCCTCCCCTTTCTTCATTAATTGGCCATAATGGAATTTCAGTAGATATATCRATTTTTTCTCTTCATTTAGCGGGGGCTTCTTCAATTATAGGGGCTATTAATTTTATTTCTACTATTTTTAATATAAATTTATTAATAATTAAAATAAATCAATTRACTTTACTTACTTGATCAATTTTAATTACRACAATTTTACTACTTTTATCTTTACCAGTTTTAGCAGGGGCAATTACAATATTACTAACAGATCGAAATTTAAACACWRRWTTTTTTGATTTTTCCGGAGGGGGAGACCCAATTTTATTTCAACACTTA.
Holotype ♀
Alajuela, Sector San Cristobal, Finca San Gabriel, 10.87766, -85.39343, 645 meters, caterpillar collection date: 23/ix/2013, wasp eclosion date: 12/x/2013. Depository:
Host data. Eutomopepla artena (Geometridae) feeding on Pleuranthodendron lindenii (Salicaceae).
Host caterpillar and holotype wasp voucher codes. 13-SRNP-4963, DHJPAR0053665.
Paratypes
None.
Other material
Four specimens in the same BIN (BMNHE897761, BMNHE897815, BMNHE897843, BMNHE897846) were collected in Belize. They were not examined.
Aleiodes bobandersoni , sp. nov.
Diagnostics
BOLD:AAA5372. Consensus barcode. AGTTTTATATTTCTTATTTGGTATATGAGCAGGAATAATTGGTATATCAATAAGRTTAATTATTCGATTAGAATTAAGAACCAGAGGTAGTATCCTAAAAAATGATCAAATTTATAATGGAATAGTAACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAACTGAYTAATTCCATTAATATTAGGAGCCCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTAATTCCTTCCCTAATACTTTTRTTAATTAGAGGAATAATCAATACAGGAGTAGGGACAGGATGAACTATATACCCTCCCTTATCTTCATTAATTGGTCATAATGGAATTTCAGTAGATATATCAATTTTTTCATTACATTTAGCTGGAGCCTCATCAATTATAGGRGCAGTTAATTTTATTTCTACTATTTTTAATATAAATTTAATAATAATTAAAATAGACCAAATTACTTTACTAATTTGATCTATTTTAATTACTACAATCTTATTATTATTATCTTTACCAGTTTTAGCAGGAGCAATTACTATATTATTAACAGATCGAAATTTAAATACAAGATTTTTTGATTTTTCAGGWGGAGGAGATCCAATTTTATTTCAACATCTTTTT.
Holotype ♀
Alajuela, Sector San Cristobal, Rio Blanco Abajo, 10.90037, -85.37254, 500 meters, caterpillar collection date: 14/vii/2000, wasp eclosion date: 28/vii/2000. Depository:
Host data. Malocampa piratica (Notodontidae) feeding on Pourouma bicolor (Urticaceae).
Host caterpillar and holotype wasp voucher codes. 00-SRNP-11970, DHJPAR0062156.
Paratypes
DHJPAR0051160, DHJPAR0029043, DHJPAR0052873, DHJPAR0062172. Depository:
Aleiodes carolinagodoyae , sp. nov.
Diagnostics
BOLD:AAW1567. Consensus barcode. ATTCTTTATTTTTTATTTGGAATATGAGCAGGAATAATTGGAATATCAATAAGTTTAATTATTCGATTAGAATTAAGATTAAG---AGGTAGAATGTTAAAAAATGATCAAATTTATAATGGTATAGTAACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTCGGAAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATTATTGTGCCTTCATTAATACTTTTATTAATAAGAGGAATTATTAATATAGGGGTAGGAACAGGATGAACTATATATCCTCCATTATCATCAACAATTGGACATAGGGGATTTTCTGTAGATATATCAATTTTTTCTTTACATTTAGCAGGAGCATCTTCAATTATAGGAGCTATCAATTTTATTTCAACTATTTTTAATATAAAAATTTTAAATATAAAAATAGATCAAATTATATTATTAATTTGATCTATTTTAATTACTACAATTTTATTATTATTATCATTACCAGTTCTTGCAGGAGCAATCACTATACTATTAACAGATCGAAATTTAAATACTAGATTCTTTGATTTTTCAGGAGGAGGAGACCCAA--------------------.
Holotype ♂
Guanacaste, Sector Pitilla, Pasmompa, 11.01926, -85.40997, 440 meters, caterpillar collection date: 26/xii/2007, wasp eclosion date: 08/i/2008. Depository:
Host data. Pleuroprucha rudimentaria (Geometridae) feeding on Inga oerstediana (Fabaceae).
Host caterpillar and holotype wasp voucher codes. 07-SRNP-34331, DHJPAR0023697.
Paratypes
None.
Aleiodes charlieobrieni , sp. nov.
Diagnostics
BOLD:ACJ4200. Consensus barcode. AATTTTATATTTTTTATTTGGTATATGATCAGGAATAATTGGATTAGGTATAAGATTAATTATTCGAATAGAATTAAGTGTTACAGGTAGAATTTTAAAAAATGATCAAATTTATAATGGTATGGTAACTATACATGCTTTTATTATAATTTTTTTTATGGTTATGCCTATTATAATAGGGGGATTTGGTAATTGATTAGTACCTTTAATATTAGGAGCTCCTGATATAGCATTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCATCTTTAATTTTATTATTAAATAGAAGATTAATTAATACAGGAATAGGGACAGGTTGAACAATATATCCTCCTTTATCTTCACTAATTGGACATATAGGTATATCAGTTGATATATCAATTTTTTCTTTACATTTAGCAGGAGCATCTTCAATTATAGGGGCAATTAATTTTATTTCTACAATTTTTAATATAAATTTAATAATAATTAAAATAGAACAATTAAGTTTATTAGTTTGATCTATTTTAATTACAGCAATTTTATTATTACTTTCTTTGCCTGTTTTAGCTGGGGCAATTACTATACTTTTAACTGATCGAAATTTAAATACAAGATTTTTTGATTTTTCTGGAGGAGGAGACCCAATTTTATTTCAGCATCTTTTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Jacobo, 10.94076, -85.3177, 461 meters, caterpillar collection date: 08/i/2013, wasp eclosion date: 26/i/2013. Depository:
Host data. Acrotomia mucia (Geometridae) feeding on Sabicea villosa (Rubiaceae)
Host caterpillar and holotype wasp voucher codes. 13-SRNP-69051, DHJPAR0051316.
Paratypes
None.
Aleiodes davefurthi , sp. nov.
Diagnostics
BOLD:ACB2701. Consensus barcode. AATTTTATATTTTTTATTTGGAATATGAGCAGGAATAATTGGAATATCAATAAGACTAATTATTCGATTAGAATTAAGAATTAGAGGTAGAATTTTAATAAATGATCAAATTTATAATGGTATAGTAACTTTACATGCCTTTATTATAATTTTCTTTATAGTTATACCAATTATAATTGGAGGTTTTGGAAATTGATTAATTCCTTTAATACTAGGAGCCCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATCCCTTCTTTATTATTCCTATTAATTAGAGGAATTATTAATACTGGAGTAGGAACAGGATGAACAATATATCCTCCTTTATCATCTTTAATTGGACATAATAGAATTTCAGTAGATATRTCAATTTTTGCTTTACATTTAGCAGGAGCTTCATCAATTATAGGAGCAATTAATTTTATTTCAACAATTTTTAATATAAATTTAATAAAAATTAAATTAGATCAAATTATATTATTAGTATGATCAGTATTAATTACTGCTATTCTATTATTACTTTCTTTACCTGTTTTAGCAGGTGCTATTACTATATTATTAACAGATCGTAATTTAAATACAAGATTTTTTGATTTCTCAGGAGGAGGAGACCCAATTTTATTTCAACATTTATTT.
Holotype ♂
Guanacaste, Sector El Hacha, Estación Los Almendros, 11.03226, -85.52776, 290 meters, caterpillar collection date: 02/i/2012, wasp eclosion date: 16/i/2012. Depository:
Host data. Herbita medonaDHJ02 (Geometridae) feeding on Hirtella racemosa (Chrysobalanaceae).
Host caterpillar and holotype wasp voucher codes. 12-SRNP-20033, DHJPAR0049349.
Paratypes
None.
Aleiodes donwhiteheadi , sp. nov.
Diagnostics
BOLD:AAA5378. Consensus barcode. TATTTTATATTTTTTATTTGGAATATGATCAGGGATAATTGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAAGAACTTCTGGCAGAGTCTTAAAAAATGATCAAATTTATAATGGTATAGTTACATTACATGCATTTATTATAATTTTTTTTATAGTTATACCTGTAATATTGGGAGGATTTGGAAATTGGTTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGGTTATTAATTCCTTCTTTAATTTTACTTTTAATAAGAGGAGTAATAAATTCAGGTGTAGGAACAGGTTGAACTATGTATCCACCTTTATCATCATTAATTGGTCATAGAGGATTTTCTGTTGATATATCTATTTTTTCTTTACATTTAGCTGGTGCATCTTCAATTATAGGATCAATTAATTTTATTTCAACAATTTTTAATATAAATTTAATAAAAATTAAGATAGATCAGATTTCATTATTTATTTGATCAATTTTAATTACAACAATTTTATTACTTCTTTCTTTACCAGTTTTAGCTGGAGCAATTACAATACTTTTAACAGATCGGAATTTAAATACAAGATTTTTTGATTTTTCAGGAGGAGGGGACCCAATTTTATTCCAGCATCTTTTT.
Holotype ♀
Guanacaste, Sector Pitilla, Estación Quica, 10.99697, -85.39666, 470 meters, caterpillar collection date: 03/xi/2010, wasp eclosion date: 08/xi/2010. Depository:
Host data. Calodesma maculifrons (Erebidae) feeding on unidentified plant.
Host caterpillar and holotype wasp voucher codes. 10-SRNP-73227, DHJPAR0042076.
Paratype
DHJPAR0029061. Depository:
Aleiodes doylemckeyi , sp. nov.
Diagnostics
BOLD:AAH8920. Consensus barcode. ATTTTGTATTTTATTTTTGGGATGTGATCGGGGATAATTGGGATATCAATAAGGTTAATTATTCGATTAGAGTTAGGAGTTTGTGGGAGGGTTTTGGGGAATGATCAAATTTATAATGGGGTGGTAACTTTACATGCTTTTGTAATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGGTTTGGAAATTGATTAATTCCTTTAATGTTAGGGTCCCCTGATATAGCTTTCCCTCGTATAAATAATATGAGGTTTTGGTTATTAGGACCTTCTTTATTATTTTTATTGATTAGGGGGTTAGTTAATAGAGGGGTAGGTACAGGGTGAACTATATATCCTCCCTTGTCTTTGGTATTAGGACATAGGGGGATATCTGTTGATATTTCTATTTTTTCATTACATTTAGCAGGGGTATCTTCTATTATGGGGGCTGTAAATTTTATTACTACTGTATTGAATATGAGTTTGTTTATAATTAAAATGGATCAAATTATATTATTTGTTTGGTCTGTAGTTATTACTGCTATATTATTATTGTTATCTTTACCAGTTTTAGCGGGGGCTATTACTATATTATTGACTGACCGA------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 16/i/2014, forest, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28676-D10.
Paratypes
None.
Aleiodes frankhovorei , sp. nov.
Diagnostics
BOLD:AAM5640. Consensus barcode. AATTTTATATTTTTTATTTGGAATATGAGCAGGAATAATTGGTATATCAATAAGATTAATTATTCGCCTAGAATTAAGAACAGGAGGGAGAATTTTAAAAAATGATCAAATCTACAATGGAATAGTAACATTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGAGCCCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTATTAATTCCTTCTCTAATACTTTTATTAATTAGAGGAATTATTAATACAGGGGTAGGTACAGGATGAACAATATATCCTCCTTTATCATCATTAATTGGTCATAATGGTATTTCAGTAGATATGTCTATTTTTTCGTTACATCTTGCTGGAGCCTCTTCAATTATAGGGGCAATTAATTTTATTTCCACTATTTTTAATATAAATCTTTTAAGAATTAAAATAGATCAAATTATATTATTAATTTGATCTATTTTAATTACCACAATTTTATTATTATTATCCCTTCCAGTTCTTGCAGGGGCAATTACAATATTATTAACAGATCGAAATTTAAATACTAGATTTTTTGAT------------------------------------------.
Holotype ♀
Guanacaste, Sector Cacao, Estación Cacao, 10.92691, -85.46822, 1150 meters, caterpillar collection date: 13/vi/2009, wasp eclosion date: 26/vi/2009. Depository:
Host data. Sabulodes arge (Geometridae) feeding on Nectandra salicifolia (flowers) (Lauraceae).
Host caterpillar and holotype wasp voucher codes. 09-SRNP-36313, DHJPAR0040077.
Paratype
DHJPAR0040076 Depository:
Aleiodes henryhowdeni , sp. nov.
Diagnostics
BOLD:ABX5209. Consensus barcode. AATTTTATATTTTTTATTTGGAATATGAGCAGGAATAATTGGTATATCAATAAGATTAATTATTCGCCTAGAATTAAGAACAGGAGGGAGAATTTTAAAAAATGATCAAATCTACAATGGAATAGTAACATTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGAGCCCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTATTAATTCCTTCTCTAATACTTTTATTAATTAGAGGAATTATTAATACAGGGGTAGGTACAGGATGAACAATATATCCTCCTTTATCATCATTAATTGGTCATAATGGTATTTCAGTAGATATGTCTATTTTTTCGTTACATCTTGCTGGAGCCTCTTCAATTATAGGGGCAATTAATTTTATTTCCACTATTTTTAATATAAATCTTTTAAGAATTAAAATAGATCAAATTATATTATTAATTTGATCTATTTTAATTACCACAATTTTATTATTATTATCCCTTCCAGTTCTTGCAGGGGCAATTACAATATTATTAACAGATCGAAATTTAAATACTAGATTTTTTGAT------------------------------------------.
Holotype ♀
Guanacaste, Sector Mundo Nuevo, Estación La Perla, 10.76737, -85.43313, 325 meters, caterpillar collection date: 27/v/2011, wasp eclosion date: 09/vi/2011. Depository:
Host data. Anticarsia gemmatalis (Erebidae) feeding on Centrosema sagittatum (Fabaceae).
Host caterpillar and holotype wasp voucher codes. 11-SRNP-55519, DHJPAR0045467.
Paratypes
DHJPAR0052871, Benisodes Poole02. DHJPAR0063146, BIOUG30149-A08. Depository:
Other material
A specimen from Belize (NHM749233) and another from Cuba (BCLDQ0693) are in the same BIN. They were not examined.
Aleiodes johnchemsaki , sp. nov.
Diagnostics
BOLD:ABX5740. Consensus barcode. AGTACTTTACTTTATATTTGGAATATGATCAGGAATAATTGGTTTATCAATAAGATTAATTATTCGATTAGAATTAAGATCAACATTAAGAATTTTAAAAAATGATCAAATTTATAATGGTATAGTAACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGGTTTGGAAATTGATTAATTCCCCTAATATTAGGAGCACCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTCTAATTCCATCTTTAATACTTTTATTAACTAGAGGAATTATTAATACAGGAGTGGGGACAGGGTGAACAATATATCCTCCATTATCATCATTAATTGGTCATAATGGAATATCAGTAGATATATCTATTTTTTCATTACATTTAGCAGGGGCCTCATCAATCATAGGAGCTATTAATTTTATTTCCACTATCTTTAATATAAATTTATTAATAATTAAATTAGATCAAATTACATTATTAATTTGATCTATTTTAATTACTACAATTTTACTATTACTATCCCTACCAGTACTTGCAGGAGCTATTACTATATTATTAACAGACCGAAATTTAAATACTAGATTTTTTGATTTT---------------------------------------.
Holotype ♀
Guanacaste, Sector Pitilla, Ingas, 11.00311, -85.42041, 580 meters, caterpillar collection date: 20/vi/2011, wasp eclosion date: 04/vii/2011. Depository:
Host data. Nemoria anae (Geometridae) feeding on Sabicea panamensis (Rubiaceae).
Host caterpillar and holotype wasp voucher codes. 11-SRNP-31717, DHJPAR0045345.
Paratypes
None.
Aleiodes johnkingsolveri , sp. nov.
Diagnostics
BOLD:AAT8850. Consensus barcode. TGTACTTTATTTCATATTTGGAATATGATCAGGAATAATTGGAATATCAATAAGTTTAATTATTCGATTAGAATTAAGATCAACTATAAGAATTTTAAAAAATGATCAAATTTATAATGGTATAGTAACAATACATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATGCTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTCTAATCCCCTCTTTAATACTTTTATTAACTAGAGGTATTATTAATTCAGGAGTAGGAACAGGATGAACAATATACCCCCCATTATCCTCCTTAATTGGTCATAATGGTATATCAGTAGATATATCTATTTTTTCATTACATTTAGCAGGTGCTTCATCAATTATAGGGTCTATCAATTTTATTTCAACTATTTTTAATATAAATTTATTAATAATTAAAATAGATCAAATTACATTACTAATTTGATCTATTTTAATTACCACAATTTTATTATTACTATCTTTACCAGTTCTCGCAGGGGCCATTACTATATTATTAACAGATCGAAATTTAAATACTAGTTTCTTTGACTTTTCCGGAGGAGGAGACCCAATTTTATTTCAACATCTTTTT.
Holotype ♀
Guanacaste, Sector Cacao, Sendero a Maritza, 1 km NW Estación Cacao, 10.92691, -85.46822, 1150 meters, caterpillar collection date: 17/v/2010, wasp eclosion date: 10/vi/2010. Depository:
Host data. Chloropteryx BioLep266 (Geometridae) feeding on Gouania lupuloides (Rhamnaceae).
Host caterpillar and holotype wasp voucher codes. 10-SRNP-35211, DHJPAR0040450.
Paratypes
None.
Aleiodes gonodontovorus
Diagnostics
BOLD:ACK7827. Consensus barcode. AATTTTATATTTTTTATTTGGTATATGATCTGGAATGRTTGGRTTATCTATAAGTTTAATTATTCGRTTAGAATTAAGAACTAYTGGAAGAATCTTAAAAAATGATCAAATTTATAATGGTATAGTTACTCTTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTGTTTTAGGAGGATTYGGTAATTGATTAATTCCTTTAATATTAGGMGCCCCAGATATAGCTTTCCCTCGTATAAATAATATAAGGTTTTGRTTATTAATTCCATCATTATTATTCTTATTRATAAGAGGWRTTGTTAATACAGGYGTGGGRACAGGRTGRACAATTTAYCCVCCTTTRTCTTCATTAATTGGTCATAGAGGAATTTCTATAGATATATCAATTTTTTCTTTACATTTRGCAGGRGCTTCTTCAATTATAGGAGCTATTAATTTTATTTCAACAATTTTAAATATAAATTTAAATCAAATTAAATTRGATCAAATATCTTTATTTATTTGATCTGTATTAATTACAGCATTTTTATTACTTTTAGCTTTACCAGTTTTGGCAGGRGCTATTACYATACTTTTAACTGATCGAAATTTAAATACTAGATTTTTTGATTTTYYKGRRGGGGGGGATCCAATTTTATTYCAGCATCTTTTT.
Imaged specimen ♀
Guanacaste, Sector Mundo Nuevo, Vado Miramonte, 10.77175, -85.43400, 305 meters, caterpillar collection date: 12/vii/2012, wasp eclosion date: 23/vii/2012. Depository:
Host data. Gonodonta incurva (Erebidae) feeding on Piper umbellatum (Piperaceae).
Host caterpillar and imaged wasp voucher codes. 12-SRNP-55790, DHJPAR0049948.
Paratypes
Hosts = Gonodonta incurva, Gonodonta fulvangula, Gonodonta correcta, Gonodonta immacula, Gonodonta uxor, Gonodonta bidens, Gonodonta pyrgo, Gonodonta nitidimacula. DHJPAR0016434, DHJPAR0021153, DHJPAR0016919, DHJPAR0016925, DHJPAR0009351, DHJPAR0029041, DHJPAR0028026, DHJPAR0028027, DHJPAR0042799, DHJPAR0045387, DHJPAR0048706, DHJPAR0048707, DHJPAR0048708, DHJPAR0049663, DHJPAR0049664, DHJPAR0052874, DHJPAR0021154, DHJPAR0021156, DHJPAR0029068, DHJPAR0021131, DHJPAR0029065, DHJPAR0028025, DHJPAR0028034, DHJPAR0028035, DHJPAR0028029, DHJPAR0028028, DHJPAR0030593, DHJPAR0030599, DHJPAR0049662. DHJPAR0021155, DHJPAR0009352, DHJPAR0021181, DHJPAR0029042, DHJPAR0029063, DHJPAR0029062, DHJPAR0045056, DHJPAR0048702, DHJPAR0048709, DHJPAR0048710, DHJPAR0051310, DHJPAR0029049. Depository:
Aleiodes manuelzumbadoi , sp. nov.
Diagnostics
BOLD:ACL0293. Consensus barcode. AGTTTTATATTTCCTATTTGGAATATGAGCAGGAATAATTGGGATATCTATAAGATTAATTATTCGATTAGAGTTAAGAGTAAGTGGAAGAATTTTAAAAAATGATCAAATTTATAATGGTATAGTAACTTTACATGCTTTCATTATAATTTTTTTCATAGTTATACCAATTATAATTGGGGGGTTTGGAAATTGATTAATTCCTTTAATATTAGGAGCCCCTGATATAGCTTTCCCCCGAATAAATAATATAAGATTCTGATTATTAATTCCCTCTTTAATACTTTTATTAATTAGAGGAGTAATTAATACAGGGGTAGGTACTGGCTGGACAATATACCCCCCATTATCTTCACTAATTGGTCATAATGGAATTTCTGTAGATATATCTATTTTTTCATTACATCTAGCTGGAGCCTCATCAATTATAGGAGCAATTAACTTTATTTCAACTATTTTTAATATAAACCTAATAACAATTAAAATAGACCAAATTATACTACTAATTTGGTCTATTTTAATTACCACAATTTTACTACTTTTATCCCTGCCCGTTCTAGCTGGAGCTATTACTATATTATTAACAGATCGAAATTTAAATACAAGATTTTTTGATTTTTCAGGAGGAGGAGACCCAATTTTATTCCAACATCTTTTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 07/i/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG09827-B10.
Paratypes
BIOUG09442-A12, BIOUG09739-H01, BIOUG17569-D12, BIOUG17575-B03, BIOUG17575-B06, BIOUG17616-A02, BIOUG28821-H03. Depository:
Aleiodes mayrabonillae , sp. nov.
Diagnostics
BOLD:AAD7873. Consensus barcode. TGTATTRTATTTTTTATTTGGTATATGAGCAGGTATAATTGGKTTATCAATAAGGTTAATTGTTCGGTTAGAGYTAGGTGTTTGTGGGAGAGTATTAGGCAATGATCAAATTTATAATGGTATAGTTACTTTGCATGCTTTTATTATAATTTTTTTTATGGTTATGCCTATTATAATTGGTGGGTTTGGGAATTGATTAATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGTTTTTGGCTATTATTACCTTCAATTTTATTACTATTATTAAGGGGTATTATTAATGTAGGGGTAGGTACTGGTTGAACAGTTTAYCCTCCTTTATCTTCATTAATTGGRCATAGTGGTATATCTGTTGATTTATCTATTTTTTCTTTACATTTAGCTGGTGTTTCTTCTATTATAGGGGCTATTAATTTTATTACAACTATTTTTAATATAAATTTATTTATAATTAAAATAGATCAAATTATATTATTTGTTTGATCTGTATTAATTACCGCTTTTTTACTTTTATTATCTTTACCTGTTTTAGCTGGTGCTATTACTATATTATTAACTGATCGTAATTTAAATACTACATTTTTTGATTTTTCGGGGGGGGGAGACCCTATTTTATTTCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Cacao, Sendero Circular, 10.92714, -85.46683, 1185 meters, caterpillar collection date: 12/ix/2012, wasp eclosion date: 18/x/2012. Depository:
Host data. Acrotomia mucia (Geometridae) feeding on Coussarea austin-smithii (Rubiaceae).
Host caterpillar and holotype wasp voucher codes. 12-SRNP-35331, DHJPAR0051376.
Paratypes
Host: same caterpillar species as holotype and same host as holotype but on Psychotria elata (Rubiaceae): DHJPAR0055814, DHJPAR0057810, DHJPAR0051676, DHJPAR0055814, DHJPAR0062993. Depository:
Aleiodes michelledsouzae , sp. nov.
Diagnostics
BOLD:ABA7287. Consensus barcode. GAGCAGGTATAATTGGGATATCAATAAGATTAATTATTCGAATAGAATTAAGGACAAGAGGGAGAATCTTAAAAAATGACCAAATTTATAATGGTATAGTAACTTTACATGCTTTCATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGGTTCGGAAATTGATTAATTCCCTTAATATTAGGGGCTCCTGATATAGCATTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCATCTTTATTATTTTTATTAATAAGAGGTGTTATTAATACAGGTGTTGGAACAGGATGAACCATATACCCCCCTTTATCATCCTTAATTGGGCATAATAGAATCTCAGTTGATATATCAATTTTTTCTTTACATTTAGCAGGAGCTTCTTCTATTATAGGGGCAATTAATTTTATTTCAACAATTTTCAATATAAATTTAATAAAAATTAAATTAGACCAAATTTCATTATTAGTCTGATCAATTTTAATTACTACTATTTTATTATTGTTATCTTTACCTGTACTAGCAGGGGCAATCACTATATTATTAACTGACCGTAACTTAAATACAAGATTTTTTGATTTTTCGGGGGGGGGGGACCCAATTTTA---------------.
Holotype ♀
Guanacaste, Pitilla, Sendero Orosilito, 10.98332, -85.43623, 900 meters, caterpillar collection date: 25/v/2011, wasp eclosion date: 28/v/2011. Depository:
Host data. Yidalpta auragalisDHJ01 (Erebidae) feeding on Securidaca sylvestris (Polygalaceae).
Host caterpillar and holotype wasp voucher codes. 11-SRNP-31492, DHJPAR0042782.
Paratypes
None.
Aleiodes mikeiviei , sp. nov.
Diagnostics
BOLD:ACS9594. Consensus barcode. AGTTTTATATTTTATGTTTGGGATATGAGCTGGTATGATTGGGCTATCAATAAGATTAATTATTCGATTAGAATTAAGAATTAG---AGGGAGAATTTTAAAGAATGATCAAATTTATAATGGTATAGTTACATTACATGCTTTTGTAATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGGTTTGGAAATTGATTGGTACCATTAATATTAGGGGCCCCTGATATAGCTTTTCCACGAATGAATAATATAAGGTTTTGATTATTAATTCCATCTTTTTTTTTATTATTAATTAGGGGGGTTATTAATGCAGGAGTTGGGACTGGATGGACAATGTATCCTCCTTTATCTTTATTAATTGGTCATAATGGTATTTCAGTTGATATATCTATTTTTTCTTTACATTTAGCTGGGGCTTCTTCAATTATAGGGGCTATTAATTTTATCACTACAATTTTTAATATAAAGTTAAAAGGGATTAAGTTAGATCAAATTTCTTTATTAATTTGATCTATTTTAATTACTACAATTTTGTTATTACTTTCATTACCTGTTTTAGCTGGTGCTATTACTATATTATTGACTGATCGTAATTTAAATACTAGATTTTTTGATTTTTCAGGGGGAGGAGACCCAG--------------------.
Holotype ♀
Guanacaste, Sector Pitilla, Bullas, 10.98670, -85.38503, 440 meters, caterpillar collection date: 31/vii/2014, wasp eclosion date: 24/viii/2014. Depository:
Host data. arctJanzen01 Janzen71434 (Erebidae) feeding on algae.
Host caterpillar and holotype wasp voucher codes. 14-SRNP-71445, DHJPAR0056334.
Paratypes
None.
Aleiodes normwoodleyi , sp. nov.
Diagnostics
BOLD:AAJ4076. Consensus barcode. AGTTTTATACTTTTTATTTGGAATATGAGCAGGAATAATTGGCTTATCAATAAGCCTAATTATTCGATTAGAATTAAGAATAAGAGGAAGTATTCTTAAAAATGATCAAATTTATAATGGTATAGTAACTTTACATGCTTTTGTTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGGTTTGGAAATTGATTAATTCCATTAATGTTAGGAGCCCCTGATATAGCTTTCCCCCGCATAAATAATATAAGATTTTGATTATTAATTCCCTCTTTAATACTTCTATTAATTAGAGGATTAATTAATACAGGAGTAGGGACAGGATGAACTATATACCCCCCATTATCATCATTAATTGGCCATAATGGAATTTGTGTAGATATATCAATTTTTTCATTACATTTAGCTGGAGCCTCATCAATTATAGGAGCAATTAATTTTATTTCAACTATTTTTAATATAAACTTAATAACTATTAAAATAGATCAAATTACATTATTAATTTGCTATTTTAATTACTACAATTTTATTATTATTATCTTTACCAGTTTAGCAGGGGCAATTACTATACTATTAACAGATCGAAATTTAAATACAAGATTTTTG---------------------------------------------. The similar Aleiodes leptocarina Fortier, 2000 was reared in Costa Rica and is also a gregarious species. Its host is unknown and described simply as a “large caterpillar.” The two species differ in numerous ways: one of the most obvious is that the antenna of A. leptocarina are “honey yellow” differing from the melanic antenna of A. normwoodleyi.
Holotype ♀
Alajuela, Sector San Cristobal, Finca San Gabriel, 10.87766, -85.39343, 645 meters, caterpillar collection date: 3/xii/2008, wasp eclosion date: 19/xii/2008. Depository:
Host data. Dysschema viuda (Erebidae) feeding on Zanthoxylum riedelianum (Rutaceae). Gregarious parasitoid with 80 specimens emerging.
Host caterpillar and holotype wasp voucher codes. 08-SRNP-6195, DHJPAR0030719.
Paratypes
None.
Etymology
Aleiodes normwoodleyi is named in honor of Norm Woodley’s long-appreciated contributions to publicity for ACG, GDFCF, and now, BioAlfa.
Aleiodes inga
Diagnostics
BOLD:AAA5377. Consensus barcode. ATTTTATATTTTTTATTTGGGATATGATCTGGAATAATCGGTTTATCAATAAGATTAATTATTCGTTTAGAATTAAGATCAACTGGGAGAGTTTTAAAGAATGATCAGATTTATAATGGAATAGTTACATTACATGCATTTATTATAATTTTTTTTATAGTTATACCTGTAATACTAGGAGGATTTGGAAATTGATTAATTCCATTAATATTAGGAGCCCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTAATTCCATCTTTATTTTTACTTTTAATAAGAGGTGTAATAAATTCAGGTGTAGGAACTGGATGAACTATATACCCTCCTTTATCTTCATTAATTGGTCATAGAGGATTTTCTATTGATATATCTATTTTTTCTTTACATTTGGCTGGAATTTCTTCAATTATGGGGTCAATTAATTTTATTTCTACTATTTTTAATATAAATTTAATGAAAATTAAGATAGATCAAATCTCTTTATTTATTTGATCAGTTTTAATTACTACTATTTTGTTACTTCTTTCTTTACCAGTTTTAGCTGGGGCAATTACTATATTATTAACTGATCGAAATTTAAATACAAGATTTTTTGATTTTTCTGGAGGAGGAGATCCAATTTTATTTCAACATCTTTTT.
Imaged specimen ♀
Alajuela, Sector Brasilia, Piedrona, 11.016, -85.359, 340 meters, caterpillar collection date: 21/ix/2010, wasp eclosion date: 30/ix/2010. Depository:
Host data. Letis mycerina (Erebidae) feeding on Inga oerstediana (Fabaceae).
Host caterpillar and imaged wasp voucher codes. 10-SRNP-65094, DHJPAR0041207.
Aleiodes pammitchellae , sp. nov.
Diagnostics
BOLD:AAD7872. Consensus barcode. GGTATTRTATTTTTTRTTTGGTATGTGATCAGGGATGGTTGGGATRTCAATAAGGTTAATTATTCGGTTAGAATTGGGAGTTTGTGGGAGAGTTTTAGGTAATGATCAAATTTATAATGGTATGGTTACTTTACATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGATCYCCGGATATAGCTTTCCCTCGTATGAATAATATGAGATTTTGAYTATTAATTCCTTCAATTTTATTATTRTTAATGAGAGGTGTTGTTAATGTTGGTGTAGGTACTGGGTGAACTATTTAYCCACCTTTATCTTCATTATTAGGTCATGGGGGGATATCTGTAGATTTGTCAATTTTTTCTATTCATTTAGCTGGGGCTTCTTCGATTATAGGAGCTATTAATTTTATTACTACAATTTTTAATATAAATTTATTTATAATTAAAATAGATCAAATTATATTATTTGTTTGATCTGTATTAATTACAGCTTTTTTATTATTATTATCTTTACCAGTTTTAGCTGGGGYAATTACAATATTATTAACAGATCGTAATATAAATACTACTTTTTTTGATTTTYCTGGAGGGGGYGATCCTATTTTATTTCAACACTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Vado Poza Salada, 10.79796, -85.64984, 8 meters, caterpillar collection date: 25/v/2006, wasp eclosion date: 24/v/2007. Depository:
Host data. Toxonprucha excavata (Erebidae) feeding on Vachellia riparia (Fabaceae).
Host caterpillar and holotype wasp voucher codes. 06-SRNP-15047, DHJPAR0062177.
Paratypes
DHJPAR0062411, DHJPAR0062410 light trapped. Depository:
Aleiodes pauljohnsoni , sp. nov.
Diagnostics
BOLD:ACM2562. Consensus barcode. AATATTATATTTTTTATTTGGAATATGAGCAGGAATAGTGGGTTCATCAATAAGATTAATTATTCGATTAGAATTAAGAACAAATGGGAGAATTTTAAAAAATGATCAAATTTATAATGGAATAGTAACATTACACGCTTTCATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTCGGAAATTGATTAATTCCATTAATATTAGGGGCTCCTGACATAGCATTCCCACGAATAAATAATATAAGATTTTGATTATTAATCCCTTCTTTAATACTTTTATTAATTAGAGGGATAATTAATACAGGTGTTGGAACTGGATGAACAATATACCCCCCATTATCATCATTAATCGGACACAATGGTCTTTCAGTAGATATATCTATTTTTTCTTTACATTTAGCAGGGGCATCATCTATTATAGGAGCAATTAATTTTATTTCAACTATTTTTAATATAAACTTAATAAAAATTAAAATAGATCAAATTATATTATTAATTTGATCTATTTTAATTACTACAATTTTATTATTATTATCATTACCAGTTTTAGCAGGAGCTATTACCATATTATTAACAGACCGAAATTTAAATACAAGATTTTTTGATTTCTCAGGAGGGGGAGACCCAATTTTATTCCAACACCTTTTT.
Holotype ♀
Guanacaste, Sector Cacao, Sendero Arenale, 10.92471, -85.46738, 1080 meters, caterpillar collection date: 31/x/2013, wasp eclosion date: 20/xi/2013. Depository:
Host data. Erosia veninotata (Uraniidae) feeding on Randia grandifolia (Rubiaceae)
Host caterpillar and holotype wasp voucher codes. 13-SRNP-36425, DHJPAR0054919.
Paratype
DHJPAR0054923. Depository:
Aleiodes rosewarnerae , sp. nov.
Diagnostics
BOLD:AAW1573. Consensus barcode. ATTTTATATTTCTTATTTGGAATATGAGCAGGTATAATTGGAATATCAATAAGATTAATTATTCGGTTAGAATTAAGATCTACTGGTAGAATTTTAAAAAATGATCAAATTTATAACAGTATAGTAACTTTACATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGGTTTGGTAATTGATTAATCCCTTTAATACTAGGAGCCCCAGATATAGCCTTCCCACGAATAAATAATATAAGATTCTGATTATTAATTCCTTCATTATTATTATTATTAATAAGAGGAATTATTAATTCAGGTGTAGGCACAGGATGAACAATATACCCTCCATTATCATCACTAATTGGTCATAGAGGTATCTCCGTAGATTTATCTATTTTTTCTTTACATTTAGCAGGAGCTTCATCTATTATAGGAGCTATTAATTTCATTTCAACTATTTTCAATATAAATTTAATAATAATTAAAATAGACCAAATTATATTATTAGTATGAGCAATTTTAATTACTACAATTTTATTATTATTATCTTTACCAGTTTTAGCAGGAGCAATTACRATATTATTAACAGACCGAAATTTAAATACAAGATTTTTTGACTTTTCTGGAGGTGGGGACCCCATTTTATTTCAACATCTCTTT.
Holotype ♂
Guanacaste, Sector Pitilla, Pasmompa, 11.01926, -85.40997, 440 meters, caterpillar collection date: 26/xii/2007, wasp eclosion date: 11/i/2008. Depository:
Host data. Pleuroprucha rudimentaria (Geometridae) feeding on Inga oerstediana (Fabaceae).
Host caterpillar and holotype wasp voucher codes. 07-SRNP-34419, DHJPAR0023522.
Paratype
Host = Pleuroprucha rudimentaria: DHJPAR0023524. Depository:
Aleiodes steveashei , sp. nov.
Diagnostics
BOLD:AAJ4092. Consensus barcode. AGTTTTATATTTCTTATTTGGTATATGAGCAGGAATAATTGGTATATCAATAAGACTAATTATTCGATTAGAATTAAGAACCAGAGGTAGAATCCTTAAAAATGATCAAATTTATAATGGAATAGTAACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAACTGATTAATCCCATTAATATTAGGAGCTCCTGATATAGCCTTCCCACGAATAAATAATATAAGGTTTTGATTATTAATTCCTTCCTTAATACTTTTATTAATTAGAGGGATAATCAATACAGGAGTAGGGACAGGATGAACTATATACCCTCCCTTATCTTCATTAATTGGTCATAATGGAATTTCAGTAGATATATCAATTTTTTCACTACATTTAGCTGGGGCCTCATCAATTATAGGAGCAGTTAATTTTATTTCTACTATTTTTAATATAAATTTAATAATAATTAAAATAGATCAAATTACCTTATTAATTTGATCTATTTTAATTACTACAATCTTATTATTATTATCTTTACCAGTTTTAGCAGGTGCAATTACTATRTTATTAACAGATCGAAATTTAAATACAAGATTTTTTGATTTTTCAGGGGGGGGAGACCCAATTTTATTTCAACATCTTTTT.
Holotype ♀
Guanacaste, Sector Cacao, Estación Cacao, 10.92691, -85.46822, 1150 meters, caterpillar collection date: 29/vii/2010, wasp eclosion date: 20/vii/2010. Depository:
Host data. Tachuda discreta (Notodontidae) feeding on Serjania schiedeana (Sapindaceae).
Host caterpillar and holotype wasp voucher codes. 10-SRNP-35614, DHJPAR0040461.
Paratypes
Hosts = Tachuda discrete, Lysana Janzen02, and Elymiotis plechelm (all Notodontidae). DHJPAR0047201. DHJPAR0021180, DHJPAR0062073, DHJPAR0062081, DHJPAR0062112. Depository:
Aleiodes terryerwini , sp. nov.
Diagnostics
BOLD:ADJ0647. Consensus barcode. TATTTTATATTTTTTATTTGGTATATGAGCAGGTATAATTGGAATATCAATAAGATTAATTATTCGATTAGAATTAAGAATTAATGGAAGAATTTTAAAAAATGATCAAATTTATAATGGAATAGTAACTTTACATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGGTTTGGAAATTGATTAATTCCATTAATATTAGGAAGACCTGATATAGCTTTCCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATACTTTTATTAATTAGAGGAATTATTAATACGGGAGTAGGAACAGGATGAACTATATACCCTCCTTTATCTTCACTTATTGGACATAATGGAATTTCCGTAGATATATCAATTTTTTCTTTACACTTAGCAGGGGCTTCATCTATTATAGGAGCAATTAATTTTATTTCAACTATTTTTAATATAAATTTAATAATAATTAAAATAGACCAAATTACATTATTAATTTGATCTATTTTAATTACTGCAATTTTATTACTATTATCTTTACCAGTCTTAGCAGGAGCTATTACTATATTATTAACAGATCGAAATCTAAACACAAGTTTTTTTGATTTTTCAGGAGGAGGAGACCCAATTTTATTCCAACATCTTTTT.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Manguera, 10.99590, -85.39842, 470 meters, caterpillar collection date: 10/v/2017, wasp eclosion date: 21/v/2017. Depository:
Host data. Paectes Poole10 (Euteliidae) feeding on Mosquitoxylum jamaicense (Anacardiaceae).
Host caterpillar and holotype wasp voucher codes. 17-SRNP-71490, DHJPAR0061494.
Paratypes
None.
Aleiodes willsflowersi , sp. nov.
Diagnostics
BOLD:AAM1704. Consensus barcode. TGTTTTATATTTTTTATTCGGATTATGAGCAGGAATAATTGGAATATCAATAAGATTAATTATTCGACTAGAGTTAAGAACTAGAGGGAGAATATTAAAAAATGATCAAATTTATAATGGAATAGTAACCTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGGTTTGGGAATTGATTAATTCCTTTAATATTAGGTGCCCCTGATATAGCCTTCCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATACTTTTATTAATTAGAGGAATAATTAATTCAGGGGTAGGGACAGGATGAACTATATATCCCCCCCTATCATCATTAATTGGTCATAACGGGGCTTCAGTAGATATATCAATTTTTTCTTTACATTTAGCGGGGGCATCCTCAATTATAGGAGCAATTAATTTTATTTCAACTATTTTTAATATAAATTTAATAAATATTAAAATAGATCAAATTATATTACTAATTTGATCTATTTTAATTACTACAATTCTATTATTACTATCATTACCAGTATTAGCGGGGGCAATCACAATACTATTAACAGACCGAAATTTAAATACAAGATTTTTTGATTTTTCAGGAGGGGGGGATCCAATTTTATTCCAACATCTTTTT.
Holotype ♀
Guanacaste, Sector Del Oro, Quebrada Oro, 11.03376, -85.47715, 290 meters, caterpillar collection date: 27/viii/2009, wasp eclosion date: 10/ix/2009. Depository:
Host data. Antaeotricha Janzen730 (Depressariidae) feeding on Cupania cinerea (Sapindaceae).
Host caterpillar and holotype wasp voucher codes. 09-SRNP-22602, DHJPAR0037966.
Paratypes
None.
Choreborogas Whitfield, 1990
The genus is restricted to the New World with nine described species, none of which is recorded from Costa Rica. Members are parasitoids of leaf-mining Lyonetiidae and Gracillariidae (
Choreborogas andydeansi , sp. nov.
Diagnostics
BOLD:ADV9625. Consensus barcode. TTATATTTTTTTTTTGGTATATGATCAGGTATGTTGGGGTTATCAATAAGATTAATTATTCGTTTTGAATTGGGGGTTCCTGGTTCATTTTTAGGTAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCTTTAGTTATAATTTTTTTTATAGTTATACCTGTTATAATTGGTGGGTTTGGAAATTGATTAATTCCCTTAATATTAGGAGCTCCTGATATGGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTATTTTAATATTATTAATTAGATCTTTGGTTAATGTTGGGGTAGGTACAGGTTGAACAATTTATCCTCCATTATCTTCTTTGATAGGTCATGGGGGTATTTCAGTTGATTTGGCTATTTTTTCTTTACATTTAGCAGGGGCATCTTCAATTATAGGGGCTATTAATTTTATTTCAACTATTTTCAATATAAATTTATTTTCAATGAAAATAGATCAAATTATATTATTAGTTTGATCTGTTTTAATTACTGCTTTCTTACTGTTGTTATCTTTACCTGTTTTGGCTGGGGCAATTACTATATTATTGTTTGATCGTAATATTAATAGAACATTTTTTGATTTTTCAGGGGGGGGGGATCCTATTTTGTTTCAACATTTATT-------------------------------------------.
Holotype ♀
Guanacaste, Sector Pailas Dos, PL12-3, 10.7631, -85.3344, 8200 meters, 18/xii/2014, Malaise trap. Depository:
Host data. None
Holotype voucher code. BIOUG44129-A07.
Paratypes
None.
Choreborogas eladiocastroi , sp. nov.
Diagnostics
BOLD:ACS0876. Consensus barcode. TTATATTTTTTTTTTGGTATATGATCAGGTATGTTGGGGTTATCAATAAGATTAATTATTCGTTTTGAATTGGGGGTTCCTGGTTCATTTTTAGGTAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCTTTAGTTATAATTTTTTTTATAGTTATACCTGTTATAATTGGTGGGTTTGGAAATTGATTAATTCCCTTAATATTAGGAGCTCCTGATATGGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTATTTTAATATTATTAATTAGATCTTTGGTTAATGTTGGGGTAGGTACAGGTTGAACAATTTATCCTCCATTATCTTCTTTGATAGGTCATGGGGGTATTTCAGTTGATTTGGCTATTTTTTCTTTACATTTAGCAGGGGCATCTTCAATTATAGGGGCTATTAATTTTATTTCAACTATTTTCAATATAAATTTATTTTCAATGAAAATAGATCAAATTATATTATTAGTTTGATCTGTTTTAATTACTGCTTTCTTACTGTTGTTATCTTTACCTGTTTTGGCTGGGGCAATTACTATATTATTGTTTGATCGTAATATTAATAGAACATTTTTTGATTTTTCAGGGGGGGGGGATCCTATTTTGTTTCAACATTTATT-------------------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 17/vii/2014, forest, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29393-G11.
Paratypes
BIOUG18822-B04, BIOUG29473-F10, BIOUG29531-D01, BIOUG30966-F05.
Choreborogas felipechavarriai , sp. nov.
Diagnostics
BOLD:ADA2251. Consensus barcode. ATATGATCTGGTATGTTAGGTCTTTCTATAAGATTGATTATTCGTTTTGAATTAAGATCTTGTGGTTCATTTTTAGGAAATGACCAAATTTATAATAGTATTGTTACAGCTCATGCCTTAGTTATAATTTTTTTTATGGTTATACCTGTTATAATTGGTGGTTTTGGTAATTGATTAATTCCTTTAATGTTAGGGGCCCCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCTATTTTTTTGTTGATAATTAGTTCTATTGTTAATGTTGGAGTAGGTACTGGTTGAACTATTTATCCTCCATTGTCTTCTTTGATGGGTCATAGTGGTATTTCTATGGATTTAGCTATTTTTTCTTTACATTTGGCTGGGGCTTCTTCAATTATAGGGGCTATTAATTTTATTACTACTATTTTTAATATAAATTTATTTTATATGAAAATAGATCAAATTGTTTTGTTAGTTTGGTCTGTTTTAATTACTGCTTTTTTATTATTGTTATCTTTACCTGTTTTAGCTGGAGCTATTACTATATTATTATTTGATCGT------------------------------------------------------------.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, rain forest/laguna, 11/xi/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28042-C10.
Paratypes
None.
Choreborogas frankjoycei , sp. nov.
Diagnostics
BOLD:ADB6582. Consensus barcode. ATATGATCTGGTATGTTAGGTTTATCTATAAGAATAATTATTCGTTTTGAATTAGGGGTTCCTGGTTCATTATTAGGTAATGATCAAATTTATAATAGGATTGTTACAGCTCATGCTTTAGTAATAATTTTTTTTATAGTTATGCCTGTAATGATTGGTGGGTTTGGTAATTGATTAATTCCTTTAATATTGGGGGCTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTATTTTTTTATTATTGATTAGTTCTATTGTTAATGTTGGGGTTGGTACTGGGTGGACTATTTATCCACCACTTTCTTCTTTATTAGGTCATGGGGGTATATCTGTTGATTTAGCTATTTTTTCCTTACATTTAGCGGGGGTTTCTTCAATTATGGGAGCTATTAATTTTATTTCAACTATTTTTAATATAAATTTATTTCATATGAAAATAGATCAAATTATATTATTAATCTGATCTGTTTTAATTACTGCTTTTTTATTATTATTGTCATTACCAGTTTTAGCAGGGGCTATTACTATATTATTATTTGATCGT----------------------------------------------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 13/ii/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29660-C05.
Paratypes
None.
Clinocentrus Haliday, 1833
The genus is worldwide in distribution with six described New World species, none of the holotypes of which are known from Costa Rica or nearby countries. Members are parasitoids of concealed caterpillars (
Clinocentrus andywarreni , sp. nov.
Diagnostics
BOLD:ACB2341. Consensus barcode. AGTTTTATATTTTATTTTTGGTATTTGATCTGGTTTAGTAGGTCTTTCTATGAGGTTAATTATTCGTTTAGAGTTGGGTATTCCTGGTAGATTATTAGGTAATGATCAACTTTATAATGTTATAGTTACTGCTCATGCTTTTGTTATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGATTTGGAAATTGGTTAATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCATCATTATTTATATTAGTAATGAGAGGTTTATTAAATGTAGGTGTAGGAACAGGATGAACAGTTTACCCTCCTTTATCTTCATTAATTGGTCATAGAGGTATTTCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCTGGAGCTTCATCAATTATGGGGGCAATTAATTTTATTTCTACTATTTTTAATATAAATTTATTAATAATTAAATTAGATCAAATAAGATTATTTATTTGATCAGTTTTAATTACAGCAGTTTTATTATTATTATCTTTACCAGTTTTAGCTGGCGCAATTACTATATTATTAACTGATCGTAATTTAAATACTACTTTTTTTGATTTTTCTGGTGGAGGGGATCCTATTTTATTTCAGCATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Corredor, 10.87868, -85.38963, 620 meters, caterpillar collection date: 23/iii/2012, wasp eclosion date: 08/iv/2012. Depository:
Host data. Antaeotricha Janzen146 (Depressariidae) feeding on Lonchocarpus oliganthus (Fabaceae).
Host caterpillar and holotype wasp voucher codes. 12-SRNP-1189, DHJPAR0049287.
Paratypes
None.
Clinocentrus angelsolisi , sp. nov.
Diagnostics
BOLD:ACL3899. Consensus barcode. AGTTTTATATTTTTTATTTGGAATTTGATCAGGATTACTAGGTTTATCAATAAGTATAATTATTCGATTAGAATTAGGTATATCAGGTAGATTATTAGGCAATGATCAAATTTATAATGTTATGGTTACAGCTCATGCTTTTGTTATAATTTTTTTTATGGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGTGCACCTGACATAGCTTTTCCCCGTATAAATAATATAAGTTTTTGGTTATTAATTCCTTCTTTAATATTATTATTATTAAGATCTATATTAAATGTTGGTGTAGGTACAGGATGGACTGTTTATCCTCCTTTATCTTCATTAATTGGTCATGGAGGGATATCTGTAGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCATCTATTATAGGTGCTATTAATTTTATTTCAACAATTTTTAATATAAGATTATTAACTTTAAAATTAGATCAAATAAGTTTATTTGTATGGTCAGTATTAATTACTGCTTTTTTATTATTATTATCTTTACCTGTATTAGCAGGTGCTATTACTATATTATTAACTGATCGTAATTTAAATACTACATTTTTTGATTTTTCTGGTGGAGGTGACCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, 28/i/2013, Malaise trap. Depository:
Holotype voucher code. BIOUG18290-G11.
Paratypes
BIOUG10016-F03, BIOUG10016-F07, BIOUG10017-C03, BIOUG10017-H11, BIOUG17569-C11, BIOUG13946-A11, BIOUG18290-B06, BIOUG18290-D09, BIOUG18398-B01, BIOUG18398-C09, BIOUG18398-C11, BIOUG18398-G06. Depository:
Cystomastax Szépligeti, 1904
The genus is restricted to the neotropics with six described species, none of which is known from Costa Rica or Central America. Members are parasitoids of concealed caterpillars (
Cystomastax alexhausmanni , sp. nov.
Diagnostics
BOLD:AAJ5011. Consensus barcode. AATTTTATATTTTATTTTTGGTATTTGAGCTGGTATATTAGGTTTGTCTATAAGAATTATTATTCGTATAGAATTAATTAATCCTATGGGT---TTAATTAAAAATGATAATATTTATAATAGTATAGTAACGTCTCATGCATTTGTAATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGATCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGGTTATTAATTCCTTCTTTATTTTTATTATTAATAAGGTCTTTTATTAATGTTGGTGTTGGTACAGGATGAACTATATATCCTCCTTTATCTTCTTTTTTAGGTCATAGTGGTATATCAGTAGATGTGGCTATTTTTTCTTTGCATTTAGCTGGTGCTTCCTCAATTATAGGTTCTATTAATTTTATTTCTACAATTTTTAATATAAAATTGTTAAATTTAAAATTAGATCAAATAAGATTGTTTATTTGGTCTGTTTTAATTACAGTGATTTTATTGTTGTTATCATTACCTGTTTTAGCTGGTGCTATTACAATGTTATTAACGGATCGTAATTTAAATACTACTTTTTTTGATTTTTCAGGGGGAGGGGATCCTATTTTATTTCAGCATTTATTT.
Holotype ♂
Guanacaste, Sector Pitilla, Calma, 11.00986, -85.39213, 412 meters, caterpillar collection date: 03/19/2014, wasp eclosion date: 04/08/2014. Depository:
Host data. Myrmecopsis strigose (Erebidae) feeding on Vailia anomala (Asclepiadaceae).
Host caterpillar and holotype wasp voucher codes. 14-SRNP-70673, DHJPAR0055780.
Paratypes
None.
Other material
A specimen (AL0391) in the same BIN was collected in Belize. It was not examined.
Cystomastax angelagonzalezae , sp. nov.
Diagnostics
BOLD:AAA5370. Consensus barcode. TGTTTTATATTTTATTTTTGGTATTTGATCTGGRATATTAGGYTTATCTATAAGAATTATTATTCGAATAGAATTAATTAATCCTGGGGGTTTTATTAAAAATGATTATATTTATAATAGAATAGTTACTTCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCTGTAATAATTGGTGGATTTGGTAATTGATTAATTCCTTTAATGTTGGGGGCTCCTGATATGGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTATTTATATTGTTAATAAGATCTTTTGTTAATGTTGGAGTTGGGACGGGKTGAACTATRTAYCCTCCTTTATCTTCTTTTTTAGGTCATGGAGGAATATCTGTAGATATATCTATTTTTTCTTTACATTTAGCAGGGTCTTCTTCTATTATAGGTGCTATTAATTTTATTTCTACAATTTTTAATATAAAATTATTTTCTTTAAAATTAGACCAATTAAATTTATTTATTTGGTCTGTTTTAATTACGGTAATTTTATTATTATTATCTTTACCAGTATTAGCTGGTGCTATTACTATGTTGTTAACTGATCGTAATTTAAATACTACTTTTTTTGATTTTYCTGGTGGGGGGGATCCTGTTTTATTTCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Pitilla, Medrano, 11.01602, -85.38053, 380 meters, collection date: 06/30/2012, wasp eclosion date: 07/16/2012. Depository:
Host data. Hypena zarabenaDHJ01 (Erebidae) feeding on Urera lianoides (Urticaceae).
Host caterpillar and holotype wasp voucher codes. 12-SRNP-71556, DHJPAR0049684.
Paratype
Host = Hypena Poole11: DHJPAR0029040. Depository:
Cystomastax ayaigarashiae , sp. nov.
Diagnostics
BOLD:AAW1603. Consensus barcode. AATTTTATATTTTATTTTTGGTATTTGATCTGGAATATTAGGTTTATCTATAAGTATTATTATTCGAATAGAATTAATTAATCCTGGTGGATTAATTAAAAATGATAATATTTATAATAATATAGTTACTTCACATGCTTTTGTAATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAATTCCTTTAATATTAGGATCTCCAGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTATTTTTATTATTAACTAGATCATTTATTAATGTTGGGGTTGGTACAGGTTGAACTATATATCCTCCTTTATCTTCTTTTTTAGGACATAGAGGAATATCTGTAGATATGGCTATTTTTTCTTTACATTTAGCTGGATCTTCTTCAATTATAGGTTCTATTAATTTTATTTCTACTATTTTTAATATAAAATTATTTTCTTTAAAATTAGATCAAATAAGTTTATTTATTTGATCTGTTTTGATTACAGTAATTTTATTATTATTATCTTTACCTGTATTAGCGGGAGCTATTACTATATTATTAACTGATCGTAATTTAAATACTACTTTTTTTGATTTTTCAGGGGGGGGAGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Laguna, 10.9888, -85.42336, 680 meters, caterpillar collection date: 06/19/2009, wasp eclosion date: 07/16/2009. Depository:
Host data. Symphlebia ipsea (Erebidae) feeding on Clethra lanata (Clethraceae).
Host caterpillar and holotype wasp voucher codes. 09-SRNP-32021, DHJPAR0035929.
Paratypes
None.
Heterogamus Wesmael, 1838
Heterogamus is nearly cosmopolitan, absent only from the Nearctic. There are 20 described species, only two of which are Neotropical, and neither is previously reported from Costa Rica. Hosts are unknown.
Heterogamus donstonei , sp. nov.
Diagnostics
BOLD:AAH8697. Consensus barcode. AATTTTATATTTTATYTTTGGGATATGGGCTGGTATAATTGGTATRTCTATAAGATTAATTATTCGAATAGAGTTGGGKGTGTGTGGAAGRYTATTAATAAATGATCAAATTTATAATGGTATAGTRACTTTACAYGCTTTTATTATRATTTTTTTTATAGTTATACCTATTATRATTGGAGGRTTTGGTAATTGAYTAATTCCTTTAATATTAGGRTCTCCTGATATAGCTTTYCCWCGAATAAATAATATAAGTTTTTGATTATTAATTCCTTCTTTAATTTTATTAATTRYTAGGGGGGTGATAAATGTRGGTGTTGGYACGGTTGAACAGTTTAYCCYCCYTTATCTTCRTTAGTAGGGCATATAGGGRTRTCTGTAGATTTATCAATTTTTTCYTTACAYTTAGCTGGGGCTTCTTCAATTATAGGGGCTATTAATTTTATTACAACTATTTTTAATATGAATTTATTTAAAATTAAAATAGATCAGATGTCTTTATTTATTTGATCTGTTTTAATTACAGCATTTTTATTATTRTTATCTTTRCCTGTATTAGCAGGGGCTATTACTATATTATTAACGGATCGAAATTTAAAYACTACATTTTTTGATTTTTCAGGGGGGARGGGGATCCTATTTTTATTYCAACATTTATTTTGATTTTTTG.
Holotype ♀
Guanacaste, Pailas Dos, PL12-9, 10.76, -85.3341, 809 meters, 17/vii/2014, forest, Malaise trap. Depository:
Host data . None.
Holotype voucher code. BIOUG29390-H04.
Paratypes
None.
Other material
In BOLD there are specimens in the same BIN from Colombia (BCLDQ0397, BCLDQ0877) and Belize (BF002031, BMNHE897721). These were not examined.
Pseudoyelicones van Achterberg, Penteado-Dias & Quicke, 1997
Pseudoyelicones is restricted to the Neotropics with four of the five previously described species recorded from Costa Rica. Here we present the first host record for the genus, a pyralid caterpillar (Pyralidae).
Pseudoyelicones bernsweeneyi , sp. nov.
Diagnostics
BOLD:AAL5801. Consensus barcode. AGTTTTATATTTTATTTTTGGTATGTGATCAGGAATTTTAGGAATATCATTAAGATTAATTATTCGAATAGAATTAGGTACTCCTGGGAAAATATTAGGAGATGATCAGTTATATAATAGTTTTGTTACTTTACATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGTTTCGGAAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTGTTAATTTCTTCTTTATTTATATTATTAATTAGAGGTATTATTAATGTAGGAACAGGAACTGGATGAACTATATATCCACCTTTATCTTCATTAATAGGTCATAGTGGAATATCTGTTGATTTATCAATTTTTTCTTTACATTTAGCTGGTGCTTCCTCTATTATAGGAGCTGTTAATTTTATTTCAACAATTTTTAATATAAAATTAACTAATATTGATATAGATAAAATTAGTTTGTTAATTTGATCAGTATTAATTACTGCTTTATTATTATTATTATCCTTACCTGTTTTGGCAGGTGCTATTACAATATTATTAACTGATCGTAACTTAAATACTACTTTTTTTGATTGTTCAGGTGGGGGCGATCCTGTTTTATTKCAACATTTRTTT. Similar to P. limonensis to which it keys in Buntika and Quicke (2004). It differs in many respects, but the easiest to observe is that the scape is yellow in P. bernsweeneyi and brown in P. limonensis.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Finca Esmeralda, 10.93548, -85.25314, 123 meters, caterpillar collection date: 09/14/2011, wasp eclosion date: 10/18/2011. Depository:
Host data. Chloropaschia mennusalis (Pyralidae) feeding on Vismia baccifera (Hypericaceae). This is the first host record for the genus. The image of the mummified pupa of the host, Chloropaschia mennusalis (Fig.
Host caterpillar and holotype wasp voucher codes. 11-SRNP-75949, DHJPAR0045816.
Paratypes
DHJPAR0035971, DHJPAR0045814, DHJPAR0063815. Depository:
Stiropius Cameron, 1911
Stiropius is restricted to the New World with most species occurring in the Neotropics. No species were previously recorded from Costa Rica. The genus has previously been recorded on larval Lyonetiidae and Gracillariidae, and here we present the first record from Crambidae. There are no revisions of the Neotropical species.
Stiropius bencrairi , sp. nov.
Diagnostics
BOLD:ACB2702. Consensus barcode. TGTATTGTATTTTTTATTTGGTATATGATCAGGAGTTTTAGGTTTATCGATAAGAATAATTATTCGATTTGAATTAAGAATTCCGGGATCTTTTTTAGGTAATGATCAAATTTATAATAGAATTGTTACATCACATGCTTTAGTAATAATTTTTTTTATAGTTATACCTGTAATAATTGGAGGGTTTGGAAATTGGTTAATTCCATTAATATTAGGAGCTCCTGATATAGCTTTTCCTCGAATAAATAATATGAGATTTTGATTATTGTTTCCTGCTATTATATTATTATTATTAAGTTCTTTTGTTAGGGTTGGAAGAGGAACAGGATGAACAATATATCCTCCATTATCTTCTTTAGTTGGTCATGGGGGAGTTTCAGTGGATTTGTCAATTTTTTCTTTACATTTAGCAGGAGTATCTTCTATTATAGGTGCTATTAATTTTATTACAACAGTTTTTAATATAAATTTATTTTATATTAAAATAGATCAGGTTATATTATTAGTTTGATCAGTATTAATTACTGCTTTTTTATTATTATTATCTTTACCTGTATTAGCAGGTGCTATTACAATATTGTTGTTTGATCGAAATATTAATACAACATTTTTTGATTTTTCTGGGGGAGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♂
Alajuela, Sector Rincon Rain Forest, San Lucas, 10.91847, -85.30338, 320 meters, caterpillar collection date: 05/01/2012, wasp eclosion date: 05/19/2012. Depository:
Host data. graciJanzen01 Janzen6864 (Gracillariidae) feeding on Protium costaricense (Burseraceae).
Host caterpillar and holotype wasp voucher codes. 12-SRNP-41777, DHJPAR0049332.
Paratypes
None.
Stiropius berndkerni , sp. nov.
Diagnostics
BOLD:ABV9879. Consensus barcode. TTTTTATATTTTTTTTTTGGTATATGATCAGGTATTTTGGGTATATCTATAAGTATAATTATTCGTTTTGAATTAAGTATTTCAGGTTCTTTTTTGGGTAATGAACAAATTTATAATAGAATTGTTACTTCTCATGCTTTAATTATAATTTTTTTTATAGTTATACCTGTAATGATTGGAGGATTTGGTAATTGGTTGATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGGTTTTGATTATTAATTCCTTCTATAATAATATTATTAATAACTTTTTTTGTTAATGTGGGGGTAGGCACTGGGTGAACTATTTATCCTCCTTTGTCTTCTTTATTAGGTCATAGAGGTATTTCTATAGATTTTGCTATTTTTTCTTTGCATTTAGCAGGAGTTTCTTCAATTATAGGTGCTATTAATTTTATTACAACAATTTTTAATATAAATTTATTTCATATAAAAATAGATCAGATTGTTTTATTTATTTGATCTGTTTTGATTACTGCTTTTTTATTATTATTATCTTTACCTGTATTAGCTGGGGCTATTACAATATTGTTATTTGATCGTAATATTAATACTACTTTTTTTGATTTTC.
Holotype ♀
Guanacaste, Sector Cacao, Sendero Cima, 10.93328, -85.45729, 1460 meters, caterpillar collection date: 06/i/2009, Malaise trap. Depository:
Host data. None.
Host caterpillar and holotype wasp voucher codes. DHJPAR0045983.
Paratypes
None.
Stiropius edgargutierrezi , sp. nov.
Diagnostics
BOLD:ACJ1403. Consensus barcode. AATTTTATATTTTATTTTTGGTTTATGGTCTGGTATGTTAGGATTATCAATAAGTTTAATTATTCGGTTTGAATTGAGGTGTCCTGGGTCATTTTTGGGGAATGATCAAATTTATAATAGAATTGTTACTGCACATGCTTTAATTATAATTTTTTTTATAGTTATACCTGTTATAATTGGGGGATTTGGAAATTGATTAATTCCTTTAATGTTAGGGGCACCTGATATAGCTTTTCCACGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTATTGTTGTTAATATTTAGATCTTTAGTTAATGTTGGGGTGGGTACTGGGTGAACAATTTATCCTCCATTATCTTCTTTATTAGGTCATGGAGGGATTTCGATAGATTTAGCTATTTTTTCTTTACATTTAGCGGGGACTTCTTCAATTATAGGGGCAATTAATTTTATTACTACTATTTTTAATATAAAATTATTTTATATAAAGTTAGATCAGTTAACTTTATTTATTTGATCAGTTTTAATTACAGCTTTTTTATTACTTTTATCTTTGCCAGTTTTAGCTGGGGCTATTACTATATTATTATTTGATCGTAATATTAATACTACATTTTTTGA.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, dry forest, 7/v/2012, forest, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG07918-G09.
Paratypes
BIOUG07457-F12, BIOUG10016-H10.
Stiropius edwilsoni , sp. nov.
Diagnostics
BOLD:ACG5440. Consensus barcode. TATTTTATATTTTTTTTTTGGTATTTGATCTGGTGTTTTAGGCATATCTATAAGGTTAATTATTCGTTTTGAGTTAGGGGTTCCTGGATCTTTTTTAGGTAATGATCAGATTTATAATAGAATTGTTACTGCTCATGCTTTGGTGATAATTTTTTTTATAGTTATACCAATTATAATTGGGGGGTTTGGAAATTGATTAATTCCTTTAATATTGGGGTGTCCTGATATAGCTTTCCCTCGTATAAATAATATGAGGTTTTGATTGTTATTRCCTTCAATTTTTTTGTTATTAATTAGTTCAATTGTTAATATTGGTGTTGGAACTGGGTGAACTATTTATCCTCCTTTATCTTCTTTAATAGGTCATAGAGGTGTTTCTTTAGATTTAGCAATTTTTTCTTTACACTTAGCTGGGGCTTCTTCAATTATAGGTGCTATTAATTTTATTACTACTATTTTTAATATGAATTTATTTTCAATAAAAATAGATCAAATTATATTAATAGTTTGATCTGTGTTAATTACTGCATTTTTATTATTATTATCTTTACCTGTTTTAGCGGGGGCAATTACTATATTATTATTTGATCGTAATATTAATACAACATTTTTTGATTTTTCTGGAGGAGGGGACCCAATTTTATTTCAACATTTGTTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, dry forest, 24/xii/2012, forest, Malaise trap. Depository:
Host data . None.
Holotype voucher code. BIOUG09739-D10.
Paratypes
BIOUG05883-A10, BIOUG09739-F06, BIOUG18290-E10.
Stiropius ehakernae , sp. nov.
Diagnostics
BOLD:ACL3545. Consensus barcode. TTTATATTTTTTTTTTGGAATATGATCTGGTATTTTGGGCTTATCTATAAGATTGATTATTCGTTTTGAAYTAGGTGTTCCAGGTTCTTTTTTGGGTAATGATCAAATTTATAATAGAATTGTTACTGCTCATGCTTTAATTATAATTTTTTTTATGGTTATACCTGTTATAATTGGGGGGTTTGGTAATTGGTTAATTCCTTTAATATTAGGGGCTCCAGATATAGCTTTTCCTCGGATAAATAATATAAGATTTTGATTATTGATTCCTTCWATTTTTTTATTATTGATTAGGTCTTTGATTAATATTGGGGTTGGTACTGGATGAACTATTTATCCTCCCTTATCATCATTAATAGGTCATAGGGGAATATCAATAGATATAGCTATTTTTTCTTTGCATTTGGCTGGTGCTTCTTCTATTATAGGTGCAATTAATTTTATTACTACTATTTTTAATATAAATTTATTTTATATAAAAATAGATCAAATTACATTATTAATTTGATCTATTTTAATTACTGCTTTTTTATTATTATTATCTTTACCTGTTTTAGCTGGGGCTATTACAATATTATTATTTGATCGTAATGTTAATACTACTTTTTTTGATTTTTCTGGGGGGGGGGA.
Holotype ♀
Guanacaste, Pailas Dos, PL12-4, 10.7629, -85.3341, 825 meters, 9/x/2014, forest, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29249-F01.
Paratype
BIOUG09826-B01. Depository:
Triraphis Ruthe, 1855
Triraphis occurs in all realms except the Australian and Antarctic. In the New World, most species are Neotropical, where several thousand remain to be described. Previous host records are Lepidoptera, including Limacodidae, Zygaenidae, Megalopygidae, Lycaenidae, and Riodinidae (van
Triraphis billfreelandi , sp. nov.
Diagnostics
BOLD:AAA5375. Consensus barcode. TGTTTTATATTTTTTATTTGGTATTTGAGCTGGTATAGTTGGTTTATCCATAAGTTTAATTATTCGTTTAGAATTAAGTATACCCGGTAGATTATTGGGTAATGACCAAATTTATAATGGTATAGTAACTGCCCATGCTTTTATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGTGGTTTTGGAAATTGATTGATTCCCTTAATATTGGGGGCTCCTGATATAGCTTTCCCTCGTATAAATAATATGAGGTTTTGATTATTAATTCCCTCATTGACATTATTAATTTTAAGAGCTGTAGTTAATGTTGGGGTAGGCACTGGGTGAACTTTGTACCCTCCTTTATCTTCTTTAGTTGGTCATAGGGGTATATCTGTAGATATGGCTATTTTTTCTTTACATTTAGCTGGTGCTTCTTCTATTATAGGGGTTGTTAATTTTATTTCTACTATTTTTAATATAAAGTTAGTATCAATTAATTTAGATCAAATTAATTTATTTGTTTGATCAGTATTAATTACAGCTGTCTTATTATTATTATCTTTACCAGTGTTAGCTGGTGCAATTACTATATTATTGACAGATCGTAATTTAAATACAACATTTTTTGATTTTTCTGGTGGGGGTGACCCTATTTTATTCCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Mismo, 10.98758, -85.41967, 680 meters, caterpillar collection date: 18/x/2012, wasp eclosion date: 05/xi/2012. Depository:
Host data. Parasa sandrae (Limacodidae) feeding on Calatola costaricensis (Icacinaceae).
Host caterpillar and holotype wasp voucher codes. 12-SRNP-31534, DHJPAR0050910.
Paratypes
DHJPAR0029047, DHJPAR0040078. Depository:
Triraphis billmclarneyi , sp. nov.
Diagnostics
BOLD:AAA7065. Consensus barcode. TGTTTTATATTTTTTATTTGGAATTTGAGCRGGTATAGTTGGATTATCTATAAGATTAATTATTCGTTTAGAATTAAGTATACCGGGGAGATTATTAGGGAATGAYCAAATTTATAAYGGTATAGTGACTGCYCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGYGGTTTTGGAAATTGATTAATTCCTTTAATATTRGGGGCYCCTGATATGGCTTTCCCTCGTATAAATAATATGAGTTTTTGGTTATTAATTCCTTCATTGACTTTATTAATTTTAAGGGCTGTAGTTAATGTTGGAGTRGGTACTGGATGAACTTTATACCCACCTTTATCTTCTTTAGTTGGTCATGGGGGTATATCTGTAGATATAGCTATTTTTTCTTTACATTTAGCTGGAGCATCTTCTATTATAGGGGTTGTTAATTTTATTTCTACTATTTTTAATATAAAATTAGTATCAATTAATTTAGATCAAATTAATTTATTTGTTTGATCAGTATTAATTACAGCTATTTTATTATTATTGTCTTTACCTGTRTTAGCTGGCGCAATTACTATATTATTAACAGATCGTAATTTAAATACAACATTTTTTGATTTTTCTGGGGGGGGKGATCCTATTTTATTTCAACATTTATTT.
Holotype ♂
Guanacaste, Sector Pitilla, Estación Pitilla, 10.98931, -85.42581, 675 meters, caterpillar collection date: 20/xi/2008, wasp eclosion date: 01/xii/2008. Depository:
Host data. zygjanzen01 Janzen23 (Zygaenidae) feeding on Doliocarpus multiflorus (Dilleniaceae).
Host caterpillar and holotype wasp voucher codes. 08-SRNP-32864, DHJPAR0030596.
Paratypes
DHJPAR0030594 DHJPAR0030595 DHJPAR0030597 DHJPAR0030598 DHJPAR0036310 DHJPAR0036311 DHJPAR0030592 DHJPAR0037873. Depository:
Other material
A specimen (BIOUG24424-E06) in the same BIN (BIOUG24424-E06) was collected in Misiones, Argentina and is deposited in Museo Argentino de Ciencias Naturales. It was not examined, but the image in BOLD is consistent with the Costa Rican specimens.
Triraphis billripplei , sp. nov.
Diagnostics
BOLD:AAB1658. Consensus barcode. AGTATTATATTTTTTATTTGGAATTTGAGCAGGTATGGTTGGATTATCTATAAGTTTAATTATTCGTTTAGAATTAAGAATACCTGGAAGTTTATTAGGTAATGATCAAATTTATAATGGTATAGTTACTGCTCACGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGGTTTGGTAATTGATTAATTCCATTAATATTGGGTGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGGTTATTAATTCCTTCATTAACATTATTAATTTTAAGGGCTGTTGTTAATGYTGGAGTAGGGACTGGGTGAACATTATATCCTCCCTTATCTTCTTTAGTTGGTCATGGGGGGATATCTGTAGATATGGCTATTTTTTCTTTACATTTAGCTGGTGCTTCTTCAATTATAGGTGTTGTTAATTTTCTTTCTACTATTTTTAATATAAAATTAGTATCTATTAATTTAGATCAAATTAATTTATTTGTCTGATCAGTATTAATTACTGCTGTTTTATTATTATTATCTTTACCAGTGTTAGCTGGTGCTATTACTATATTATTGACAGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGAGGGGACCCCATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Horizontes, Vado Esperanza, 10.78938, -85.55098, 85 meters, caterpillar collection date: 15/vi/2009, wasp eclosion date: 16/vi/2009. Depository:
Host data. Norape Janzend03 (Megalopygidae) feeding on Mimosa pigra (Fabaceae).
Host caterpillar and holotype wasp voucher codes. 09-SRNP-13855, DHJPAR0036305.
Paratypes
DHJPAR0021201, DHJPAR0028329, DHJPAR0028330. Depository:
Triraphis bobandersoni , sp. nov.
Diagnostics
BOLD:AAB8975. Consensus barcode. TGTGTTATATTTTTTATTTGGGATTTGATCAGGTATAGTTGGTTTATCTATAAGTTTAATTATTCGTTTAGAATTAAGAATACCCGGGAGTTTATTGGGTAATGATCAAATTTATAATGGTATAGTTACTGCTCATGCCTTTATTATAATTTTTTTTATGGTTATACCTATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTCCCACGTATAAATAACATAAGATTTTGATTATTAATTCCTTCATTAACATTATTAATTTTAAGGGCTGTAGTTAATGTTGGGGTAGGTACTGGATGAACTTTATATCCTCCTTTATCTTCTTTAGTTGGGCACGGYGGGATATCCGTTGATATAGCTATTTTTTCTTTACATTTGGCTGGGGCTTCTTCAATTATAGGTGTTGTTAATTTTATTTCTACTATTTTTAATATAAAATTAGTGTCAATTAATTTAGATCAAATTAATTTATTTGTTTGATCAGTATTAATTACTGCTGTTTTATTATTATTATCTTTACCAGTATTGGCAGGGGCTATTACTATATTGTTAACAGATCGTAATTTAAATACAACATTTTTTGATTTTTCTGGTGGTGGTGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Memos, 10.98171 -85.42785, 740 meters, caterpillar collection date: 18/iii/2013, wasp eclosion date: 03/iv/2013. Depository:
Host data. Epiperola vaferella (Limacodidae) feeding on Cyathea multiflora (Cyatheaceae).
Host caterpillar and holotype wasp voucher codes. 13-SRNP-30462, DHJPAR0051666.
Paratypes
DHJPAR0017286, DHJPAR0023518, DHJPAR0023519, DHJPAR0023527, DHJPAR0023532, DHJPAR0023533, DHJPAR0023689, DHJPAR0023690, DHJPAR0023694, DHJPAR0023698, DHJPAR0023702, DHJPAR0023704, DHJPAR0023716, DHJPAR0023722, DHJPAR0023701, DHJPAR0030619, DHJPAR0038384, DHJPAR0038385, DHJPAR0038386, DHJPAR0037964, DHJPAR0037969, DHJPAR0037970, DHJPAR0037972, DHJPAR0037973, DHJPAR0051315. Depository:
Triraphis bobrobbinsi , sp. nov.
Diagnostics
BOLD:AAD2033. Consensus barcode. TGTTTTATATTTTTTATTTGGAATTTGAGCARGRATATTRGGTTTATCAATGAGATTAATTATTCGATTAGAGTTAAGTATACCAGGAAGTTTATTAGGTAATGATCAAATTTATAATGGTATAGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGGGGATTTGGTAATTGGTTGATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTTCCTCGGATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAACTTTGTTAATTTTAAGTGCTGTAGTTAATGTTGGGGTTGGTACTGGTTGAACTATATATCCTCCTTTATCTTCTTTAGTTGGTCATGGGGGTATATCTGTTGATATAGCTATTTTTTCTTTACATTTGGCGGGTGCATCATCTATTATAGGTGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAATTTCTATTAGATTAGATCAAATTAATTTATTTGTTTGATCAGTTTTAATTACAGCTGTATTATTATTATTATCTTTACCTGTTTTAGCTGGTGCAATTACTATATTACTTACTGATCGTAATTTAAATACGACTTTTTTTGATTTTTCTGGTGGTGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Estación Llanura, 10.93332, -85.25331, 135 meters, caterpillar collection date: 25/v/2013, wasp eclosion date: 07/vi/2013. Depository:
Host data. Thisbe irenea (Riodinidae) feeding on Croton morifolius (Euphorbiaceae).
Host caterpillar and holotype wasp voucher codes. 13-SRNP-76430, DHJPAR0052094.
Paratypes
DHJPAR0021182, DHJPAR0021183, DHJPAR0021185, DHJPAR0021187, DHJPAR0021190, DHJPAR0021192, DHJPAR0021193, DHJPAR0021195, DHJPAR0040352, DHJPAR0040353, DHJPAR0040354, DHJPAR0040355, DHJPAR0040356, DHJPAR0040357, DHJPAR0041144, DHJPAR0048705, DHJPAR0056005. Depository:
Triraphis bradzlotnicki , sp. nov.
Diagnostics
BOLD:AAD7334. Consensus barcode. TGTATTATATTTTTTATTTGGTATTTGGGCTGGTATGGTTGGTTTATCTATAAGATTAATTATTCGATTAGAATTAAGAATGCCAGGTAGTTTATTAGGTAATGATCAAATTTATAATGGYATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGGTTTGGAAATTGGTTAATTCCATTAATGTTAGGGGCYCCTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAACTTTATTAATTTTAAGTGCTGTAGTTAATGTRGGAGTTGGAACAGGATGAACTATRTATCCTCCATTATCTTCTTTAGTTGGTCATGGGGGAATATCGGTTGATATAGCTATTTTTTCTTTACATTTGGCTGGTGCYTCATCTATTATAGGTGTTGTTAATTTTATTTCTACTATTTTTAATATAAAATTGGTATCTATTAATTTGGATCAAATTAATTTATTTGTTTGGTCTGTTTTAATTACTGCTGTTTTATTATTATTGTCTTTACCGGTTTTGGCTGGTGCTATTACTATATTATTAACTGATCGTAATTTAAATACAACATTTTTTGATTTTTCAGGTGGGGGAGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Naciente, 10.98705, -85.42816, 700 meters, caterpillar collection date: 11/iv/2009, wasp eclosion date: 22/iv/2009. Depository:
Host data. Podalia orsilocha (Megalopygidae) feeding on Chimarrhis parviflora (Rubiaceae).
Host caterpillar and holotype wasp voucher codes. 09-SRNP-31273, DHJPAR0035258.
Paratypes
Hosts = Podalia orsilocha and Venadicodia caneti (Limacodidae). DHJPAR0035536, DHJPAR0035545, DHJPAR0035546, DHJPAR0035251. DHJPAR0045688, DHJPAR0062195. Depository:
Other material
Four specimens in the same BIN were collected in Jalisco, Mexico (BIOUG20632-B06, BIOUG20863-E05, BIOUG20444-H09, BIOUG20584-D04). They were not examined, but the one specimen that has an image on BOLD is somewhat lighter in coloration than the Costa Rican specimens.
Triraphis brianbrowni , sp. nov.
Diagnostics
BOLD:AAE0329. Consensus barcode. TGTTTTATATTTTTTATTTGGAATTTGAGCAGGAATAGTRGGTTTGTCAATAAGATTAATTATTCGATTRGAATTAAGTATACCAGGTAGTTTATTAGGTAATGATCAAATTTATAATGGTATAGTAACAGCKCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATYATAATTGGGGGGTTTGGTAATTGGTTRATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTYCCTCGTATAAATAATATGAGTTTTTGATTATTAATTCCTTCATTAACTTTATTAATTTTAAGGGCTGTAGTWAATGTTGGGGTTGGTACTGGTTGAACTATRTATCCTCCTTTGTCTTCTTTAGTTGGTCATGGGGGTATGTCTGTTGATATAGCTATTTTTTCTTTACATTTAGCTGGTGCATCATCAATTATAGGTGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAGTTTCTATTAGATTGGATCAAATTAATTTATTTGTTTGATCAGTTTTAATTACAGCTGTATTATTATTATTATCTTTACCTGTTTTAGCTGGTGCAATTACTATATTACTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGTGGAGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Mundo Nuevo, Quebrada Tibio Perla, 10.76261, -85.42979, 330 meters, caterpillar collection date: 16/xii/2013, wasp eclosion date: 03/i/2014. Depository:
Host data. Panthiades bitias (Lycaenidae) feeding on Inga vera (Fabaceae).
Host caterpillar and holotype wasp voucher codes. 13-SRNP-57666, DHJPAR0054548.
Paratypes
DHJPAR0028327, DHJPAR0061496, BIOUG29630-A05, BIOUG09733-H06. Depository:
Other material
Five specimens from the Chiquibul Rainforest in Belize are in the same BIN and are likely conspecific (BMNHE897790, BMNHE897797, BMNHE897810, BMNHE897828, BMNHE897833). They were not examined.
Triraphis brianlaueri , sp. nov.
Diagnostics
BOLD:ACL0916. Consensus barcode. TGTATTATATTTTTTATTTGGTATTTGATCAGGTATGGTTGGTTTATCTATGAGGGTAATTATTCGGTTAGAATTAAGGATACCTGGTAGTTTATTAGGTAATGATCAAATTTATAATGGAATAGTTACTGCTCATGCACTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGATTTGGTAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATGGCTTTCCCACGTATGAATAATATAAGATTTTGATTGTTAATTCCTTCATTAATTTTATTAATTTTGAGGGCGGTAGTAAATGTTGGAGTTGGGACAGGKTGAACAATTTATCCTCCTTTATCTTCTTTAATAGGTCATGGGGGGATATCTGTTGATATAGCTATTTTTTCTTTACATTTAGCAGGGGCTTCTTCTATTATGGGAGTAATTAATTTYATTTCTACTATTTTTAATATAAAATTAATTTCAATTAAGTTAGATCAAATTAATTTATTTGTTTGATCAGTTTTAATTACTGCGGTGYTATTATTATTATCTTTACCTGTTTTGGCAGGGGCTATYACTATATTATTAACTGATCGTAACTTAAATACTACTTTTTTTGATTTTTCTGGTGGAGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, dry forest, 31/xii/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG17956-H11.
Paratypes
BIOUG09430-G03, BIOUG09740-C12, BIOUG09827-C08, BIOUG17566-G07, BIOUG18248-A06, BIOUG28779-A10, BIOUG29092-C07, BIOUG30950-C06. Depository:
Triraphis briannestjacquesae , sp. nov.
Diagnostics
BOLD:AAF7900. Consensus barcode. AGTTTTATATTTTTTATTTGGTATTTGAGCGGGAATAGTTGGTTTATCTATAAGTTTGATTATTCGACTAGAATTAAGAATACCTGGTAGTTTATTAGGTAATGATCAAATTTATAATGGAATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGTGGATTTGGAAATTGGTTAATTCCTTTAATATTAGGAGCACCTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAACATTATTAATTTTAAGAGCTGTAGTTAATGTTGGGGTAGGAACTGGTTGAACATTGTATCCTCCTTTATCTTCTCTAGTGGGTCATGGGGGTATATCTGTAGATATAGCTATTTTTTCTTTACATTTAGCTGGGGTTTCTTCTATTATAGGTGTTGTAAATTTTATTTCTACTATTTTTAATATAAAATTAATTTCTATTAATTTAGATCAAATTAATTTATTTGTTTGATCAGTATTAATTACTGCTGTTTTATTATTATTATCTTTACCTGTATTGGCAGGGGCTATTACTATATTATTAACAGATCGTAATTTAAATACAACATTTTTTGATTTTTCTGGAGGAGGAGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector Rincon Rain Forest, Estación Caribe, 10.90187, -85.27495, 415 meters, caterpillar collection date: 07/vii/2014, wasp eclosion date: 18/vii/2014. Depository:
Host data. Trosia nigropunctigera (Megalopygidae) feeding on Miconia xalapensis (Melastomataceae).
Host caterpillar and holotype wasp voucher codes. 14-SRNP-43092, DHJPAR0055967.
Paratypes
DHJPAR0045388 DHJPAR0055965 DHJPAR0055966 DHJPAR0055968. Depository:
Triraphis camilocamargoi , sp. nov.
Diagnostics
BOLD:AAJ3968. Consensus barcode. TGTTTTATATTTTTTATTTGGTATTTGAGCTGGTATAGTGGGATTATCAATAAGTTTAATTATTCGTTTAGAATTAAGTATACCAGGTAGTTTATTAGGTAATGATCAAATTTATAATGGAATAGTAACGGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGTAATTGATTAATTCCTTTAATATTGGGGGCACCTGATATAGCTTTTCCACGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAACTTTATTAATTTTAAGTGCTGTAGTAAATGTTGGGGTAGGGACTGGGTGAACTATATATCCTCCTTTATCTTCTTTAGTTGGTCATGGGGGGATATCTGTTGATATGGCAATTTTTTCTTTACATTTAGCAGGGGCATCTTCTATTATAGGGGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAATTTCTATTAGTTTAGATCAAATTAATTTATTTGTTTGATCTGTTTTAATTACAGCAGTATTATTATTATTATCTTTACCTGTTTTAGCTGGGGCTATTACTATATTACTTACTGATCGTAATTTGAATACAACTTTTTTTGACTTTTCTGGTGGGGGGGATCCTATTTTATTTCAACATTTATTT. Triraphis donbrighti occupies the same BIN as T. camilocamargoi but can be separated by multiple morphological characters, the most obvious of which is the single terminal melanic band in the forewing of T. donbrighti which in T. camilocamargoi is accompanied by a second band near midlength of the forewing.
Holotype ♀
Alajuela, Sector San Cristobal, Sendero Huerta, 10.93050, -85.37223, 527 meters, caterpillar collection date: 02/vii/2012, wasp eclosion date: 15/vii/2012. Depository:
Host data. Anteros formosus (Riodinidae) feeding on Miconia xalapensis (Melastomataceae).
Host caterpillar and holotype wasp voucher codes. 12-SRNP-2680, DHJPAR0049947.
Paratypes
Hosts = same as holotype. DHJPAR0017285, DHJPAR0049949, DHJPAR0051362. Depository:
Triraphis carlosherrerai , sp. nov.
Diagnostics
BOLD:AAW1546. Consensus barcode. TGTTTTATATTTTTTATTTGGTATTTGAGCAGGAATAGTGGGATTATCAATAAGTTTAATTATTCGTTTAGAGTTAAGAATACCAGGTAGTTTATTAGGTAATGATCAAATTTATAATAGAATAGTAACAGCTCATGCTTTTATTATAATTTTTTTTATGGTTATACCTATTATAATTGGGGGGTTTGGTAATTGATTAATTCCATTAATATTGGGGGCYCCTGATATAGCTTTTCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTGACTTTATTAATTTTAAGGGCTATAGTTAATGTTGGTGTAGGGACTGGTTGAACTATATATCCTCCTTTATCTTCTTTAGTTGGTCATGGTGGCATATCAGTTGATATAGCAATTTTTTCTTTACATTTAGCAGGTATTTCATCTATTATAGGTGTAGTTAATTTTATTTCTACAATTTTTAATATAAAATTAATTTCTATTGGTTTAGATCAAATTAATTTATTTGTTTGGTCTGTTTTAATTACAGCAGTATTATTATTATTATCTTTACCTGTTTTGGCAGGGGCTATTACTATATTACTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGGGGTGGAGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Puente Palma, 10.9163, -85.37869, 460 meters, caterpillar collection date: 04/xi/2008, wasp eclosion date: 11/xi/2008. Depository:
Host data. Emesis mandana (Riodinidae) feeding on Hieronyma alchorneoides (Phyllanthaceae). Gregarious parasitoid with 27 specimens emerging from the host.
Host caterpillar and holotype wasp voucher codes. 08-SRNP-6211, DHJPAR0030715.
Paratypes
Host = same as holotype. DHJPAR0050030. Depository:
Etymology
Triraphis carlosherrerai is named to acknowledge Carlos Herrera’s contributions to publicity for ACG, GDFCF, and now, BioAlfa.
Triraphis carolinepalmerae , sp. nov.
Diagnostics
BOLD:ABA9319. Consensus barcode. TGTTTTATATTTTTTATTTGGTATTTGGGCTGGTATAGTTGGTTTATCTATAAGGTTAATTATTCGTTTAGAATTAAGTATACCTGGTAGATTACTGGGTAATGATCAAATTTATAATGGTATAGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGCGGGTTTGGGAATTGATTAATTCCTTTAATATTGGGGGCACCTGATATAGCTTTTCCTCGTATAAATAATATGAGTTTTTGATTATTAATTCCTTCATTAACATTATTAATTTTAAGGGCTGTAGTTAATGTTGGGGTAGGTACTGGGTGAACTTTATATCCACCTTTATCTTCTTTAGTTGGTCATGGGGGCATATCTGTAGATATAKCTATTTTTTCTTTACATTTAGCTGGTGCTTCTTCTATTATGGGGGTTGTTAATTTTATTTCTACTATTTTTAATATAAAATTAGTATCAATTAATTTAGATCAAATTAATTTATTTGTTTGATCAGTATTAATTACAGCTGTTTTATTATTATTGTCTTTACCAGTATTAGCTGGTGCAATTACTATATTATTGACAGATCGTAATTTAAATACAACATTTTTTGATTTTTCTGGTGGGGGGGA-CCTATTTTA-TTCAACACTTATTT.
Holotype ♂
Guanacaste, Sector Pitilla, Sendero Laguna, 10.9888, -85.42336, 680 meters, caterpillar collection date: 06/vi/2011, wasp eclosion date: 20/vi/2011. Depository:
Host data. Isochaetes dwagsi (Limacodidae) feeding on Solanaceae 16453 (Solanaceae).
Host caterpillar and holotype wasp voucher codes. 11-SRNP-31583, DHJPAR0045383.
Paratype
BIOUG27999-A10. Depository:
Triraphis charlesmorrisi , sp. nov.
Diagnostics
BOLD:ABU8062. Consensus barcode. TGTTTTATATTTTTTATTTGGAATTTGATCAGGTATAGTTGGTTTGTCAATAAGATTAATTATTCGTTTAGAATTAAGAATACCAGGTAGTTTATTAGGTAATGATCAAATTTATAATGGTATAGTTACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGTGGGTTTGGTAATTGATTAATTCCTTTAATATTAGGTGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGTTTTTGGTTATTAATTCCTTCTTTAACTTTATTAATTTTAAGAGCTGTAGTTAATGTTGGGGTTGGTACTGGATGAACTATATATCCTCCTTTATCTTCTTTAGTTGGTCATGGTGGAATATCTGTTGATATAGCAATTTTTTCTTTACATTTGGCTGGGGTATCTTCTATTATGGGGGTAATTAATTTTATTTCAACTATTTTTAATATAAAATTAATTTCTATTAAATTAGATCAAATTAATTTATTTGTTTGATCAGTTTTAATTACAGCTTTTTTATTATTATTATCTTTACCTGTTTTAGCAGGTGCTATTACTATATTATTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCGGGTGGAGGAGATCCAATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Sendero Laguna, 10.9888, -85.42336, 680 meters, caterpillar collection date: 23/ix/2011, wasp eclosion date: 10/x/2011. Depository:
Host data. Eurybia lycisca (Riodinidae) feeding on Calathea lasiostachya (Marantaceae).
Host caterpillar and holotype wasp voucher codes. 11-SRNP-32841, DHJPAR0045793.
Paratypes
Host = same as holotype. DHJPAR0045790, DHJPAR0045791, DHJPAR0045792, DHJPAR0045794. Depository:
Triraphis chigiybinellae , sp. nov.
Diagnostics
BOLD:ACK5778. Consensus barcode. TGTTTTATATTTTTTATTTGGAATTTGAGCAGGAATATTAGGTTTATCAATAAGGTTAATTATTCGATTAGAATTAAGTATACCT---GGTAGTTTATTAGGTAATGATCAAATTTATAATGGTATAGTTACAGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGTTTTGGAAATTGATTAATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAACTTTATTGATTTTAAGTGCTGTAGTTAATGTTGGGGTTGGTACTGGTTGAACTATATATCCTCCTTTGTCTTCTTTAGTT---GGTCATGGGGGAATATCTGTTGATATGGCAATTTTTTCTTTACATTTAGCTGGTGCGTCATCTATTATAGGAGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAATTTCTATTAGATTAGATCAAATTAATTTATTTGTTTGATCAGTTTTAATTACAGCTATATTATTATTATTATCTTTACCTGTATTAGCTGGTGCAATTACTATATTACTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGTGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Alajuela, Sector San Cristobal, Finca San Gabriel, 10.87766, -85.39343, 645 meters, caterpillar collection date: 23/ix/2013, wasp eclosion date: 08/x/2013. Depository:
Host data. Metacharis victrix (Riodinidae) feeding on Heisteria costaricensis (Olacaceae).
Host caterpillar and holotype wasp voucher codes. 13-SRNP-4931, DHJPAR0053602.
Paratypes
None.
Triraphis christerhanssoni , sp. nov.
Diagnostics
BOLD:ADB1219. Consensus barcode. AGTATTATATTTTTTATTTGGAATTTGGGCAGGTATAGTTGGATTATCTATAAGTTTAATTATTCGTTTAGAATTAAGAATGCCTGGAAGTTTACTAGGTAATGATCAAATTTATAATGGGATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGTTTTGGTAATTGACTAATTCCATTAATATTGGGGGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGGCTATTAATTCCTTCATTAACATTATTAATTTTAAGAGCTGTTGTTAATGTTGGAGTAGGGACTGGGTGAACATTATATCCTCCCTTATCTTCTTTAGTTGGTCATGGTGGGATATCTGTAGATATAGCTATTTTTTCTTTACATTTAGCTGGTGCTTCTTCAATTATAGGGGTTGTTAATTTTCTTTCTACTATTTTTAATATAAAATTAGTATCTATTAATTTAGATCAAATTAATTTATTTGTTTGATCAGTATTAATTACTGCTGTTTTATTATTATTATCTTTACCAGTATTAGCTGGGGCTATTACTATATTATTGACAGATCGTAATTTAAATACAACCTTTTTTGATTTTTCTGGTGGAGGAGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Cacao, Sendero Arenales, 10.92471, -85.46738, 1080 meters, caterpillar collection date: 04/xi/2015, wasp eclosion date: 21/xi/2015. Depository:
Host data. megaJanzen01 98-SRNP-3934 (Megalopygidae) feeding on Inga punctata (Fabaceae).
Host caterpillar and holotype wasp voucher codes. 15-SRNP-35615, DHJPAR0058772.
Paratypes
Hosts = megaJanzen01. DHJPAR0058770, DHJPAR0058771, DHJPAR0058773, DHJPAR0058774, DHJPAR0058775. Depository:
Triraphis christhompsoni , sp. nov.
Diagnostics
BOLD:ACK7801. Consensus barcode. TGTATTATATTTTTTATTTGGAATTTGATCAGGTATAGTTGGGTTATCTATAAGTTTAATTATTCGTTTAGAATTAAGAATACCTGGAAGTTTATTGGGGAATGATCAAATTTATAATGGTATAGTGACTGCCCATGCTTTTATTATAATTTTTTTTATGGTTATACCTATTATAATTGGTGGGTTTGGAAATTGATTAATTCCCYTAATATTAGGGGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAACTTTAYTAATTTTAAGGGCTGTAGTTAATGTTGGGGTAGGAACAGGATGAACTTTATATCCACCTTTATCTTCTTTAGTTGGACATGGTGGAATATCTGTTGATATAGCTATTTTTTCTTTACATTTGGCTGGGGCGTCTTCAATTATAGGGGTTGTTAATTTTATTTCTACTATTTTTAATATAAAATTAGTATCAATTAATTTAGATCAAATTAATTTATTTGTTTGATCAGTATTAATTACTGCTGTTTTATTATTATTATCTTTACCAGTATTGGCAGGTGCTATTACTATATTGTTAACAGATCGTAATTTAAATACAACATTTTTTGATTTTTCTGGTGGTGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector El Hacha, Estación Los Almendros, 11.03226, -85.52776, 290 meters, caterpillar collection date: 29/ix/2011, wasp eclosion date: 10/x/2011. Depository:
Host data. zygJanzen01 Janzen04 (Zygaenidae) feeding on Davilla nitida (Dilleniaceae).
Host caterpillar and holotype wasp voucher codes. 11-SRNP-22833, DHJPAR0045818.
Paratypes
Hosts = same as holotype. DHJPAR0029048, DHJPAR0029064, DHJPAR0029059, DHJPAR0048107, DHJPAR0050069, DHJPAR0059698. Depository:
Other material
A specimen from Mexico (BIOUG20849-A08) and one from Belize (NHM749275) are in the same BIN and are likely conspecific with T. christhompsoni. They were not examined.
Etymology
The species epithet is a patronym for Chris Thompson, a noted North American dipterist, and acknowledges his significant contributions to the inventory of ACG, and Costa and Rica in general, through the identification of Diptera and assisting with the curation of the collection formerly known as INBio.
Triraphis conniebarlowae , sp. nov.
Diagnostics
BOLD:ADJ3083. Consensus barcode. TGTTTTATATTTTTTATTTGGAATTTGAGCAGGAATAGTAGGTTTATCAATGAGATTAATTATTCGATTAGAATTAAGTATACCGGGAAGTTTATTAGGTAATGATCAAATTTATAATGGTATAGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGTAATTGGTTGATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAACTTTATTAATTTTAAGGGCTGTAGTTAATGTTGGGGTTGGGACTGGTTGGACTATATATCCTCCTTTATCTTCTTTAGTTGGTCATGGGGGCATATCTGTTGATATGGCTATTTTTTCTTTACATTTAGCGGGTGCATCATCTATTATAGGTGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAGTTTCTATTAGATTGGATCAAATTAATTTATTTGTTTGATCAGTTTTAATTACAGCTGTATTATTATTATTATCTTTACCTGTTTTAGCCGGTGCAATTACTATACTACTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGTGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Medrano, 11.01601, -85.38052, 380 meters, caterpillar collection date: 22/vii/2017, wasp eclosion date: 07/viii/2017. Depository:
Host data. Caria rhacotis (Riodinidae) feeding on Celtis iguanaea (Ulmaceae).
Host caterpillar and holotype wasp voucher codes. 17-SRNP-72605, DHJPAR0061495.
Paratype
Host = same as holotype. DHJPAR0061498. Depository:
Triraphis craigsimonsi , sp. nov.
Diagnostics
BOLD:ADM7982. Consensus barcode. TGTTTTATATTTTTTATTTGGAATTTGAGCAGGAATAGTAGGTTTATCAATGAGATTAATTATTCGATTAGAATTAAGTATACCAGGAAGTTTATTAGGTAATGATCAGATTTATAATGGTATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGTAATTGGTTAATTCCTTTAATATTAGGGGCTCCTGACATAGCTTTTCCTCGTATAAATAATATGAGGTTTTGATTATTAATTCCTTCATTAACTTTATTAATTTTAAGAGCTGTAGTTAATGTTGGAGTTGGTACTGGTTGAACTATATATCCTCCTTTATCTTCTTTGGTTGGTCATGGGGGTATATCTGTTGATATAGCTATTTTTTCTTTACATTTAGCGGGTGCATCATCTATTATAGGTGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAATTTCTATTGGATTGGATCAAATTAATTTATTTGTTTGATCAGTTTTAATTACAGCTGTATTATTATTATTATCTTTACCTGTTTTGGCTGGTGCAATTACTATATTACTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGTGGGGACCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Loaiciga, 11.01983, -85.41342, 445 meters, caterpillar collection date: 19/iii/2007, wasp eclosion date: 09/iv/2007. Depository:
Host data. Napaea eucharila (Riodinidae) feeding on Vriesea sanguinolenta.
Host caterpillar and holotype wasp voucher codes. 07-SRNP-31839, DHJPAR0062174.
Paratypes
None.
Triraphis danielhubi , sp. nov.
Diagnostics
BOLD:AAM9596. Consensus barcode. AGTATTATATTTTTTATTTGGTATTTGATCYGGTATGGTRGGATTATCTATGAGTTTAATTATTCGTTTAGAATTAAGAATACCYGGTAGTTTATTGGGTAATGATCAAATTTACAATGGRATAGTTACTGCTCATGCTTTAATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGTGGGTTTGGAAATTGATTAGTACCYTTAATATTGGGGTCCCCCGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTAATTCCYTCTTTAATTTTATTAATATTAAGAGCTGTAGTAAATATTGGGGTTGGCACTGGTTGAACTATATAYCCTCCTTTATCTTCTTTAATTGGTCATGGGGGTATATCTGTTGATATAGCAATTTTTTCTTTACATTTAGCCGGTATTTCATCTATTATAGGGGTAATTAATTTTATTTCTACTATTTTTAATATGAAATTAGTTTCAATTAAGTTGGATCAAATTAATTTATTTATTTGATCTGTTTTAATTACTGCTATATTATTATTATTATCTTTACCTGTTTTAGCGGGAGCAATTACTATATTATTGACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGGGGAGGAGATCCTATTTTATTTCAGCATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, dry forest, 01/x/2012, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG17468-D07.
Paratypes
All Malaise-trapped. BIOUG08905-F04, BIOUG08911-B10, BIOUG08911-B11, BIOUG17494-D05. Depository:
Other material
A specimen from Belize (BMNHE897776) is in the same BIN and is likely conspecific.
Etymology
Triraphis danielhubi is named in honor of Daniel Hub’s long-appreciated contributions to publicity for ACG, GDFCF, and now, BioAlfa.
Triraphis davidduthiei , sp. nov.
Diagnostics
BOLD:ACF6933. Consensus barcode. TGTTTTATATTTTTTATTTGGTATTTGAGCGGGTATAGTGGGTTTATCAATGAGGTTAATTATTCGTTTAGAATTAAGTATGCCAGGTAGTTTATTAGGTAATGATCAGATTTATAATGGTATAGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAACTTTATTAATTTTAAGAGCTGTAGTTAATGTTGGGGTTGGGACTGGTTGAACTATGTATCCTCCTTTGTCTTCTTTAATTGGTCATGGGGGGATATCTGTTGATATAGCAATTTTTTCTTTACATTTAGCGGGGGYATCATCTATTATAGGTGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAATCTCTATTAGATTAGATCAAATTAATTTATTTGTTTGATCAGTTTTAATTACAGCTGTATTATTATTATTATCTTTACCTGTTTTAGCTGGTGCAATTACTATATTACTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGTGGAGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Pailas Dos, PL12-6, 10.7637, -85.333, 853 meters, 23/i/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG28804-H04.
Paratypes
All Malaise-trapped. BIOUG05181-G05, BIOUG07453-D06, BIOUG17330-A05, BIOUG18040-B11, BIOUG29481-H06, BIOUG29677-D09.
Triraphis davidwahli , sp. nov.
Diagnostics
BOLD:AAM9598. Consensus barcode. TGTYTTATATTTTATATTTGGTATTTGAGCAGGRATAGTTGGYTTATCAATGAGATTAATTATTCGGTTAGAATTAAGCATACCAGGTAGTTTATTAGGTAATGATCAAATTTATAATGGYATAGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGGGGGTTTGGTAATTGGTTGATTCCTTTGATATTAGGGGCTCCTGATATAGCTTTCYCTCGTATAAATAACATAAGTTTTTGAYTATTAATTCCTTCATTAACTTTATTAGTTTTAAGAGCTGTAGTTAATGTAGGGGTTGGAACTGGATGGACTATATATCCTCCTTTATCTTCTTTAATTGGTCATGGTGGAATGTCTGTTGATATAGCTATTTTTTCTTTACATTTAGCGGGYGYATCATCKATTATAGGTGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAATTTCTATTAAATTGGATCAAATTAATTTATTTGTTTGATCAGTTTTAATTACAGCTGTATTATTATTATTATCTTTACCTGTTTTAGCTGGYGCAATTACTATATTACTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGTGGTGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, dry forest, 25/ii/2013, Malaise trap. Depository:
Host data. No data for holotype, but see below for paratype.
Holotype voucher code. BIOUG18433-B04.
Paratypes
BIOUG05082-G10, BIOUG08078-H08, BIOUG08539-D09, BIOUG09433-G08, BIOUG09826-F07, BIOUG09740-D11, BIOUG09740-D11, DHJPAR0055202 [Host = Panthiades bitias (Lycaenidae)], BIOUG17616-C02, BIOUG17714-E06, BIOUG17759-D05. Depository:
Other material
A specimen from Belize (BMNHE897830) is in the same BIN and is likely conspecific. A specimen from French Guiana (CCDB-07375 D01) is in the same BIN, and the image on BOLD is consistent with those from Costa Rica. Neither was examined.
Triraphis defectus
Diagnostics
BOLD:AAA5374. Consensus barcode. TGTATTGTAT--------TTTTTATTTGGAATTTGATCAGGTATAGTTGGGYTATCTATAAGTTTAATTATTCGRTTAGAATTAAGAATACCTGGGAGTTTATTGGGTAATGATCAAATTTATAATGGTATAGTGACTGCTCATGCCTTTATTATAATTTTTTTTATRGTTATGCCTATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATG--TTAGGAGCTCCTG---ATATAGCTTTYCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAACTTTATTAATTTTAAGGGCYGTAGTTAATGTTGGGGTAGGGACAGGATGAACTTTATATCCWCCTTTATCTTCTTTAGTTGGACATGGKGGTATATCTGTTGATATAGCTATTTTTTCTTTACATTTGGCAGGGGCYTCTTCAATTATAGGGGTTGTTAAYTTTATTTCTACTATTTTTAATATAAAATTAGTATCAATTAATTTAG------------ATCAAATTAATTTATTTGTTTGATCA-GTATTAATTACTGCTGTTTTATTATTATTATCT---TTACCAGTA---TTGGCRGGGGCTATTACTATATTGTTAACAGATCGTAATTTAAACACAACATTTTTTGATTTTTCTGGTGGTGGGGATCCTATTTTATTTCAACATTTATTT.
Specimens examined ♀
Guanacaste, Sector Santa Rosa, Bosque San Emilio, 10.8438, -85.6138, 300 meters, dry forest, 16/vii/2012, Malaise trap. DHJPAR0021194, DHJPAR0029046, BIOUG05279-H01. Depository:
Host data
.
Imaged specimen voucher code . BIOUG05386-B06.
Triraphis federicomatarritai , sp. nov.
Diagnostics
BOLD:ACX5348. Consensus barcode. GTATTATATTTTTTATTTGGAATTTGGGCGGGTATAGTTGGATTATCAATAAGTTTAATTATTCGTTTAGAATTAAGTATGCCGGGTAGTTTATTGGGGAATGATCAAATTTATAATGGCATGGTAACTGCTCATGCTTTTATTATAATTTTTTTTATGGTTATACCCATTATAATTGGGGGATTTGGGAATTGATTGATTCCCTTAATGTTGGGGGCCCCCGACATAGCTTTCCCTCGTATAAATAATATGAGATTTTGGTTATTAATTCCCTCATTAACGTTATTAATTTTAAGGGCTGTGGTTAATGTCGGGGTAGGAACAGGGTGAACTTTATATCCCCCCCTATCTTCCCTAGTTGGTCATGGGGGTATATCTGTGGATATGGCTATTTTTTCTTTACATTTAGCTGGGGCTTCTTCAATTATAGGAGTTGTAAATTTTATTTCTACTATTTTTAATATAAAATTAGTATCAATTAATTTAGATCAAATTAATTTATTTGTTTGATCAGTATTAATTACTGCTGTTTTATTATTATTATCTTTGCCTGTATTAGCTGGGGCAATTACCATATTATTAACAGATCGT---------------------------------------------.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, rain forest/laguna, 23/ix/2013, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG19932-A06.
Paratypes
None.
Triraphis ferrisjabri , sp. nov.
Diagnostics
BOLD:ADA2501. Consensus barcode. GTTTTATATTTTTTATTCGGGATTTGAGCGGGTATGATTGGGCTATCTATAAGTATAATTATTCGGTTAGAATTAAGTATACCGGGCAGATTTTTGGGGAATGATCAAATTTATAATGGCATAGTGACTGCACATGCTTTTATTATAATCTTTTTTATAGTTATGCCCATTATAATTGGCGGTTTTGGAAATTGATTAATTCCCCTAATGTTAGGGGCTCCCGATATAGCTTTCCCTCGTATAAATAATATGAGATTTTGATTATTAATTCCCTCTTTGACGTTGCTTATTTTAAGGGCAGTAGTTAATGTTGGGGTAGGTACTGGCTGAACTTTATACCCCCCTTTATCTTCATTAGTTGGTCATGGGGGCATATCTGTAGATATAGCAATTTTTTCTTTACATCTAGCTGGGGCCTCTTCTATTATAGGGGTTGTTAATTTTATTTCTACTATTTTTAATATAAAATTAGTATCAATTAATTTAGATCAAATTAATTTATTTGTTTGATCTGTATTAATTACAGCTGTTTTATTATTATTATCTTTGCCGGTATTAGCTGGTGCCATCACTATATTATTGACA------------------------------------------------.
Holotype ♀
Guanacaste, Sector San Cristobal, Estación San Gerardo, 10.8801, -85.389, 575 meters, rain forest/laguna, 8/ix/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG27980-B02.
Paratypes
None.
Triraphis mariobozai , sp. nov.
Diagnostics
BOLD:AAB1652. Consensus barcode. GTTTTATATTTTTTATTTGGAATTTGAGCTGGTATAGTTGGGTTATCTATAARATTAATTATTCGATTAGAATTAAGTATACCTGGTAGATTATTGGGGAATGATCAAATTTATAATGGTATGGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGTTTTGGAAATTGATTAATTCCTTTAATATTGGGGGCTCCCGATATAGCTTTTCCTCGTATAAATAATATGAGGTTTTGATTATTAATTCCTTCATTGACATTATTAATTCTAAGGGCTGTAGTTAATGTTGGGGTAGGGACTGGATGAACTTTATATCCACCTTTATCTTCTTTAGTTGGTCATGGTGGGATATCTGTGGATATAGCTATTTTTTCTTTACATTTAGCTGGGGCTTCTTCTATTATGGGGGTTGTTAATTTTATTTCTACTATTTTTAATATAAAATTAGTATCAATTAATTTAGATCAAATTAATTTATTTGTTTGATCAGTATTAATTACAGCTGTTTTATTATTATTATCTTTACCAGTATTAGCTGGTGCAATTACTATATTATTGACAGATCGTAATTTAAATACAACATTTTTTGATTTTTCTGGGGGGGGTGACCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Santa Elena, Casa Potrero Grande, 10.84918, -85.77315, 17 meters, caterpillar collection date: 29/xii/2005, wasp eclosion date: 23/i/2005. Depository:
Host data. Megalopyge Janzen06 (Megalopygidae) feeding on Inga vera (Fabaceae).
Host caterpillar and holotype wasp voucher codes. 05-SRNP-64341, DHJPAR0021178.
Paratypes
All with host data the same as holotype: DHJPAR0021189, DHJPAR0021172, DHJPAR0021173, DHJPAR0021174, DHJPAR0021175, DHJPAR0021176, DHJPAR0021177, DHJPAR0021179, DHJPAR0021186, DHJPAR0028756. Depository:
Triraphis martindohrni , sp. nov.
Diagnostics
BOLD:ABZ7672. Consensus barcode. TGTTTTATATTTTTTATTTGGAATTTGAGCAGGAATAGTAGGTTTATCAATGAGRTTAATTATTCGATTAGARTTAAGTATACCAGGAAGTTTATTAGGTAATGATCAAATTTATAATGGTATAGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGATTTGGTAATTGGTTRATTCCTTTAATATTAGGGGCTCCTGATATAGCTTTYCCTCGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAACTTTGTTAATTTTAAGAGCTGTAGTTAATGTTGGGGTTGGTACTGGTTGAACTATATATCCTCCYTTATCTTCTTTAGTTGGTCATGGGGGGATATCTGTTGATATAGCTATTTTTTCTTTACATTTGGCGGGTGCATCATCTATTATAGGTGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAATTTCTATTARATTGGAYCAAATTAATTTATTTGTTTGATCAGTTTTAATTACAGCTGTATTATTATTATTATCTTTACCTGTTTTAGCTGGTGCAATTACTATATTACTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCTGGTGGTGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Del Oro, Bosque Aguirre, 11.004, -85.441, 571 meters, caterpillar collection date: 20/i/2011, wasp eclosion date: 5/ii/2011. Depository:
Host data. Mesosemia grandis (Riodinidae) feeding on Psychotria lamarinensis (Rubiaceae).
Host caterpillar and holotype wasp voucher codes. 05-SRNP-64341, DHJPAR0042388.
Paratypes
None.
Other material
A specimen from French Guiana (CCDB-07376 H09) is in the same BIN. The image on BOLD shows an almost entirely straw-colored specimen, suggesting that it is probably not conspecific with T. martindohrni.
Triraphis matssegnestami , sp. nov.
Diagnostics
BOLD:ABU7454. Consensus barcode. TGTATTGTATTTTTTATTTGGTATTTGGGCGGGTATGGTTGGTTTATCTATAAGATTAATTATTCGATTAGAATTAAGAATGCCGGGTAGTTTATTGGGTAATGATCAAATTTATAATGGTATAGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGGTTTGGAAATTGATTAATTCCATTAATGTTAGGGGCTCCTGATATGGCTTTTCCACGTATAAATAATATAAGATTTTGGTTATTAATTCCTTCATTAACTTTATTAATTTTAAGTGCTGTAGTTAATGTAGGAGTTGGAACAGGATGAACTATATACCCTCCATTATCTTCTTTAGTTGGTCATGGGGGTATATCAGTTGATATGGCTATTTTTTCTTTACATTTGGCGGGGGCTTCATCTATTATAGGTGTTGTTAATTTTATTTCTACTATTTTTAATATAAAATTGGTATCTATTAATTTAGATCAAATTAATTTATTTGTTTGATCTGTTTTAATTACTGCTGTTTTACTATTATTGTCTTTACCGGTTTTGGCTGGTGCTATTACTATATTATTAACTGATCGTAATTTAAATACAACATTTTTTGATTTTTCAGGTGGGGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Orosi, Quebrada Las Yeguitas, 10.96160, -85.49585, 560 meters, caterpillar collection date: 28/ix/2011, wasp eclosion date: 9/x/2011. Depository:
Host data. Venadicodia caneti (Limacodidae) feeding on Mespilodaphne veraguensis (Lauraceae).
Host caterpillar and holotype wasp voucher codes. 05-SRNP-64341, DHJPAR0045708.
Paratypes
None.
Triraphis mehrdadhajibabaei , sp. nov.
Diagnostics
BOLD:AAJ2715. Consensus barcode. TGTTTTATATTTTTTATTTGGTATTTGAGCTGGTATAGTAGGATTGTCAATAAGATTAATTATTCGGTTAGAATTAAGAATGCCGGGAAGTTTATTAGGTAATGATCAAATTTATAATGGTATAGTAACAGCCCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGGGGGTTTGGTAATTGATTAATTCCTTTAATATTAGGGGCACCTGATATAGCTTTCCCACGTATAAATAATATAAGATTTTGATTATTAATTCCTTCATTAACTTTATTAATTTTAAGTGCGGTAGTAAATGTTGGGGTAGGAACTGGTTGGACTATGTATCCTCCTTTATCTTCTTTAGTGGGTCATGGRGGGATATCTGTTGATATAGCAATTTTTTCTTTACATTTAGCAGGYGCATCTTCTATTATAGGGGTAGTTAATTTTATTTCTACTATTTATAATATAAAATTAGTTTCTATTAGGTTAGATCAAATTAATTTATTTGTTTGATCTGTTTTAATTACTGCAGTATTATTATTATTATCTTTACCAGTTTTRGCTGGRGCTATTACTATATTACTTACTGATCGTAATTTAAATACAACTTTTTTTGATTTTTCWGGCGGGGGRRRMCCTATTTTATTCCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Bullas, 10.98670, -85.38503, 440 meters, caterpillar collection date: 15/iv/2011, wasp eclosion date: 30/iv/2011. Depository:
Host data. Ancyluris inca (Riodinidae) feeding on Miconia argentea (Melastomataceae).
Host caterpillar and holotype wasp voucher codes. 11-SRNP-65193, DHJPAR0042798.
Paratypes
None.
Triraphis ollieflinti , sp. nov.
Diagnostics
BOLD:ABU5849. Consensus barcode. TGTTTTATATTTTTTATTTGGTATTTGAGCTGGTATAGTGGGTTTATCAATAAGGTTGATTATTCGTTTAGAATTAAGAATACCAGGTAGTTTATTAGGTAATGATCAAATTTATAATGGGATAGTAACGGCTCATGCTTTTATTATAATTTTCTTTATAGTTATACCTATTATAATTGGGGGATTTGGTAATTGATTAATTCCCTTAATATTGGGGGCACCTGATATAGCTTTTCCACGTATAAATAATATAAGTTTTTGATTATTAATTCCTTCATTAACTTTATTAATTTTAAGTGCTGTAGTAAATGTTGGGGTAGGGACTGGGTGAACTATTTATCCTCCTTTATCTTCTTTAGTTGGTCATGGAGGTATATCTGTTGATATAGCAATTTTTTCTTTACATTTAGCAGGGGCATCTTCTATTATAGGAGTAGTTAATTTTATTTCTACTATTTTTAATATAAAATTAATTTCTATTAGTTTAGATCAAATTAATTTATTCGTTTGATCTGTTTTAATTACAGCAGTATTATTATTATTATCTTTACCTGTTTTAGCTGGGGCTATTACTATATTACTTACTGATCGTAATTTGAATACAACTTTTTTTGATTTTTCTGGTGGGGGGGATCCTATTTTATTTCAACATTTATTT.
Holotype ♀
Guanacaste, Sector Pitilla, Pasmompa, 11.01926, -85.40997, 440 meters, caterpillar collection date: 21/x/2010, wasp eclosion date: 9/xi/2010. Depository:
Host data. ereJanzen 10-SRNP-32223 (Erebidae) feeding on algae scraped off the surface of a palm leaf.
Host caterpillar and holotype wasp voucher codes. 10-SRNP-32223, DHJPAR0041604.
Paratypes
None.
Triraphis tildalauerae , sp. nov.
Diagnostics
BOLD:ADB6156. Consensus barcode. GTGTTATATTTTTTGTTTGGTATTTGAGCGGGTATGTTGGGGTTATCAATAAGTTTAATTATTCGATTAGAGTTGAGAATGCCTGGGAGATTATTAGGTAATGATCAAATTTATAATGGTATAGTTACAGGGCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGTTTTGGAAATTGATTAATTCCTTTGATGTTAGGGGCCCCTGATATAGCTTTTCCTCGTATAAATAATATGAGATTTTGATTATTAATTCCTTCGTTAATTTTATTAATTTTAAGGGCTGTAGTTAATGTTGGGGTTGGGACTGGGTGGACTATATATCCTCCTTTATCTTCTTTAGTTGGTCATGGAGGGATATCTGTTGATATGGCGATTTTTTCTTTACATTTAGCTGGGGCATCTTCTATTATAGGGGTAATTAATTTTATTTCTACTATTTTTAATATAAAATTAATTTCTATTAAATTGGATCAAGTTAATTTATTTGTGTGATCAGTTTTAATTACGGCTATTTTATTATTATTATCATTGCCTGTATTAGCTGGGGCTATTACTATATTACTTACT------------------------------------------------.
Holotype ♀
Guanacaste, Pailas Dos, PL12-3, 10.7631, -85.3344, 820 meters, 09/i/2014, Malaise trap. Depository:
Host data. None.
Holotype voucher code. BIOUG29480-H10.
Paratypes
None.
Yelicones Cameron, 1887
Members of Yelicones are found worldwide. There are 85 described species in the New World. Members are recorded as parasitoids of Pyralidae and Crambidae.
Yelicones dirksteinkei , sp. nov.
Diagnostics
BOLD:AAT9023. Consensus barcode. TTTATTATATTTTTTTTTTGGTATTTGATCAGGTTTATTAGGTTTATCGTTAAGGTTAGTTATTCGTATAGAATTGAGGAATCCTGGAAGTTTGTTAGGGAGTGATCAATTATATAATTTAATAGTAACAATTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAATTCCATTAATATTAGGTTCACCTGATATGGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTAATTCCTTCTTTAATATTATTATTATTAAGAGGATTTACTAATATGGGTGTAGGCACAGGTTGAACTATATATCCTCCTTTAAGATCTTTATCTGGACATCCTGGGATTTCTGTTGATATAGCTATTTTTTCTTTACATTTAGCAGGGGTATCATCTATTATAGGGGCTGTTAATTTTATTACTACTATTTTTAATATAAAATTGTATAAGTTAAAATTAGATCAATTAAGATTATTTGTATGATCTGTTTTAATTACTGCATTTTTATTATTGTTATCTTTACCTGTTTTGGCAGGTGGGATTACTATATTATTAACTGATCGTAATTTAAATACTAGGTTTTTTGATTTTGCTGGGGGAGGAGATCCAATTTTATTTCAACATTTATTT. It is distinguished from all other described New World Yelicones by the uniquely colored mesoscutum and scutellum, which are piceous-black laterally and pale yellow-cream medially. In the key by
Holotype ♂
Alajuela, Sector Rincon Rain Forest, Rio Francia Arriba, 10.89666, -85.29003, 400 meters, caterpillar collection date: 16/v/2012, wasp eclosion date: 18/vi/2012. Depository:
Host data. pyrabiolep01 biolep29 (Pyralidae) feeding on Croton billbergianus (Euphorbiaceae).
Host caterpillar and holotype wasp voucher codes. 12-SRNP-42090, DHJPAR0049352.
Paratypes
Host = phyBioLep01 BioLep757 (Pyralidae): DHJPAR0049307, DHJPAR0049352. Depository:
Yelicones markmetzi , sp. nov.
Diagnostics
BOLD:ADD0488. Consensus barcode. GGATTAGTAGGATTATCTTTAAGAGTGATTATTCGATTAGAATTAAGTAATTTGGGGAGGTTACTAGGAAGAGACCAGATTTATAATGTAGTAGTAACTATACATGCCTTTATTATAATTTTTTTTATAGTGATACCAATTATAATAGGAGGGTTTGGGAATTGGTTAGTTCCATTAATGATAGGGGGGCCAGATATAGCTTTTCCACGGATGAATAATATGAGGTTTTGGTTATTATTACCCTCATTTTTTTTGATAATATTAAGAGGATTTATAAATGTAGGGGGAGGAACGGGGTGGACTATTTACCCTCCTTTAAGGTCTTTAATAGGGCATATAGGAGTTTCTGTTGATATAATAATTTTTTCTTTGCATTTAGCAGGGGTTCTTCTATTATAGGGGCTGTAAATTTTATTACTACTATTTTTAATATAAATTTATTTTTACAGATGGATCAAATTGTATTATTTTCATGGTCGGTATTAATTACTGCTTTTTTATTATTAATATCTTTGCCAGTTTTAGCAGGAGGGATTACGATGTTATTAACAGATCGAAATTTAAATACTAGGTTTTTTGATTATTCGGGGGGGGGG. Keys to couplet 40 in
Holotype ♂
Guanacaste, Sector Mundo Nuevo, Vado Agria, 10.75876, -85.37543, 560 meters, caterpillar collection date: 7/vii/2008, wasp eclosion date: 25/viii/2008. Depository:
Host data. Accinctapubes albifasciata (Pyralidae) feeding on Damburneya salicifolia (Lauraceae).
Host caterpillar and holotype wasp voucher codes. 08-SRNP-56572, DHJPAR0028135.
Paratypes
None.
Yelicones monserrathvargasae , sp. nov.
Diagnostics
BOLD:ADQ1035. Consensus barcode. TTTATTATACTTTATTTTTGGTATTTGATCAGGTTTATTAGGATTATCTTTTAGTATAATTATTCGGTTAGAATTAAATAACCCTGGGAGTTTATTAGGTAATGATCAAATTTATAATTTAATGGTTACTATTCATGCTTTTATTATGATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGGAATTGATTGATCCCTTTAATATTAGGGGCTCCTGATATAGCATTTCCTCGAATAAATAATATAAGATTTTGGTTATTGATTCCTTCTTTATTTTTGATAATATTGAGTGGTTTTATAAATGTAGGGGTAGGAACAGGTTGAACAATATATCCTCCTTTAAGATCTTTAATAGGTCATAGAGGATTTTCTGTAGATTTTATAATTTTTTCTTTACATTTAGCGGGGTTTTCTTCAATCATGGGTGCTATTAATTTTATTACTACTATTTTTAATATAAAATTAATTTTTATAAAATTAGATCAAATAAGTTTATTTATTTGATCTGTTCTAATTACAGCTTTTTTATTATTATTATCATTACCTGTATTAGCAGGTGGAATTACTATATTATTAACTGATCGAAATTTAAATACTAGTTTTTTTGATTTTTCTGGGGGGGGGGATCCTATTTTGTTTCAACATTTATTT. This species keys to couplet 69 in
Holotype ♂
Guanacaste, Sector Pitilla, Sendero Memos, 10.9817, -85.4278, 740 meters, caterpillar collection date: 10/i/2018, wasp eclosion date: 13/iii/2018. Depository:
Host data. Monoloxis flavicintalisDHJ02 (Pyralidae) feeding on Lacistema aggregatum (Lacistemataceae).
Host caterpillar and holotype wasp voucher codes. 18-SRNP-30118, DHJPAR0062960.
Paratypes
None.
Yelicones tricolor
Diagnostics
BOLD:ACB2775. Consensus barcode. ATTATTATATTTTATTTTTGGTATTTGATCTGGATTATTAGGTTTATCTTTTAGGATAATTATTCGTATAGAATTAAATAATCCTGGTAGGTTATTAGGTAATGATCAAATTTATAATTTAATAGTGACAATTCATGCTTTTATTATAATTTTTTTTATGGTAATACCTATTATAATTGGTGGATTTGGAAATTGATTAATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTYCCTCGTATAAATAATATGAGGTTTTGATTATTAATTCCTTCTTTATTTTTAATAATATTAAGAGGATTTAATAATGTAGGGGTAGGTACTGGTTGGACTATATATCCACCTCTTAGATCATTGGTAGGTCATAGAGGATTTTCTGTTGATTTTATAATTTTTTCATTACATTTAGCAGGAGTTTCTTCTATTATAGGGGCTATTAATTTTATTACTACAATTTTTAATATAAAATTAATATTTATTAAATTGGATCAAATAAGATTATTTATTTGGTCTGTTTTAATCACAGCTATTTTATTATTATTATCTTTACCTGTATTAGCAGGTGGTATCACAATATTATTAACGGATCGAAATTTAAATACTAGATTTTTTGATTTTTCAGGAGGGGGAGATCCAGTTTTATTTCAACATTTATTT.
Material examined
♀: Alajuela, Sector Rincon Rain Forest, Sendero Anonas, 10.90528, -85.27882, 405 meters, caterpillar collection date: 01/iii/2012, wasp eclosion date: 06/iv/2012. Depository:
Host data. chryJanzen01 Janzen165 (Pyralidae) feeding on Piper21150 (Piperaceae).
Host caterpillar and holotype wasp voucher codes. 12-SRNP-40868, DHJPAR0049366.
Yelicones woldai
Diagnostics
BOLD:ACR8199. Consensus barcode. TGTTTTATATTTTTTGTTTGGTATTTGATGTGGGTTATTAGGATTATCTTTAAGAATTATTATTCGATTAGAGTTAAGGAATTTAGGGAGATTATTGGGAAATGATCAAATTTATAATGTAATTGTTACGATACATGCTTTTGTAATAATTTTTTTTATAGTAATACCTATTATAATTGGGGGGTTTGGAAATTGATTGATTCCTTTAATATTAGGGTGTCCTGATATAGCCTTCCCTCGTATAAATAATATAAGATTTTGGTTATTGATTCCTTCTTTAATTTTAATGATTATAAGAGGGTTTATTATAGTAGGGAGAGGAACGGGTTGGACAATATATCCTCCTTTAAGTTCTTTAATTGGTCATGGAGGGTTTTCAGTTGATATAGTTATTTTTTCTTTACATTTAGCAGGGGTATCTTCCATTATAGGAGCTATTAATTTTATTACAACAATTTTTAATATAAAATTAATGTTAAAATTGGATCAAGTTATATTGTTTGTATGATCTGTTTTGATTACTGCTTTTTTATTATTATTATCTTTGCCTGTGTTAGCAGGAGGAATTACTATATTATTAACAGATCGTAATTTAAATACTTCTTTTTTTGATTTTTCAGGGGGAGGAGATCCTATTTTATTTCAGCATTTATTT.
Illustrated specimen ♀
Alajuela, Sector San Cristobal, Puente Palma, 10.9163, -85.37869, 460 meters, caterpillar collection date: 11/ix/2011, wasp eclosion date: 21/x/2011. Depository:
Host data. Chloropaschia mennusalis (Pyralidae) feeding on Clusia cylindrica (Clusiaceae).
Host caterpillar and holotype wasp voucher codes. 11-SRNP-3557, DHJPAR0045811.
Acknowledgements
We thank reviewers of the manuscript, who may or may not agree with our methods: Lyubomir Penev, Frank Krell, Jose Fernandez-Triana, James Whitfield, Thomas Pape, and Yves Braet. We gratefully acknowledge the unflagging support of the team of ACG parataxonomists who collected and reared the specimens used in this study, and the team of biodiversity managers who protect and manage the ACG forests that are home to these wasps and their caterpillar hosts. The study has been supported by U.S. National Science Foundation grants BSR 9024770 and DEB 9306296, 9400829, 9705072, 0072730, 0515699, and grants from the Wege Foundation, International Conservation Fund of Canada, Jessie B. Cox Charitable Trust, Blue Moon Fund, Guanacaste Dry Forest Conservation Fund, Permian Global, individual donors, and University of Pennsylvania (DHJ&WH). This study has been supported by the Government of Canada through its ongoing support to the Canadian National Collection, and by grants from Genome Canada and Ontario Genomics to PDNH in support of the Centre for Biodiversity Genomics at the University of Guelph, and to the Natural Sciences and Engineering Research Council of Canada. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA; USDA is an equal opportunity provider and employer.
References
- Achterberg C van (1979) A revision of the subfamily Zelinae auct. (Hymenoptera: Braconidae). Tijdschrift voor Entomologie 122: 241–479.
- Achterberg C van (1987) Revisionary notes on the subfamily Orgilinae (Hymenoptera, Braconidae) (Hymenoptera: Braconidae). Zoologische Verhandelingen, Leiden 242: 1–111. https://repository.naturalis.nl/pub/317720/ZV1987242001.pdf
- Achterberg C van (1989) Pheloura gen. nov., a Neotropical genus with an extremely long pseudo-ovipositor (Hymenoptera: Braconidae). Entomologische Berichten, Amsterdam 49: 105–108.
- Achterberg C van (1990a) Revision of the subtribe Mesocoelina Viereck (Hymenoptera: Braconidae). Zoologische Mededelingen, Leiden 64(4): 31–57. https://repository.naturalis.nl/pub/318412/ZM1990064004.pdf
- Achterberg C van (1990b) Revision of the western Palaearctic Phanerotomini (Hymenoptera, Braconidae). Zoologische Verhandelingen Leiden 255: 3–106.
- Achterberg C van (1991) Revision of the genera of the Afrotropical and W. Palaearctical Rogadinae Foerster (Hymenoptera: Braconidae). Zoologische Verhandelingen Leiden 273: 1–102. https://repository.naturalis.nl/pub/317741/ZV1991273001.pdf
- Areekul B, Quicke DLJ (2004) Two new species of Pseudoyelicones (Braconidae: Rogadinae) from Costa Rica. Journal of Hymenoptera Research 13(2): 207–213.
- Areekul B, Quicke DLJ (2006) Systematics of the parasitic wasp genus Yelicones Cameron (Hymenoptera: Braconidae: Rogadinae) and revision of the genus from the New World. Systematics and Biodiversity (3): 255–376. https://doi.org/10.1017/S1477200005001866
- Belokobylskij SA (1986) Five new species of braconids (Hymenoptera: Braconidae) from the Asiatic part of the USSR. In: Ler PA, Belokobylskij SA, Storozheza NA (Eds) Hymenoptera of Eastern Siberia and the Far East. Collected Works. Academy of Sciences USSR. Far East Science Centre, Vladivostok [152 pp.], 28–40. [In Russian]
- Belokobylskij SA (1996) A new Palaearctic genus of the subfamily Ichneutinae (Hymenoptera: Braconidae). Zoosystematica Rossica 4: 307–310.
- Bertrand C, Janzen DH, Hallwachs W, Burns JM, Gibson JF, Shokralla S, Hajibabaei M (2014) Mitochondrial and nuclear phylogenetic analysis with Sanger and next-generation sequencing shows that, in Área de Conservación Guanacaste, northwestern Costa Rica, the skipper butterfly named Urbanus belli (family Hesperiidae) comprises three morphologically cryptic species. BMC Evolutionary Biology 14: 153–171. https://doi.org/10.1186/1471-2148-14-153
- Bortoni MA, Penteado-Días AM (2015) New species of Plesiocoelus van Achterberg and Mesocoelus Schulz (Hymenoptera, Braconidae) from Brazil. Journal of Hymenoptera Research 46: 61–70. https://doi.org/10.3897/JHR.46.6661
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN (2007) DNA barcodes of closely related (but morphologically and ecologically distinct) species of skipper butterflies (Hesperiidae) can differ by only one to three nucleotides. Journal of the Lepidopterists Society 61: 138–153.
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN (2008) DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservación Guanacaste, Costa Rica. Proceedings of the National Academy of Sciences 105: 6350–6355. https://doi.org/10.1073/pnas.0712181105
- Chacon IA, Janzen DH, Hallwachs W, Sullivan JB, Hajibabaei M (2013) Cryptic species within cryptic moths: new species of Dunama Schaus (Notodontidae, Nystaleinae) in Costa Rica. ZooKeys 264: 11–45. https://doi.org/10.3897/zookeys.264.4440
- Chen X-X, Achterberg C van (2019) Systematics, Phylogeny and evolution of braconid wasps: 30 years of progress. Annual Review of Entomology 64: 335–358. https://doi.org/10.1146/annurev-ento-011118-111856
- Dadelahi SD, Shaw SR, Aguirre H, De Almeida LFV (2018) A taxonomic study of Costa Rican Leptodrepana with the description of twenty-four new species (Hymenoptera, Braconidae, Cheloninae). ZooKeys 750: 59–130. https://doi.org/10.3897/zookeys.750.23536
- Decree 41767 (2019) Government of Costa Rica, La Gaceta Diario Oficial, no. 143, 31 julio 2019. Declaratoria de interes publico del Proyecto BioAlfa, 5–7. https://www.imprentanacional.go.cr/pub/2019/07/31/COMP_31_07_2019.pdf
- Enderlein G (1920) Zur Kenntnis aussereuropäischer Braconiden. Archiv für Naturgeschichte 84(A) 11(1918): 51–224. https://doi.org/10.5962/bhl.part.13627
- Espinoza BA, Janzen DH, Hallwachs W (2017) 17 new species hiding in 10 long-named gaudy tropical moths (Lepidoptera: Erebidae: Arctiinae). Tropical Lepidoptera Research 27 (Supplement 1): 1–29.
- Favret C, Sieracki JM (2016) Machine vision automated species identification scaled towards production levels. Systematic Entomology 41: 133–143. https://doi.org/10.1111/syen.12146
- Fernandez-Triana JL, Whitfield JB, Rodriguez JJ, Smith MA, Janzen DH, Hallwachs W, Hajibabaei M, Burns JM, Solis MA, Brown J, Cardinal S, Goulet H, Hebert PDN (2014) Review of Apanteles sensu strictu (Hymenoptera, Braconidae, Microgastrinae) from northwestern Costa Rica, with keys to all described species from Mesoamerica. ZooKeys 383: 1–565. https://doi.org/10.3897/zookeys.383.6418
- Fernandez-Triana J, Shaw MR, Boudreault C, Beaudin M, Broad GR (2020) Annotated and illustrated world checklist of Microgastrinae parasitoid wasps (Hymenoptera, Braconidae). ZooKeys 920: 1–1089. https://doi.org/10.3897/zookeys.920.39128
- Fleming AJ, Wood DM, Smith MA, Hallwachs W, Janzen DH (2014) Revision of the New World species of Houghia Coquillett (Diptera, Tachinidae) reared from caterpillars in Area de Conservación Guanacaste, Costa Rica. Zootaxa 3858(1): 001–090. https://doi.org/10.11646/zootaxa.3858.1.1
- Gupta A, Achterberg C van, Chitrala M (2016) A new species of Crinibracon Quicke (Hymenoptera: Braconidae) parasitic on pupae of Hasora chromus (Cramer) (Lepidoptera: Hesperiidae) from India. Zootaxa 4158(2): 281–291. https://doi.org/10.11646/zootaxa.4158.2.9
- Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN (2006) DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences 103: 968–971. https://doi.org/10.1073/pnas.0510466103
- Hansson C, Smith MA, Janzen DH, Hallwachs W (2015) Integrative taxonomy of New World Euplectrus Westwood (Hymenoptera, Eulophidae), with focus on 55 new species from Area de Conservación Guanacaste, northwestern Costa Rica. ZooKeys 485: 1–236. https://doi.org/10.3897/zookeys.485.9124
- Hanson PE, Gauld ID (2006) Hymenoptera de la Region Neotropical. The American Entomological Institute, Gainesville, 994 pp.
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America 101(41): 14812–14817. https://doi.org/10.1073/pnas.0406166101
- ICZN (1999) International Code of Zoological Nomenclature. 4th edn. The International Trust for Zoological Nomenclature, London, 306 pp. https://www.iczn.org/the-code/the-international-code-of-zoological-nomenclature/the-code-online/
- Ivanova NV, Dewaard JR, Hebert PD (2006) An inexpensive, automation‐friendly protocol for recovering high‐quality DNA. Molecular Ecology Resources 6(4): 998–1002. https://doi.org/10.1111/j.1471-8286.2006.01428.x
- Ivanova NV, Grainger CM (2007) CCDB protocols, COI amplification. [Available from] http://ccdb.ca/site/wp-content/uploads/2016/09/CCDB_Amplification.pdf [accessed 1 July 2019]
- Janzen DH (1999a) Gardenification of tropical conserved wildlands: multitasking, multicropping, and multiusers. PNAS 96(11): 5987–5994. https://doi.org/10.1073/pnas.96.11.5987
- Janzen DH (1999b) La sobrevivencia de las areas silvestres de Costa Rica por medio de su jardinificación. Ciencias Ambientales 16(1): 8–18.
- Janzen DH, Sharkey MJ, Burns JM (1998) Parasitization biology of a new species of Braconidae (Hymenoptera) feeding on larvae of Costa Rican dry forest skippers (Lepidoptera: Hesperiidae: Pyrginae). Journal of Neotropical Lepidoptera 9: 33–41. https://journals.flvc.org/troplep/article/view/90141
- Janzen DH, Hajibabaei M, Burns JM, Hallwachs W, Remigio E, Hebert PDN (2005) Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Philosophical Transactions of the Royal Society B 360(1462): 1835–1845. https://doi.org/10.1098/rstb.2005.1715
- Janzen DH, Burns JM, Cong Q, Hallwachs W, Dapkey T, Manjunath R, Hajibabaei M, Hebert PDN, Grishin NV (2017) Nuclear genomes distinguish cryptic species suggested by their DNA barcodes and ecology. Proceedings of the National Academy of Sciences of the United States of America 114(31): 8313–8318. https://doi.org/10.1073/pnas.1621504114
- Janzen DH, Hallwachs W, Blandin P, Burns JM, Cadiou J, Chacon IS, Dapkey T, Deans AR, Epstein ME, Espinoza B, Franclemont JG, Haber WA, Ibabaei MH, Hall JP, Hebert PDN, Gauld ID, Harvey DJ, Hausmann A, Kitching IJ, Lafontaine D, Landry J-F, Lemaire C, Miller JE, Miller JS, Miller L, Miller SE, Montero J, Munroe E, Green SR, Ratnasingham S, Rawlins JE, Robbins RK, Rodriguez JJ, Rougerie R, Sharkey MJ, Smith MA, Solis MA, Sullivan JB, Thiaucourt P, Wahl DB, Weller SJ, Whitfield JB, Willmott KR, Wood DM, Woodley NE, Wilson JJ (2009) Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity. Molecular Ecology Resources 9(Suppl. 1): 1–26. https://doi.org/10.1111/j.1755-0998.2009.02628.x
- Janzen DH, Hallwachs W, Burns JM, Hajibabaei M, Bertrand C, Hebert PDN (2011) Reading the complex skipper butterfly fauna of one tropical place. PLoS ONE 6(8): e19874. https://doi.org/10.1371/journal.pone.0019874
- Janzen DH, Hallwachs W, Harvey DJ, Darrow K, Rougerie R, Hajibabaei M, Smith MA, Bertrand C, Gamboa IC, Espinoza B, Sullivan JB, Decaens T, Herbin D, Chavarria LF, Franco R, Cambronero H, Rios S, Quesada F, Pereira G, Vargas J, Guadamuz A, Espinoza R, Hernandez J, Rios L, Cantillano E, Moraga R, Moraga C, Rios P, Rios M, Calero R, Martinez D, Briceño D, Carmona M, Apu E, Aragon K, Umaña C, Perez J, Cordoba A, Umaña P, Sihezar G, Espinoza O, Cano C, Araya E, Garcia D, Ramirez H, Pereira M, Cortez J, Pereira M, Medina W, Hebert PDN (2012) What happens to the traditional taxonomy when a well-known tropical saturniid moth fauna is DNA barcoded? Invertebrate Systematics 26: 478–505. https://doi.org/10.1071/IS12038
- Janzen DH, Burns JM, Cong Q, Hallwachs W, Dapkey T, Manjunath R, Hajibabaei M, Hebert PDN, Grishin NV (2017) Nuclear genomes distinguish cryptic species suggested by their DNA barcodes and ecology. Proceedings of the National Academy of Sciences 114(31): 8313–8318. https://doi.org/10.1073/pnas.1621504114
- Janzen DH, Hallwachs W (1994) Ethical aspects of the impact of humans on biodiversity. In: Marini-Bettolo GB (Ed.) Man and his environment. Tropical forests and the conservation of species. Pontificiae Academiae Scientiarum Scripta Varia 84: 227–255.
- Janzen DH, Hallwachs W (2011) Joining inventory by parataxonomists with DNA barcoding of a large complex tropical conserved wildland in northwestern Costa Rica. PLoS ONE 6(8): e18123. https://doi.org/10.1371/journal.pone.0018123
- Janzen DH, Hallwachs W (2016) DNA barcoding the Lepidoptera inventory of a large complex tropical conserved wildland, Area de Conservacion Guanacaste, northwestern Costa Rica. Genome 59: 641–660. https://doi.org/10.1139/gen-2016-0005
- Janzen DH, Hallwachs W (2019a) Perspective: where might be many tropical insects? Biological Conservation 233: 102–108. https://doi.org/10.1016/j.biocon.2019.02.030
- Janzen DH, Hallwachs W (2019b) How a tropical country can DNA barcode itself. iBOL Barcode Bulletin October 2, 2019, 1–6. https://doi.org/10.21083/ibol.v9i1.5526
- Janzen DH, Hallwachs W (2020a) Área de Conservación Guanacaste, northwestern Costa Rica: Converting a tropical national park to conservation via biodevelopment. Biotropica 00: 1–20. https://doi.org/10.1111/btp.12755
- Janzen DH, Hallwachs W (2020b) As insectometers, it is clear to us that insect decline in our Costa Rican tropics is real, so let’s be kind to the survivors. PNAS (in press).
- Janzen DH, Hallwachs W, Pereira G, Blanco R, Masis A, Chavarria MM, Chavarria F, Guadamuz A, Araya M, Smith MA, Valerio J, Guido H, Sanchez E, Bermudez S, Perez K, Manjunath R, Ratanasingham S, St Jacques B, Milton M, DeWaard JR, Zakharov E, Naik S, Hajibabaei M, Hebert PDN, Hasegawa M (2020b) Using DNA-barcoded Malaise trap samples to measure impact of a geothermal energy project on the biodiversity of a Costa Rican old-growth rain forest. Genome 24: 1–30. https://doi.org/10.1139/gen-2020-0002
- Jones M, Ghoorah A, Blaxter M (2011) jMOTU and taxonerator: turning DNA barcode sequences into annotated operational taxonomic units. PLoS ONE 6(4): e19259. https://doi.org/10.1371/journal.pone.0019259
- Kang I, Chapman EG, Janzen DH, Hallwachs W, Dapkey T, Smith MA, Sharkey MJ (2017) Revision of the species of Lytopylus from Area de Conservación Guanacaste, northwestern Costa Rica (Hymenoptera, Braconidae, Agathidinae). ZooKeys 721: 93–158. https://doi.org/10.3897/zookeys.721.20287
- Kittel RN, Austin AD, Klopfstein S (2015) Molecular and morphological phylogenetics of chelonine parasitoid wasps (Hymenoptera: Braconidae), with a critical assessment of divergence time estimations. Molecular Phylogenetics and Evolution 101: 224–241. https://doi.org/10.1016/j.ympev.2016.05.016
- Kittel RN (2018) A taxonomic review of the parasitoid wasp genera Furcidentia Zettel and Pseudophanerotoma Zettel (Hymenoptera: Braconidae: Cheloninae) from the Neotropics with the description of four new species. Zootaxa 4486(2): 146–160. https://doi.org/10.11646/zootaxa.4486.2.4
- Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30: 3276–3278. https://doi.org/10.1093/bioinformatics/btu531
- Leathers J, Sharkey MJ (2003) Taxonomy and Life History of Costa Rican Alabagrus (Hymenoptera: Braconidae), with a key to world species. Contributions in Science 497: 1–82. http://sharkeylab.org/sharkeylab/Misc/pdf/v26753.pdf
- Long KD, Achterberg C van (2016) A new species of the genus Plesiocoelus van Achterberg (Hymenoptera: Braconidae: Agathidinae) from Vietnam. [Tap chi Sinh hoc] Academia Journal of Biology 38(3): 304–309. https://doi.org/10.15625/0866-7160/v38n3.8722
- López-Martínez V, Delfín-Gonzalez H, Achterberg C van, Alia-Tejacal I (2011) A new species of the genus Exasticolus van Achterberg (Hymenoptera: Braconidae: Homolobinae) from Mexico. Studies on Neotropical Fauna and Environment 46(1): 59–62. https://doi.org/10.1080/01650521.2010.543022
- Lovejoy TE, Brouillet L, Doolittle WF, Gonzalez A, Green D, Hall P, Hebert P, Herrmann TM, Hyde D, Lee J, Maddison WP (2010) Canadian taxonomy: exploring biodiversity, creating opportunity. The Expert Panel on Biodiversity Science Council of Canadian Academies, Ottawa, 126 pp. https://cca-reports.ca/wp-content/uploads/2018/10/biodiversity_report_final_e.pdf
- Meierotto S, Sharkey MJ, Janzen DH, Hallwachs W, Chapman EG, Smith MA, Hebert PDN (2019a) A revolutionary protocol to describe understudied hyperdiverse taxa and overcome the taxonomic impediment. Deutsche Entomologische Zeitschrift 66(2): 119–145. https://doi.org/10.3897/dez.66.34683.figure14
- Meierotto S, Sharkey M, Achterberg C van (2019b) Taxonomic review of the genera Zelomorpha Ashmead and Hemichoma Enderlein (Hymenoptera, Braconidae, Agathidinae) with new combination assignments. Zootaxa 4565: 131–137. https://doi.org/10.11646/zootaxa.4565.1.11
- Ohnesorge B (1960) Der Einfluß der Temperatur auf die Entwicklung der Kleinen Fichtenblattwespe, Pristiphora abietina (Christ), im Kokon (Hymenoptera: Tenthredinidae). Beiträge zur Entomologie 10: 854–871.
- Papp J (2012) Five new braconid species from Colombia (Hymenoptera, Braconidae) Journal of Hymenoptera Research 28: 67–84. http://dx.doi.org/10.3897/JHR.28.2023
- Papp J (2016) First survey of the Neotropical species of Microchelonus Szépligeti with descriptions of twenty-five new species (Hymenoptera: Braconidae: Cheloninae). Acta Zoologica Academiae Scientiarum Hungaricae 62: 217–344. https://doi.org/10.17109/AZH.62.3.217.2016
- Penteado-Dias AM, Figueiredo MLC, Dias MM, Osório TC, Cruz I (2006) First host records for Exasticolus fuscicornis (Cameron, 1887) (Hymenoptera: Braconidae: Homolobinae). Zoologische Mededelingen Leiden: 80: 109–111. https://www.alice.cnptia.embrapa.br/bitstream/doc/489833/1/Firsthost.pdf
- Phillips-Rodriguez E, Powell JA, Hallwachs W, Janzen DH (2014) A synopsis of the genus Ethmia Hubner in Costa Rica: biology, distribution, and description of 22 new species (Lepidoptera, Gelechioidea, Depressariidae, Ethmiinae), with emphasis on the 42 species known from Area de Conservación Guanacaste. ZooKeys 461: 1–86. https://doi.org/10.3897/zookeys.461.8377
- Rakotoarison A, Scherz MD, Glaw F, Köhler J, Andreone F, Franzen M, Glos J, Hawlitschek O, Jono T, Mori A, Ndriantsoa SH, Raminosoa NR, Riemann JC, Rödel M-O, Rosa GM, Vieites DR, Crottini A, Vences M (2017) Describing the smaller majority: integrative taxonomy reveals twenty-six new species of tiny microhylid frogs (genus Stumpffia) from Madagascar. Vertebrate Zoology 67: 271–398.
- Quicke DLJ (1988) Four new genera of the Plesiobracon Cameron group (Insecta, Hymenoptera, Braconinae). Zoologica Scripta 17: 411–418. https://doi.org/10.1111/j.1463-6409.1988.tb00116.x
- Quicke DLJ (1997) Braconinae. In: Wharton RA, Marsh PM, Sharkey MJ (Eds) Manual of the New World genera of the family Braconidae (Hymenoptera). Special Publication of the International Society of Hymenopterists (1), 149–174.
- Quicke DLJ, Huddleston T (1991) The extraordinary genus Gnathobracon Costa (Hym., Braconidae, Braconinae) with the description of a new species from Peru. Entomologist’s Monthly Magazine 127: 191–195.
- Quicke DLJ, Smith MA, Janzen DH, Hallwachs W, Fernandez-Triana J, Laurenne NM, Zaldivar-Riveron A, Shaw MR, Broad GR, Klopfstein S, Shaw SR, Hrcek J, Hebert PDN, Miller SE, Rodriguez JJ, Whitfield JB, Sharkey MJ, Sharanowski B, Jussila R, Gauld ID, Chesters D, Vogler AP (2012) Utility of the DNA barcoding gene fragment for parasitic wasp phylogeny (Hymenoptera: Ichneumonoidea): data release and new measure of taxonomic congruence. Molecular Ecology Resources 12: 676–685. https://doi.org/10.1111/j.1755-0998.2012.03143.x
- Ratnasingham S, Hebert PD (2013) A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE 8(7): e66213. https://doi.org/10.1371/journal.pone.0066213
- Rodriguez JJ, Fernandez-Triana JL, Smith MA, Janzen DH, Hallwachs W, Erwin TL, Whitfield JB (2013) Extrapolations from field studies and known faunas converge on dramatically increased estimates of global microgastrine parasitoid wasp species richness (Hymenoptera: Braconidae). Insect Conservation and Diversity 6: 530–536. https://doi.org/10.1111/icad.12003
- Scatolini D, Penteado-Dias AM, Achterberg C van (2002) Pseudorhysipolis gen. nov.(Hymenoptera: Braconidae: Rhysipolinae), with nine new species from Brazil, Suriname and Panama. Zoologische Mededelingen, Leiden 76: 109–131. http://repository.naturalis.nl/document/45091
- Sharkey MJ (1990) A revision of Zacremnops Sharkey and Wharton (Hymenoptera: Braconidae: Agathidinae). Proceedings of the Entomological Society of Washington 92: 561–570. http://sharkeylab.org/sharkeylab/Misc/pdf/v15005.pdf
- Sharkey MJ (1997) Key to the New World subfamilies of the Braconidae. Pages 39–64. In: Wharton RA, Marsh PM, Sharkey MJ (Eds) Manual of the New World genera of Braconidae (Hymenoptera). Special Publication of the International Society of Hymenopterists 1: 1–439.
- Sharkey MJ, Clutts Stoelb SA, Tucker EM, Janzen D, Hallwachs W, Dapkey T, Smith MA (2011a) Lytopylus Förster (Hymenoptera, Braconidae, Agathidinae) species from Costa Rica, with an emphasis on specimens reared from caterpillars in Area de Conservación Guanacaste. ZooKeys 130: 379–419. https://doi.org/10.3897/zookeys.130.1569
- Sharkey MJ, Parys KS, Clutts S (2011b) A new genus of Agathidinae with the description of a new species parasitic on Samea multiplicalis (Guenée). Journal of Hymenoptera Research 23: 43–53. https://doi.org/10.3897/jhr.23.1100
- Sharkey MJ, Chapman EG (2015) The Nearctic genera of Agathidinae (Hymenoptera: Braconidae) with a phylogenetic analysis, illustrated generic key and the description of three new genera. Zootaxa 4000: 49–72. https://doi.org/10.11646/zootaxa.4000.1.2
- Sharkey MJ, Chapman EG, Iza de Campos GY (2016) Revision of Aerophilus Szépligeti (Hymenoptera, Braconidae, Agathidinae) from eastern North America, with a key to Nearctic species north of Mexico. Contributions in Science 524: 51–109. http://sharkeylab.org/sharkeylab/docs/posts/web/Sharkey_etal_2016_Aerophilus.pdf
- Sharkey MJ, Meierotto S, Chapman EG, Janzen DJ, Hallwachs W, Dapkey T, Solis MA (2018) Alabagrus Enderlein (Hymenoptera, Braconidae, Agathidinae) species of Costa Rica, with an emphasis on specimens reared from caterpillars in Area de Conservación Guanacaste. Contributions in Science 526: 31–180. http://sharkeylab.org/sharkeylab/docs/posts/web/Sharkey_etal_2018_Alabagrus.pdf
- Sharkey MJ, Wharton RA (1994) A revision of the genera of Ichneutinae (Hymenoptera: Braconidae). Journal of Natural History 28: 873–912. https://doi.org/10.1080/00222939400770471
- Sharanowski BJ, Dowling AP, Sharkey MJ (2011) Molecular phylogenetics of Braconidae (Hymenoptera: Ichneumonoidea), based on multiple nuclear genes, and implications for classification. Systematic Entomology 36: 549–572. https://doi.org/10.1111/j.1365-3113.2011.00580.x
- Shaw S (1997) Cheloninae. In: Wharton RA, Marsh PM, Sharkey MJ (Eds) Manual of the New World genera of the family Braconidae (Hymenoptera). Special Publication of the International Society of Hymenopterists 1: 192–201.
- Shaw MR (1983) On evolution of endoparasitism: The biology of some genera of Rogadinae (Braconidae). Contributions of the American Entomological Institute 20: 307–328.
- Shaw MR, Sims I (2015) Notes on the biology, morphology, nomenclature and classification of Pseudavga flavicoxa Tobias, 1964 (Hymenoptera, Braconidae, Rhysipolinae), a genus and species new to Britain parasitizing Bucculatrix thoracella (Thunberg) (Lepidoptera, Bucculatricidae). Journal of Hymenoptera Research 42: 21–32. https://doi.org/10.3897/JHR.42.8935
- Shaw MR, Huddleston T (1991) Classification and biology of braconid wasps (Hymenoptera: Braconidae). Handbooks for the Identification of British Insects 7(11): 1–126.
- Shimbori EM, Bortoni MA, Shaw SR, Souza-Gessner CDaS, Cerântola PdeCM, Penteado-dias AM (2019) Revision of the New World genera Adelius Haliday and Paradelius de Saeger (Hymenoptera: Braconidae: Cheloninae: Adeliini). Zootaxa 4571: 151–200. https://doi.org/10.11646/zootaxa.4571.2.1
- Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN (2006) DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proceedings of the National Academy of Sciences 103: 3657–3662. https://doi.org/10.1073/pnas.0511318103
- Smith MA, Wood DM, Janzen DH, Hallwachs W, Hebert PDN (2007) DNA barcodes affirm that 16 species of apparently generalist tropical parasitoid flies (Diptera, Tachinidae) are not all generalists. Proceedings of the National Academy of Sciences 104: 4967–4972. https://doi.org/10.1073/pnas.0700050104
- Smith MA, Rodriguez JJ, Whitfield JB, Deans AR, Janzen DH, Hallwachs W, Hebert PDN (2008) Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proceedings of the National Academy of Sciences 105: 12359–12364. https://doi.org/10.1073/pnas.0805319105
- Smith MA, Bertrand C, Crosby K, Eveleigh ES, Fernandez-Triana J, Fisher BL, Gibbs J, Hajibabaei M, Hallwachs W, Hind K, Hrcek J, Huang D-W, Janda M, Janzen DH, Li Y, Miller SE, Packer L, Quicke D, Ratnasingham S, Rodriguez J, Rougerie R, Shaw MR, Sheffield C, Stahlhut JK, Steinke D, Whitfield J, Wood M, Zhou X (2012a) Wolbachia and DNA Barcoding Insects: Patterns, Potential, and Problems. PLoS ONE 7(5): e36514. https://doi.org/10.1371/journal.pone.0036514
- Smith MA, Fernández-Triana JL, Eveleigh E, Gómez J, Guclu C, Hallwachs W, Hebert PDN, Hrcek J, Huber JT, Janzen DH, Mason PG, Miller S, Quicke DLJ, Rodriguez JJ, Rougerie R, Shaw MR, Várkonyi G, Ward D, Whitfield JB, Zaldivar-Riveron A (2012b) DNA barcoding and the taxonomy of Microgastrinae wasps (Hymenoptera, Braconidae): impacts after 8 years and nearly 20,000 sequences. Molecular Ecology Resources 13(2): 168–176. https://doi.org/10.1111/1755-0998.12038
- Solís-Lemus C, Knowles LL, Ané C (2015) Bayesian species delimitation combining multiple genes and traits in a unified framework. Evolution 69: 492–507. https://doi.org/10.1111/evo.12582
- Valerio AA, Shaw S (2015) Thirteen new Costa Rican species belonging to the genus Triraphis Ruthe (Braconidae: Rogadinae) with their host records. Zootaxa 3904(4): 501–540. https://doi.org/10.11646/zootaxa.3904.4.2
- Wharton RA (1984) The status of certain Braconidae (Hymenoptera) cultured for biological control programs, and description of a new species of Macrocentrus. Proceedings of the Entomological Society of Washington 86(4): 902–912.
- Whitfield JB (1990) Phylogenetic review of the Stiropius group of genera (Hymenoptera: Braconidae, Rogadinae) with description of a new neotropical genus. Proceedings of the Entomological Society of Washington 92: 36–43.
- Yu DSK, Achterberg C van, Horstmann K (2016) Taxapad, Ichneumonoidea. Vancouver. http://www.taxapad.com.
- Zamani A, Vahtera V, Sääksjärvi IE, Scherz MD (2020) The omission of critical data in the pursuit of “revolutionary” methods to accelerate the description of species. Systematic Entomology [early view]. https://doi.org/10.1111/syen.12444
- Zettel H (1990) Beitrage zur Kenntnis neotropischer Arten der Gattung Phanerotoma Wesmael: 4. Die Ph. fuscovaria-Gruppe (Hymenoptera: Braconidae, Cheloninae). Linzer Biologische Beitraege 22: 3–19.
Supplementary materials
Agathidinae
Data type: molecular data
Braconinae
Data type: molecular data
Cheloninae
Data type: molecular data
Homolobinae
Data type: molecular data
Hormiinae
Data type: molecular data
Ichneutinae and Proteropinae
Data type: molecular data
Macrocentrinae
Data type: molecular data
Orgilinae
Data type: molecular data
Rhysipolinae
Data type: molecular data
Rogadinae
Data type: molecular data