Citation: Marchán DF, Fernández R, Novo M, Cosín DJD (2014) New light into the hormogastrid riddle: morphological and molecular description of Hormogaster joseantonioi sp. n. (Annelida, Clitellata, Hormogastridae). ZooKeys 414: 1–17. doi: 10.3897/zookeys.414.7665
The earthworm family Hormogastridae shows a remarkable disjunction in its distribution in the Iberian Peninsula, with the Hormogaster elisae species complex isolated from the rest of the species. Hormogaster joseantonioi sp. n., a new species found in the intermediate area between the main ranges (in Teruel, Aragón), was described following the integrative approach, as it is suitable for earthworms due to their highly homoplasic morphology. The phylogenetic analysis of the molecular markers placed the new species as a sister taxon to H. elisae, thus showing the colonizing lineage of Central Iberian Peninsula could have originated near the H. joseantonioi sp. n. current range. External morphological characters revealed some degree of overlap with previously described species, but internal characters presented configurations/states unknown from other members of the family. These traits make the new species a key piece to understand the evolution of Hormogastridae.
Species description, earthworm, integrative taxonomy, phylogeny, disjunct distribution
The increasing availability of molecular and ecological data has placed the integrative taxonomy (as defined by
Taxonomic characters traditionally used for the study of earthworms are few and sometimes present high intraspecific variability (
On the other hand, it seems that most key characters used for hormogastrid traditional taxonomy and phylogeny (notably the shape, number and position of the spermathecae) are highly homoplasic, showing little or no phylogenetic signal across the family.
Due to its relevance for this subject, the intermediate area between the main ranges of hormogastrids in Spain has been subject to recent sampling campaigns. Both Zaragoza and Teruel (Aragón, Spain) were suitable regions as they have been poorly sampled for earthworms unlike the surrounding provinces. While no success was met in Zaragoza, a population assignable to a new species of Hormogastridae was recently found in Teruel.
This paper focuses on the description of Hormogaster joseantonioi sp. n. from an integrative point of view, following the example of
Specimens were collected by hand and fixed in the field in ca. 96% EtOH, with subsequent alcohol changes. Once in the laboratory, specimens were preserved at -20 °C.
The studied material includes 10 specimens (five mature specimens, one semimature specimen with tubercula pubertatis and four immatures) collected in a cleared holm-oak wood at the foothill of Sierra de Oriche, road A-2514 between Huesa del Común and Rudilla, Teruel (Spain) (41°0'55.68"N, 0°58'55.98"W) (Figure 1).
Map of the Iberian Peninsula showing the collection site of Hormogaster joseantonioi sp.n. (indicated by the white star). The northeastern hormogastrid range is shown in green, Hormogaster elisae range is shown in pink and Xana omodeoi known location is indicated in yellow.
Specimens have been deposited in the Oligochaete collection of the Departamento de Zoología y Antropología Física, Universidad Complutense de Madrid (UCMLT), Spain with vouchers UCMLT 00001-00010.
Specimens available from previous studies (
Total genomic DNA was extracted from ventral integument tissue samples using the DNeasy Tissue Kit (QIAGEN) with two consecutive steps of elution (70 µl of buffer). Seven molecular regions were amplified: mitochondrial subunit 1 of cytochrome c oxidase (COI), 16S rRNA and tRNA Leu, Ala, and Ser (16S t-RNAs), one nuclear ribosomal gene (a fragment of 28S rRNA) and one nuclear protein-encoding gene (histone H3). Primer sequences, polymerase chain reactions (PCR) and sequencing reactions are the same as in
Holo- and paragenetypes (sensu
| Specimen | Voucher | COI | 16S-tRNAs | 28S rRNA | H3 |
|---|---|---|---|---|---|
| HRUD1 | UCMLT 00001 | KJ632674 | KJ632684 | KJ632686 | KJ632688 |
| HRUD2 | UCMLT 00002 | KJ632675 | KJ632685 | KJ632687 | KJ632689 |
| HRUD3 | UCMLT 00003 | KJ632676 | |||
| HRUD4 | UCMLT 00004 | KJ632677 | |||
| HRUD5 | UCMLT 00005 | KJ632678 | |||
| HRUD6 | UCMLT 00006 | KJ632679 | |||
| HRUD7 | UCMLT 00007 | KJ632680 | |||
| HRUD8 | UCMLT 00008 | KJ632681 | |||
| HRUD9 | UCMLT 00009 | KJ632682 | |||
| HRUD10 | UCMLT 00010 | KJ632683 |
The new sequences were combined with all the hormogastrid information available from previous studies (
Sequences of each individual gene were aligned in MAFFT (
Bayesian Inference (BI) of the phylogeny was estimated with MRBAYES v.3.1.2 (
Uncorrected pairwise differences for the mitochondrial regions were calculated between Hormogaster joseantonioi and the most closely related species with Arlequin 3.5 (
Type-species. Hormogaster redii Rosa, 1887.
Holotype. Adult (UCMLT 00003), 41°0'55.68"N, 0°58'55.98"W, from a cleared holm-oak wood on the foothill of Oriche mountains, road A-2514 between Huesa del Común and Rudilla, Teruel (Spain), collectors D. Fernández Marchán and J.A. Fernández Fernández.
Paratypes. Nine individuals (UCMLT 00001, 00002, 00004-00010), with the same collection data of the holotype.
Other material examined. 16 hormogastrid species and several subspecies belonging to the UCMLT collection.
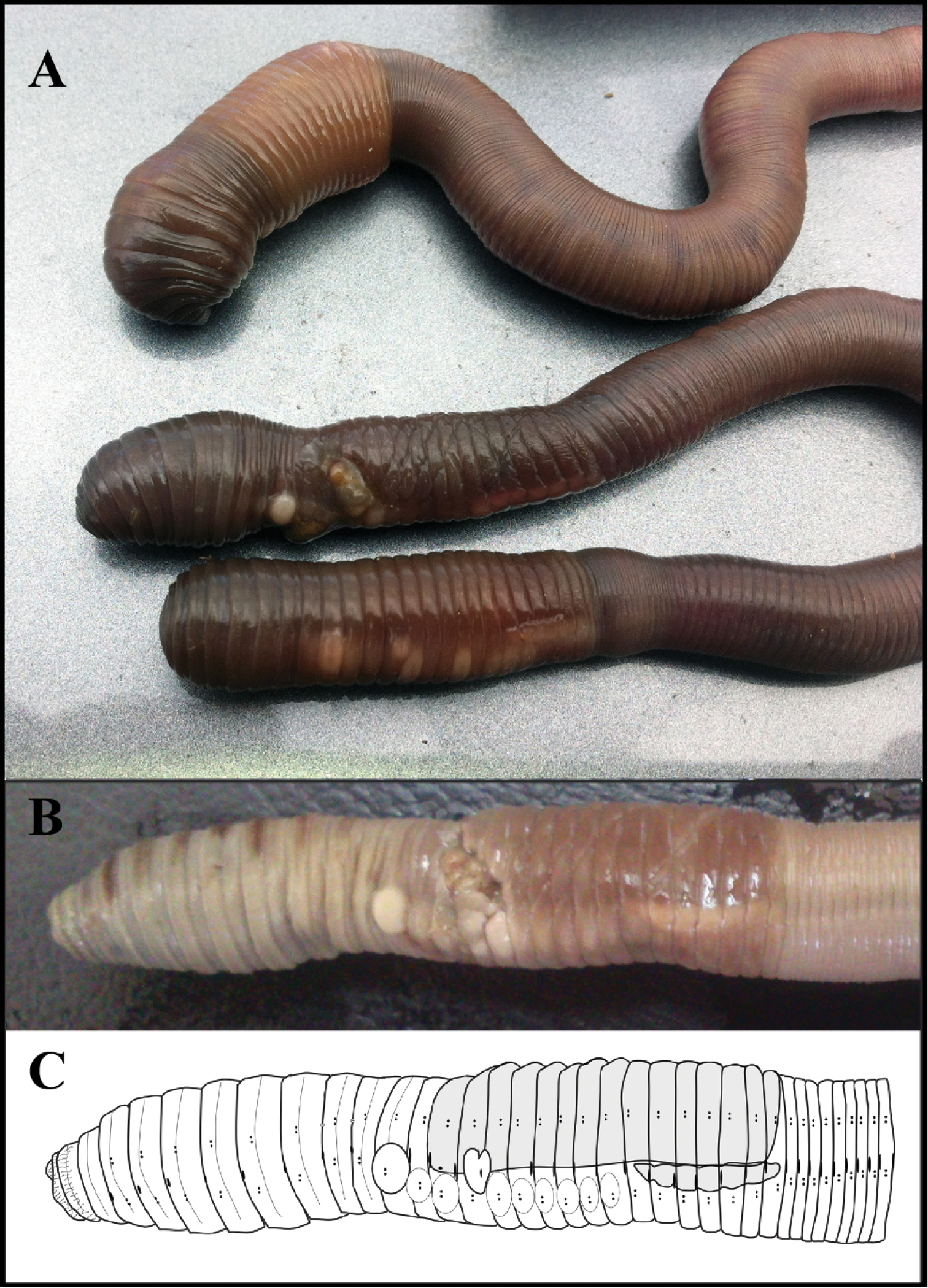
External morphology (Figure 2). *Measures taken on the two only complete specimens, one being the holotype.
(A) Live specimens of Hormogaster joseantonioi sp.n. External morphology of a fixed specimen, shown in a picture (B) and diagram (C).
Length of mature specimens*: 178–180 mm.
Maximum diameter (pre-clitellar, clitellar, post-clitellar) of mature specimens: 8–10, 9–11, 7–10 mm.
Number of segments*: 305–369.
Weight (fixed specimens)*: 7.05–11.57 g.
Colour: From light brown to dark chocolate brown varying between individuals, with orangeish-brown clitellum of a lighter shade on living specimens (Figure 2a). Beige with brown stripes or patches, mainly on the anterior end, with darker clitellum on fixed specimens (Figure 2b).
Prostomium prolobic, longitudinal striation on segments 1 and 2.
Closely paired chaetae; interchaetal ratio at segment 40, aa: 33, ab: 1.3, bc: 6, cd: 1, dd: 27. Nephridial pores in a row between chaetae b and c (very close to b), visible on fixed specimens as a brownish line.
Spermathecal pores at intersegments 9/10 and 10/11 at the level of cd.
Male pores open over chaetae ab at the intersegment 15/16, surrounded by heart-shaped porophores which extend over most of segment 15 and at least half of 16. Female pores in segment 14 at the same level as male pores.
Clitellum saddle-shaped extending over segments (13) 14–28. Tubercula pubertatis on 1/n 22-27(1/n 28) as a continuous line. Papillae of chaetae ab in variable positions, usually between segments 12 and 28: papillae on 12 always showing an unusual degree of development in mature individuals, being very conspicuous both in live and fixed specimens (Figure 2a).
Funnel shaped, strongly thickened septa in 6/7, 7/8 and 8/9, septum 9/10 slightly thickened. The latter’s attachment to the dorsal body wall is displaced two segments backwards, creating a mismatch between inner and outer segmentation with an internally very wide segment 9.
Last pair of hearts in segment 11. Three shiny, strongly muscular gizzards in 6, 7 and 8. Not apparent Morren’s glands, even though small wrinkles exist in the oesophageal wall between segments 10 and 16.
A posterior gizzard is not well differentiated. There is a slight dilatation of the oesophagus between 14 and 16, but it lacks the muscular wall and reinforcements of a true gizzard. First section of the intestine is not dilated.
Typhlosole begins around segments 20 and 21 with seven lamellae, which around segments 26–27 increase to nine. From there they decrease gradually in number until segments 80–105, where they fuse in a single lamella. The latter extends until segments 218-230, where the typhlosole ends.
Fraying testes and iridescent seminal funnels in 10 and 11. Two pairs of voluminous, grainy seminal vesicles in 11 and 12. Ovaries and female funnels in 13, ovisacs in 14.
Two pairs of spermathecae in intersegments 9/10 and 10/11 (but apparently contained in segment 9 due to septum’s backward displacement), the posterior pair bigger. They are sessile and disc-shaped, with multiple inner chambers which open to the exterior through a common pore, in the intersegments 9/10 and 10/11. Some individuals show double spermathecae (each multicameral and with own pore), either in 9/10 or 10/11 (Figure 3a).
A) Spermathecae in segments 9 and 10. Note the double spermathecae in segment 10 of this specimen. B) Nephridial bladder of segment 7.
Anterior nephridial bladders U-shaped with very close branches and no apparent cecum (Figure 3b). Bladders gradually flatten towards the end of the body, taking the usual elongated shape.
Known only from its type locality.
The specimens were collected at 10–20 cm deep in the soil in a cleared holm-oak wood, at the border between a dense forest of Quercus rotundifolia and a dryland farm. The soil had the following characteristics: 23.03% coarse sand, 8.06% fine sand, 5.33% coarse silt, 60.74% fine silt, and 2.84% clay, constituting a silty loam soil, carbon (C): 2.40%, nitrogen (N): 0.24%, C/N: 10.18, pH: 7.98. Mean annual temperature is 12.7 °C and mean annual precipitation is 447.2 mm, as indicated by the nearest weather station (in Herrera de Los Navarros, Zaragoza-23 km away http://www.aragon.es/DepartamentosOrganismosPublicos/Organismos/InstitutoAragonesEstadistica/AreasTematicas/14_Medio_Ambiente_Y_Energia/ci.05_Clima_Datos_climatologicos.detalleDepartamento?channelSelected=ea9fa856c66de310VgnVCM2000002f551bacRCRD#section1).
The species is named after Jose Antonio Fernández Fernández, father of the first author Daniel Fernández Marchán and important contributor during the sampling campaign in which this species was discovered.
Analyses were conducted on sequences from loci COI (10 individuals), 16S (2 individuals), 28S (2 individuals) and H3 (2 individuals) of the new species, combined with similar sequences from other hormogastrid species.
The resulting Bayesian inference of the phylogenetic tree is shown in Figure 4. Its topology was congruent with that of the Maximum Likelihood inferred tree, except for the different placement of Xana omodeoi. Hormogaster joseantonioi sp.n. was recovered as a monophyletic clade, with the Hormogaster elisae species complex as a sister clade.
Bayesian inference of the phylogenetic tree on the concatenated sequence. Numbers above branches indicate posterior probability/bootstrap (of the Maximum Likelihood analysis) support values higher than 0.9/70 (shown as asterisks on terminal branches). Black rectangles show clades not recovered in both analyses (the alternative is shown with a dashed line). The cryptic species included in Hormogaster elisae are numbered from 1 to 5 (following
Uncorrected pairwise distances for the genes COI and 16S-tRNA for Hormogaster joseantonioi and the species within the same clade (with Hormogaster elisae divided into its five cryptic species) are shown in Table 2.
Uncorrected pairwise distances for the genes COI (below the diagonal) and 16S-tRNA (above the diagonal) for Hormogaster joseantonioi and the species on the same clade. XAN – Xana omodeoi, HPRE – Hormogaster pretiosa, HNAJ – Hormogaster najaformis, HEM – two populations of Hemigastrodrilus monicae. Intraspecific divergence for COI/16S is shown in the diagonal.
| HJOS | HE3 | HE1 | HE2 | HE5 | HE4 | XAN | HPRE | HNAJ | HEM* | HEM** | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HJOS | 0.14/0 | 13.10 | 14.20 | 12.50 | 19.41 | 13.50 | 14.23 | 14.28 | 15.31 | 17.40 | 16.07 |
| HE3 | 18.10 | 0.29/0 | 9.87 | 9.96 | 17.18 | 12.34 | 14.37 | 15.93 | 16.69 | 17.54 | 15.57 |
| HE1 | 17.77 | 15.51 | 10.03/4.10 | 7.97 | 17.83 | 12.95 | 15.54 | 17.73 | 17.54 | 17.26 | 16.56 |
| HE2 | 16.47 | 14.16 | 15.13 | 1.75/0.67 | 17.03 | 13.38 | 14.93 | 16.62 | 18.18 | 16.70 | 16.70 |
| HE5 | 16.83 | 16.28 | 17.48 | 16.36 | 0.34/0 | 16.37 | 21.04 | 21.55 | 22.37 | 22.28 | 21.32 |
| HE4 | 19.08 | 15.67 | 17.37 | 16.86 | 10.38 | 3.75/1.75 | 15.49 | 18.06 | 17.51 | 17.81 | 16.53 |
| XAN | 18.30 | 18.26 | 18.36 | 18.96 | 17.01 | 18.49 | 0.37/0.19 | 11.60 | 13.58 | 14.34 | 12.66 |
| HPRE | 18.61 | 20.17 | 20.34 | 19.74 | 18.92 | 19.52 | 17.76 | 0/2.14 | 10.74 | 16.47 | 13.69 |
| HNAJ | 18.92 | 18.39 | 19.77 | 18.19 | 18.64 | 19.17 | 19.92 | 17.31 | 0.10/0.18 | 16.69 | 14.86 |
| HEM* | 18.38 | 18.52 | 19.17 | 20.45 | 17.06 | 18.58 | 20.45 | 19.67 | 19.92 | 3.50/1.97 | 8.76 |
| HEM** | 18.11 | 18.19 | 18.10 | 17.79 | 16.14 | 16.55 | 18.31 | 19.24 | 18.93 | 17.63 | 6.30/2.07 |
Both morphological and molecular characters of Hormogaster joseantonioi sp.n. separate it clearly from all known hormogastrid species, the number of typhosole lamellae and the kind and location of the spermathecae being particularly distinctive. Those characters, while failing to resolve internal relationships within Hormogastridae, have been shown to be suitable for species diagnosis (
The species Hormogaster riojana, while distantly related according to molecular phylogeny, shows many similarities in morphology to Hormogaster joseantonioi (Table 3). However, Hormogaster joseantonioi differ by its lower number of lamellae in its typhlosole and shorter tubercula pubertatis. Moreover it is longer and heavier. While the two species share a very similar position and shape of the spermathecae, some Hormogaster joseantonioi individuals show an additional spermatheca in segment 10 (on the right or left side). These cases don’t seem to be teratologic, as the supernumerary spermathecae have their own pore in the body surface and contain sperm, thus being fully functional.
Comparison of the morphological characters of Hormogaster joseantonioi sp. n. and some of the phylogenetically closest species (Hormogaster elisae, Xana omodeoi and Hormogaster najaformis Qiu & Bouché, 1998) plus the distantly related Hormogaster riojana and Hormogaster castillana Qiu & Bouché, 1998. N. segments: number of segments. N. typhlosole lamellae: number of typhlosole lamellae. Body length, weight and number of segments refer to adult specimens.
| Hormogaster joseantonioi | Hormogaster elisae | Xana omodeoi | Hormogaster najaformis | Hormogaster riojana | Hormogaster castillana | |
|---|---|---|---|---|---|---|
| Colour | Brownish | Colourless | Colourless | Slightly greyish | Dark brownish | Brownish grey |
| Clitellum | (13)14–28 | (13)14(15)–26(27)28 | 14–26 | 13–31 | 13, 14, 17–27, 28 | 1/14, 15–29, 1/2 30 |
| Tubercula pubertatis | 1/n 22–27 (1/n 28) | 22(23)–25(26) | 23–26 | 20–26 | (20)21–27 | 22–28 |
| Length (mm) | 178–180 | 92–200 | 20–161 | 188–230 | 154 | 200–325 |
| N. segments | 305–369 | 205–300 | 190–230 | 395–523 | 243–278 | 320–429 |
| Weight (g) | 7.05–11.57 | 1.96–9.67 | 0.59–4.23 | 22.6–31.4 | 6.57 | 12.85–29.38 |
| Spermathecae position (pores) and appearance | 9 (see text) (9/10, 10/11) Simple(double) Multicameral, disc shaped | 9, 10 (9/10, 10/11) Simple Tubular | 10, 11 (9/10, 10/11) Simple Small, globular | 10, 11 (10/11, 11/12) Multiple Small, globular | 9, 10 (9/10, 10/11) Simple Multicameral, disc shaped | 9, 10 (9/10, 10/11) Simple Globular |
| N. typhlosole lamellae | 9 | 5 | 12 | 15–17 | 15 | 21–23 |
| Thickened septa | 6/7, 7/8, 8/9, (9/10) | 6/7, 7/8, 8/9, (9/10) | (6/7), 7/8, 8/9, 9/10, (10/11) | 6/7, 7/8, 8/9, (9/10) | 7/8, 8/9, 9/10, (10/11) | 7/8, 8/9, 9/10, (10/11) |
Other hormogastrid species possess double or multiple spermathecae, but never of the multicameral, disc shaped kind.
The geographically closest species, Hormogaster castillana (from Puerto Querol, Castellón), is neither morphologically nor phylogenetically closely related (Table 3).
Hormogaster joseantonioi sp. n. appears nested on a weakly supported clade on the phylogenetic tree, consisting in Hemigastrodrilus monicae, Xana omodeoi, Hormogaster pretiosa from Villamassargia, Hormogaster najaformis (and HPA from Omodeo, see
The Hormogaster elisae morphospecies was recovered as sister clade to Hormogaster joseantonioi sp. n. with high support. From a morphological point of view, most of their external characters overlap, except for a slightly longer clitellum and tubercula pubertatis, bigger average size and stronger pigmentation in Hormogaster joseantonioi sp. n. However, internal characters are very different and these species match neither in the number of lamellae in the typhlosole (five versus nine) nor in the structure of the spermathecae, which are tubular in Hormogaster elisae and disc-shaped and multicameral in Hormogaster joseantonioi. It’s worth noting that Hormogaster elisae shares the backwardly displaced disposition of the 9/10 septum.
Based on their phylogenetic and morphological relatedness, an origin of Hormogaster elisae from a common ancestor with Hormogaster joseantonioi sp. n. seems likely. This scenario is sensible from a biogeographical point of view, as the locality of the new species is intermediate between the ranges of Hormogaster elisae and the northeastern main hormogastrid range. A connection of emerged lands would have been possible from the Cretaceous-Tertiary boundary onwards (
While Hormogaster joseantonioi status as a good species and its phylogenetic relationships seem quite clear, generic assignment is a more problematic matter.
Based on its distinctive morphology and geographic range, high genetic divergence and consistent recovery as a well-defined clade,
At this stage it is more conservative to assign Hormogaster joseantonioi to the genus Hormogaster until the revision of the family is completed, which will allow to establish (if possible) a well-founded genera system on Hormogastridae. This work narrows the discontinuity between the North-Eastern and Central ranges of the Spanish hormogastrids. At the same time it highlights the importance of an intensive sampling of the area between Teruel and the center of the Iberian Peninsula (mainly zones of Soria and Guadalajara) to hopefully find new species along the hypothetical colonization route.
We are indebted to Jose Antonio Fernández Fernández for his help during the sampling campaign. Subject Editor Rob Blakemore and two anonymous reviewers helped to improve the manuscript. This research was funded by project CGL2010-16032 from the Spanish Government.
GenBank accession numbers for all sequences used in the phylogenetic analysis, including outgroups. RF: sequences provided by Rosa Fernández.