Citation: Martin-Scheel D, Slater GJ, Kolokotronis S-O, Potter CW, Rotstein DS, Tsangaras K, Greenwood AD, Helgen KM (2014) Biogeography and taxonomy of extinct and endangered monk seals illuminated by ancient DNA and skull morphology. ZooKeys 409: 1–33. doi: 10.3897/zookeys.409.6244
Extinctions and declines of large marine vertebrates have major ecological impacts and are of critical concern in marine environments. The Caribbean monk seal, Monachus tropicalis, last definitively reported in 1952, was one of the few marine mammal species to become extinct in historical times. Despite its importance for understanding the evolutionary biogeography of southern phocids, the relationships of M. tropicalis to the two living species of critically endangered monk seals have not been resolved. In this study we present the first molecular data for M. tropicalis, derived from museum skins. Phylogenetic analysis of cytochrome b sequences indicates that M. tropicalis was more closely related to the Hawaiian rather than the Mediterranean monk seal. Divergence time estimation implicates the formation of the Panamanian Isthmus in the speciation of Caribbean and Hawaiian monk seals. Molecular, morphological and temporal divergence between the Mediterranean and “New World monk seals” (Hawaiian and Caribbean) is profound, equivalent to or greater than between sister genera of phocids. As a result, we classify the Caribbean and Hawaiian monk seals together in a newly erected genus, Neomonachus. The two genera of extant monk seals (Monachus and Neomonachus) represent old evolutionary lineages each represented by a single critically endangered species, both warranting continuing and concerted conservation attention and investment if they are to avoid the fate of their Caribbean relative.
Ancient DNA, extinction, mitochondrial DNA, Panamanian Seaway, Phocidae, systematics
“… he discovered a group of islets abounding with sea-fowl and marine animals. On one of them his sailors, in the course of a single night … took fourteen sea-wolves, and killed a vast quantity of pelicans and other birds.”
“Cruise of Ponce de Leon in search of the Fountain of Youth”
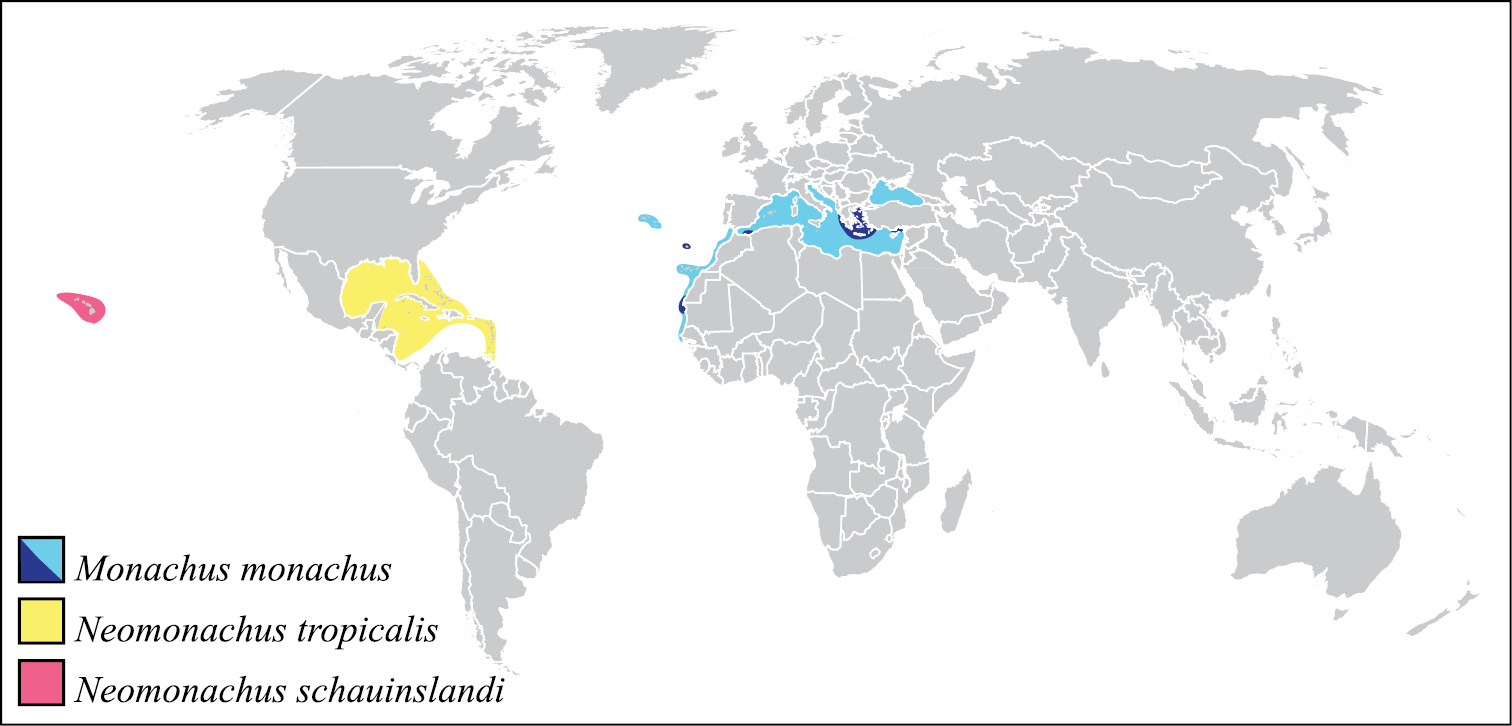
The Caribbean monk seal, Monachus tropicalis (Gray, 1850), first referenced on the New World voyages of Columbus in 1494 and Ponce de Leon in 1513, was one of the few large mammals to become extinct in the twentieth century. Until relatively recently, Monachus tropicalis was widely distributed in the Caribbean region (Figure 1), including along the Caribbean coasts of North, Central, and South America, and in the Bahamas and the Greater and Lesser Antilles (
Distributions of the three monk seal species. The range for the Caribbean monk seal is taken from
There are two species of extant monk seals, both also classified in the genus Monachus Fleming, 1822, and both recognized by the IUCN as Critically Endangered species. The Mediterranean monk seal, Monachus monachus (Hermann, 1779), occurred historically throughout the Mediterranean and Black Seas and along the Atlantic Coast of northern Africa (Figure 1). Today, a heavily fragmented global population of 350–450 individuals remains, dispersed throughout the Mediterranean and south-eastern North Atlantic (
The relationship of the Caribbean monk seal to the two living species of Monachus has long been of interest to paleontologists, evolutionary biologists, and biogeographers (
The initial objective of this study was to determine the phylogenetic placement of Monachus tropicalis and to estimate the timing of its divergence from the Hawaiian and Mediterranean monk seals, as well as from other phocids. Using ancient DNA methods (
We examined all specimens of the three Monachus species in the collections of the USNM, which houses the only large collection of Monachus tropicalis (44 specimens, mostly represented by skins and/or skulls, most of which were collected by E.W. Nelson and E.A. Goldman at the turn of the twentieth century;
Several grams of dry skin were collected from six Caribbean monk seal museum specimens. All were originally collected in the Triangle Keys, a remote and tiny group of islands in Campeche, off the Yucatan Coast of Mexico. Two specimens (USNM 83711 and 83712) from the Triangle Keys were originally kept as captive animals on display at the National Zoological Park (Washington, D.C.), and transferred to the museum upon their death in 1897. The remaining specimens were collected by E.W. Nelson and E.A. Goldman, in June 1900 (
Specimen information for Caribbean monk seals sampled in this study.
| USNM Number | Sex | Locality | Year | Collector |
|---|---|---|---|---|
| 83711 | Female | Bay of Campeche, Mexico | 1897 | National Zoological Park |
| 83712 | Male | Bay of Campeche, Mexico | 1897 | National Zoological Park |
| 100358 | Male | Triangle Keys, Bay of Campeche, Mexico | 1900 | EW Nelson and EA Goldman |
| 100359 | Female | Triangle Keys, Bay of Campeche, Mexico | 1900 | EW Nelson and EA Goldman |
| 102527 | Female | Triangle Keys, Bay of Campeche, Mexico | 1900 | EW Nelson and EA Goldman |
| 102535 | Female | Triangle Keys, Bay of Campeche, Mexico | 1900 | EW Nelson and EA Goldman |
Historical DNA experiments were conducted in an ancient DNA laboratory dedicated to extractions and other pre-PCR procedures involving museum samples. The laboratory was equipped with plexiglass UV PCR hoods for extraction and PCR setup. All reagents and equipment in the lab were exclusively dedicated to ancient DNA experimental use. Modern DNA and PCR products never entered the room. All workstations were regularly UV-irradiated to avoid contamination. Extractions were performed with the Geneclean Kit for Ancient DNA (MP Biomedicals) as described by
List of primers used in this study. Asterisks indicate that the sequence extends over the 3’ or 5’ ends of the cytb gene. Amplicon sizes are shown in base pairs. The primer combinations are designated by numbers, e.g. P7.
| Primer | Name | Sequence (5’–3’) | Length (bp) |
|---|---|---|---|
| P7 | CYTB-MT-F1 CYTB-MT-R1 |
ATG ACC AAC AT(C/T) CGA AAA AC AAA GGC TGT (A/G)GT TGT GTC TG |
149 |
| P10 | CYTB-MT-F4 CYTB-MT-R4 |
(T/C)TA CCA TGA GGA CAA AT(G/A) TC TG(T/G) ACT GCT (A/G)CT AGT GCT |
153 |
| P16 | CYTB-MT-F1 CYTB-MT-R4 |
ATG ACC AAC AT(C/T) CGA AAA AC TG(T/G) ACT GCT (A/G)CT AGT GCT |
549 |
| P20 | Flank-MT-F CYTB-FFla-R |
CCA CCG TTG TAA TTC AAC TA GAT GAG TGA GTT ATT GAT AA |
48* |
| P21 | CYTB-MT-F9 CYTB-MT-R9 |
TAT TCC TAG CTA TAC ACT AC GTG AAT GTG TAG GAG CCG TA |
144 |
| P22 | CYTB-MT-F10 CYTB-MT-R10 |
TAT CTG CTT ATA TAT ACA CGT A AGT AGA TTG GTG ATG ACG GT |
135 |
| P23 | CYTB-MT-F11 CYTB-MT-R11 |
CAC TTC ATT ATA CCC TTC AT AAT GGG ATT TTG TCT GAG T |
77 |
| P24 | CYTB-MT-F11 CYTB-MT-R5 |
CAC TTC ATT ATA CCC TTC AT CC(C/T) AGA ATG TCT TTA ATT GT |
109 |
| P32 | CYTB-MT-F13 CYTB-MT-R13 |
CTC AGA CAA AAT CCC ATT TC GGG TTT GAT ATG TGG TGG |
131 |
| P40 | CYTB-MT-F11 CYTB-MT-R13 |
CAC TTC ATT ATA CCC TTC AT GGG TTT GAT ATG TGG TGG |
229 |
| P42 | CYTB-MT-F16 Flank-MT-R3 |
AGA CCC TGA CAA CTA TAC C GGT CTT GTA AAC CAA AAA CG |
413* |
We integrated the Caribbean monk seal cytb sequence into an alignment of 35 previously sequenced caniform carnivore cytb sequences containing all extant phocid species (n = 18) and 17 outgroup taxa spanning 5 families (Table 3). We performed thorough phylogenetic analyses under the Maximum Likelihood (ML) optimality criterion using the POSIX-threads build of RAxML v7.5.3 (
List of species and associated GenBank accession information used in phylogenetic analysis of cytb sequence data.
| Family | Binomial | Common name | GenBank accession no. | Source |
|---|---|---|---|---|
| Phocidae | Phoca largha | Spotted seal | X82305 | |
| Phoca vitulina | Harbor seal | X82306 | ||
| Pusa sibirica | Baikal seal | AY140977 | ||
| Pusa hispida | Ringed seal | X82304 | ||
| Pusa caspica | Caspian seal | AY140978 | ||
| Halichoerus grypus | Gray seal | NC001602 | ||
| Cystophora cristata | Hooded seal | X82294 | ||
| Histriophoca fasciata | Ribbon seal | X82302 | ||
| Pagophilus groenlandicus | Harp seal | X82303 | ||
| Erignathus barbatus | Bearded seal | AY170104 | ||
| Hydrurga leptonyx | Leopard seal | AY377323 | ||
| Leptonychotes weddellii | Weddell seal | AY377324 | ||
| Ommatophoca rossii | Ross seal | AY377322 | ||
| Lobodon carcinophagus | Crabeater seal | AY377321 | ||
| Neomonachus schauinslandi | Hawaiian monk seal | X72209 | ||
| Neomonachus tropicalis | Caribbean monk seal | JX853967 | this study | |
| Monachus monachus | Mediterranean monk seal | AY377327 | ||
| Mirounga leonina | Southern elephant seal | AY377326 | ||
| Mirounga angustirostris | Northern elephant seal | AY377325 | ||
| Otariidae | Arctocephalus australis | South American fur seal | AY377329 | |
| Arctocephalus forsteri | New Zealand fur seal | X82293 | ||
| Arctocephalus gazella | Antarctic fur seal | X82292 | ||
| Otaria flavescens | South American sea lion | AY377328 | ||
| Zalophus californianus | California sea lion | X82310 | ||
| Eumetopias jubatus | Steller sea lion | DQ145021 | ||
| Callorhinus ursinus | Northern fur seal | NC008415 | ||
| Odobenidae | Odobenus rosmarus | Walrus | X82299 | |
| Mustelidae | Meles meles | Eurasian badger | NC011125 | |
| Gulo gulo | Wolverine | NC009685 | ||
| Mustela nivalis | Least weasel | HM106319 | ||
| Ursidae | Ursus americanus | American black bear | NC003426 | |
| Ursus arctos | Brown bear | HQ685963 | ||
| Ailuropoda melanoleuca | Giant panda | NC009492 | ||
| Canidae | Vulpes vulpes | Red fox | NC008434 | |
| Nyctereutes procyonoides | Raccoon dog | GU256221 | ||
| Canis lupus | Gray wolf | AY170103 |
We estimated divergence times using BEAST v1.7.4 (
Some authors have questioned the appropriateness of using taxa with incomplete sequences in phylogenetic and divergence time inferences (
We performed joint estimation of topology and divergence times in BEAST v1.7.4 (
The Caribbean monk seal cytb gene was generated from a 112-year-old specimen (USNM 100358) from multiple extractions and overlapping PCR amplicons (Suppl. material 1). The consensus sequence of the 1140 bp of cytb represents at least duplicate coverage of every base by an independent amplicon. The results were consistent across all experiments, except for two nucleotide positions that differed in more than one PCR. Those were G-A changes and are most likely due to template deamination on the complementary strand (
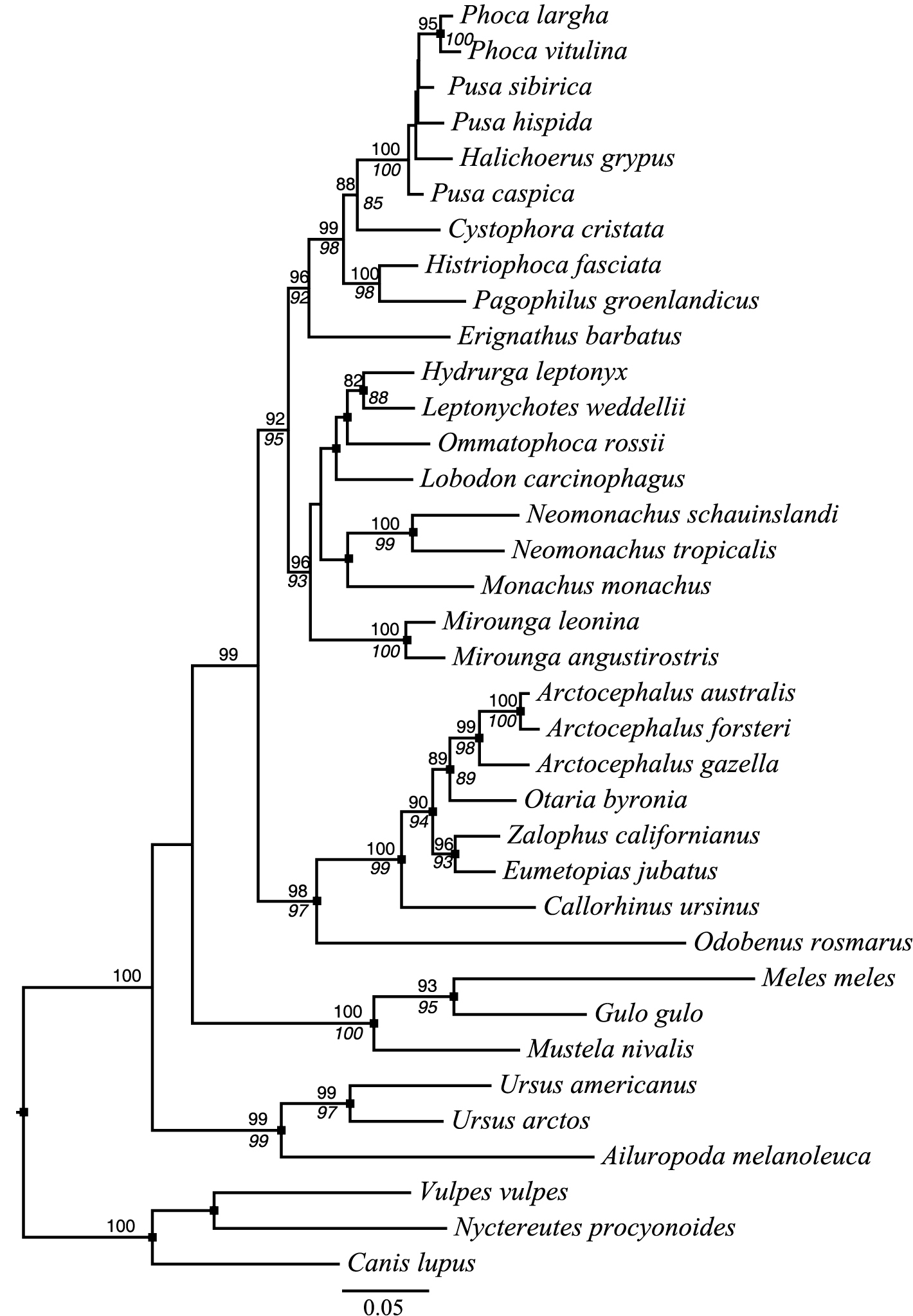
Phylogenetic analyses of the complete cytb sequences under MP, ML, and BI (Figure 2, Suppl. material 4) recovered a monophyletic monk seal clade (BS-MP = 45%, BS-ML = 70%, SH-aLRT = 0.78, PP = 0.93), with a well-supported subclade of New World species (BS-MP = 96%, BS-ML = 100%, SH-aLRT = 0.99, PP = 1.00). Our analyses therefore indicate that the Caribbean and Hawaiian monk seals are more closely related to each other than either is to the Mediterranean monk seal, supported by 9 synapomorphic, non-synonymous changes. The rest of our cytb tree topology is generally consistent with earlier studies of phocid relationships. One notable exception concerns relationships at the base of Monachinae, where we recovered Mirounga (the elephant seals) as the sister lineage to other monachines (Figure 2). Recent studies utilizing both nuclear and mitochondrial loci have revealed that Monachus is sister to Mirounga + Lobodontini (
Maximum likelihood phylogram inferred from cytb sequence data using the GTR + Γ4 substitution model. Node support is expressed as the percent proportion of 1000 bootstrap pseudoreplicates that agree with the bipartitions on the best ML tree (above internode branches) as well as the aLRT SH-like score (below internode branches). Support values above 80% for both measures are shown. Black boxes indicate nodes recovered with >0.88 posterior probability in Bayesian analyses. The scale bar indicates the number of substitutions per site.
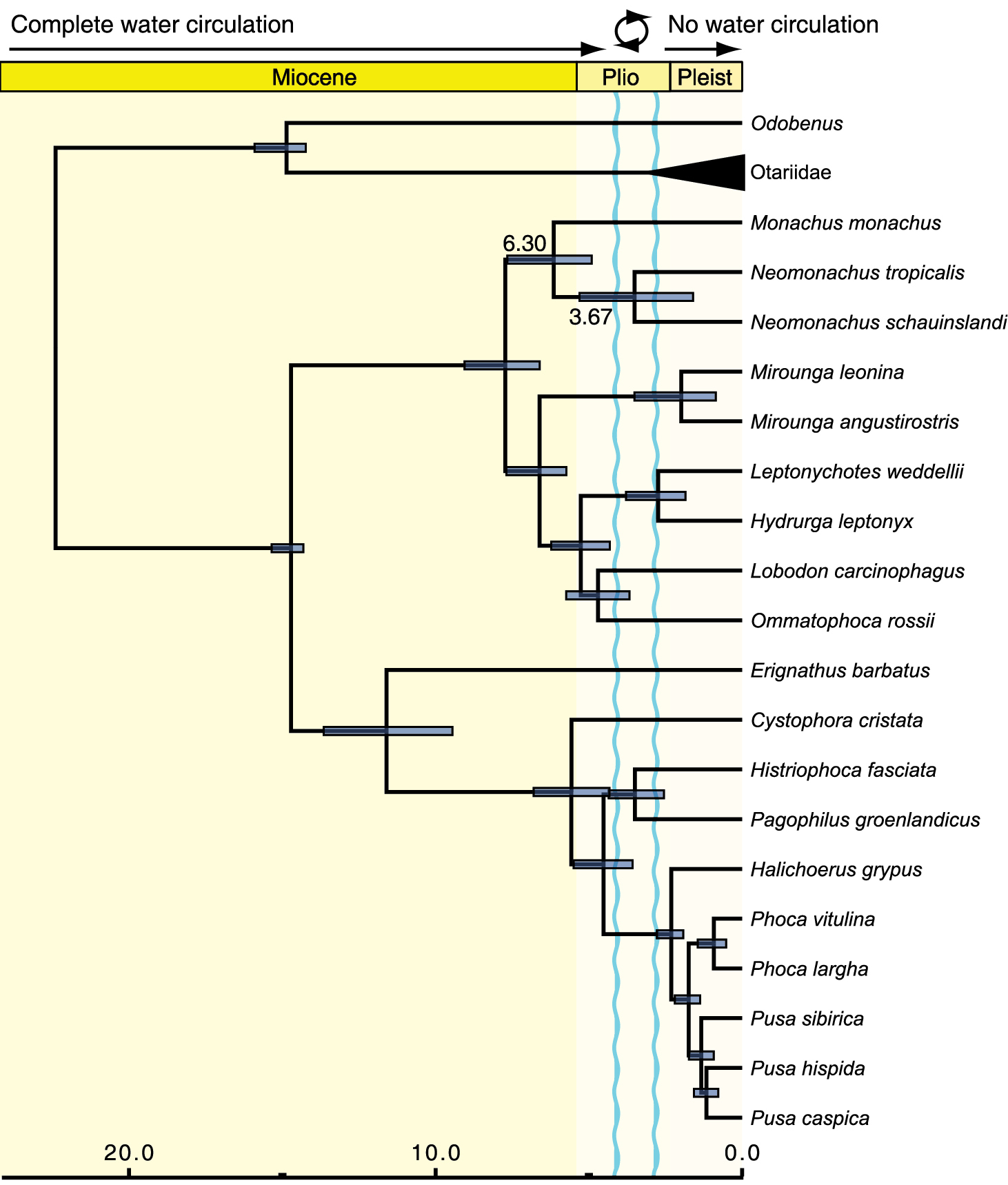
We estimated the divergence of the Caribbean and Hawaiian monk seals at 3.67 Mya (95% HPD = 1.90–5.45 Mya). Node age estimates throughout the rest of the chronogram derived from analysis of the nuclear + mitochondrial genome data are consistent with those recovered by the original analysis of
Time-calibrated phylogeny of the seals estimated from combined nuclear and mitochondrial data. Time scale is in millions of years before present. Note that the chronogram has been pruned to show only true seals and immediate pinniped outgroups. Node bars show the 95% HPD intervals for divergence time estimates and mean ages are labeled for the two divergence times within the monk seals. Labeling at the top indicates water circulation through the Central American Seaway, the circle and associated wavy blue lines indicate a period during which water circulation periodically ceased and resumed but a shallow seaway remained open.
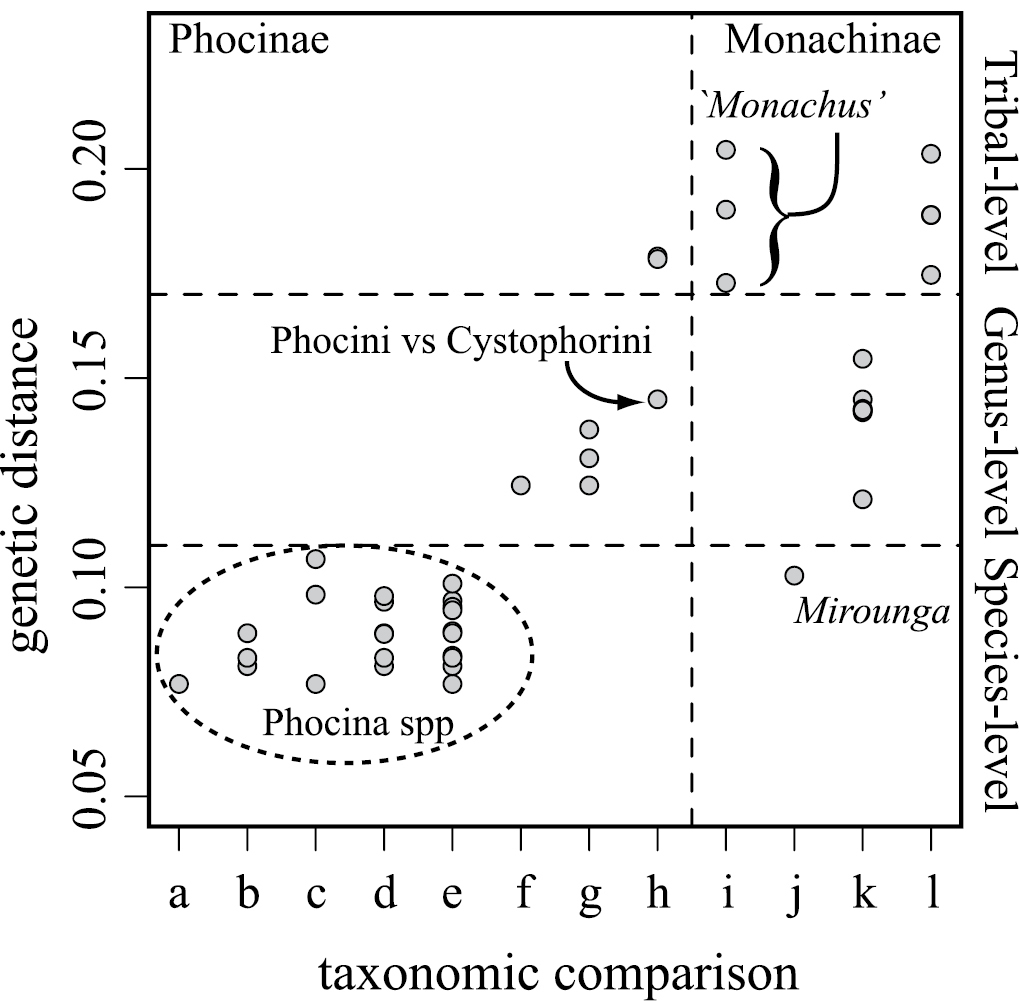
Pairwise genetic distances incorporating the Caribbean monk seal (Figure 4) confirm the findings of
Genetic distances between currently recognized taxonomic units within Phocidae derived from logdet distances for cytb. Distances within: a Phoca b Pusa c Phoca versus Halichoerus d Pusa versus Halichoerus e Phoca versus Pusa f Histriophoca versus Pagophilus g Phocini h Phocinae i Monachus j Mirounga k Lobodontini, and l Monachini.
Our results provide the first molecular evidence for the phylogenetic placement of the Caribbean monk seal. The monophyly of Monachus has been questioned on the basis of morphology (
The exact relationships of the extinct Caribbean monk seal have previously been unclear, with sister relationships to both extant monk seal species suggested on the basis of morphology (
Our analyses confirm that the closure of the Central American seaway after the completion of the Panamanian Isthmus could have played a prominent role in explaining the evolution and distributions of the two New World monk seals. Although the phylogenetic placement of the Caribbean monk seal has been uncertain until now, this significant geological and biotic event has traditionally been invoked to explain the divergence of New World Monachus species through vicariance (e.g.,
Though all three monk seals are currently classified in a single genus, Monachus, the split between New World monk seals and the Mediterranean monk seal is far older than the basal divergence within any other currently recognized modern seal genus (Figure 3) and genetic distances between Monachus species exceed those among other phocid tribes (Figure 4).
All previous generic-level names applied to monk seals have as their type species the Mediterranean monk seal, Monachus monachus (Hermann, 1779), and are thus synonyms of Monachus Fleming, 1822, the earliest generic name erected with that species as its type. These synonyms include Pelagios F. Cuvier, 1824 (including its various subsequent spellings and the replacement name Rigoon Gistel, 1848), Pelagocyon Gloger, 1841, and Heliophoca Gray, 1854, as well as
Monachus schauinslandi Matschie, 1905 (endemic to the Hawaiian Islands).
A second species, Monachus tropicalis (Gray, 1850) (endemic to the Caribbean region, recently extinct). We note here, as an aside, that an earlier specific epithet, antillarum Gray, 1849, has often been included in the synonymy of tropicalis, where it is identified either as a partial synonym (e.g.,
The new generic name combines the Greek Neo- (new), with Monachus, the genus name previously used for all monk seals. The name references both the recognition of a new genus within the monk seals and its New World (Western Hemisphere) distribution.
Species of Neomonachus can be distinguished from Monachus in their smaller average body size and in lacking a white ventral patch on the pelage (in both adults and young) (
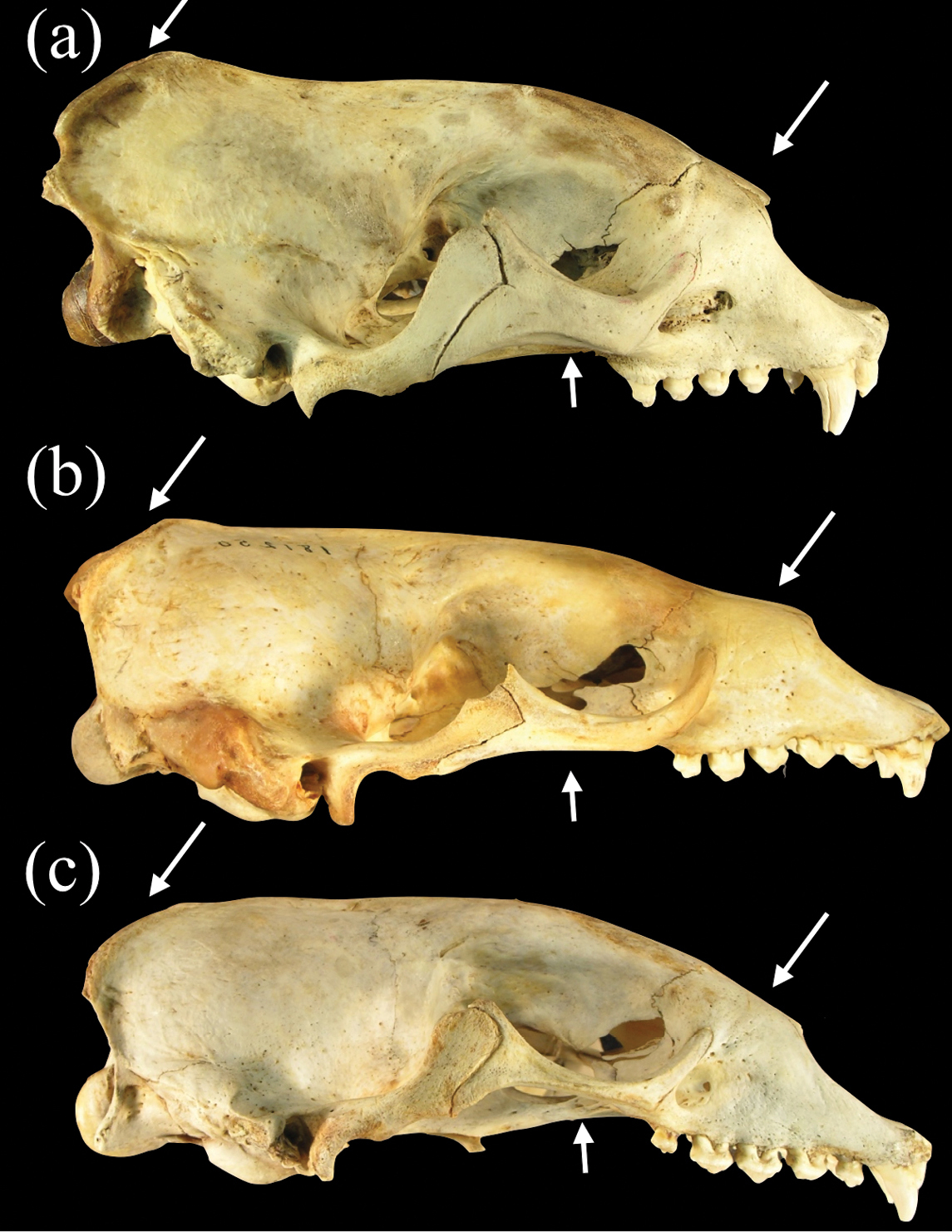
Lateral views of crania of a Monachus monachus b Neomonachus schauinslandi, and c Neomonachus tropicalis. Arrows indicate the more developed occipital crest and zygomatic arches, and deeper snout of Monachus compared to Neomonachus species.
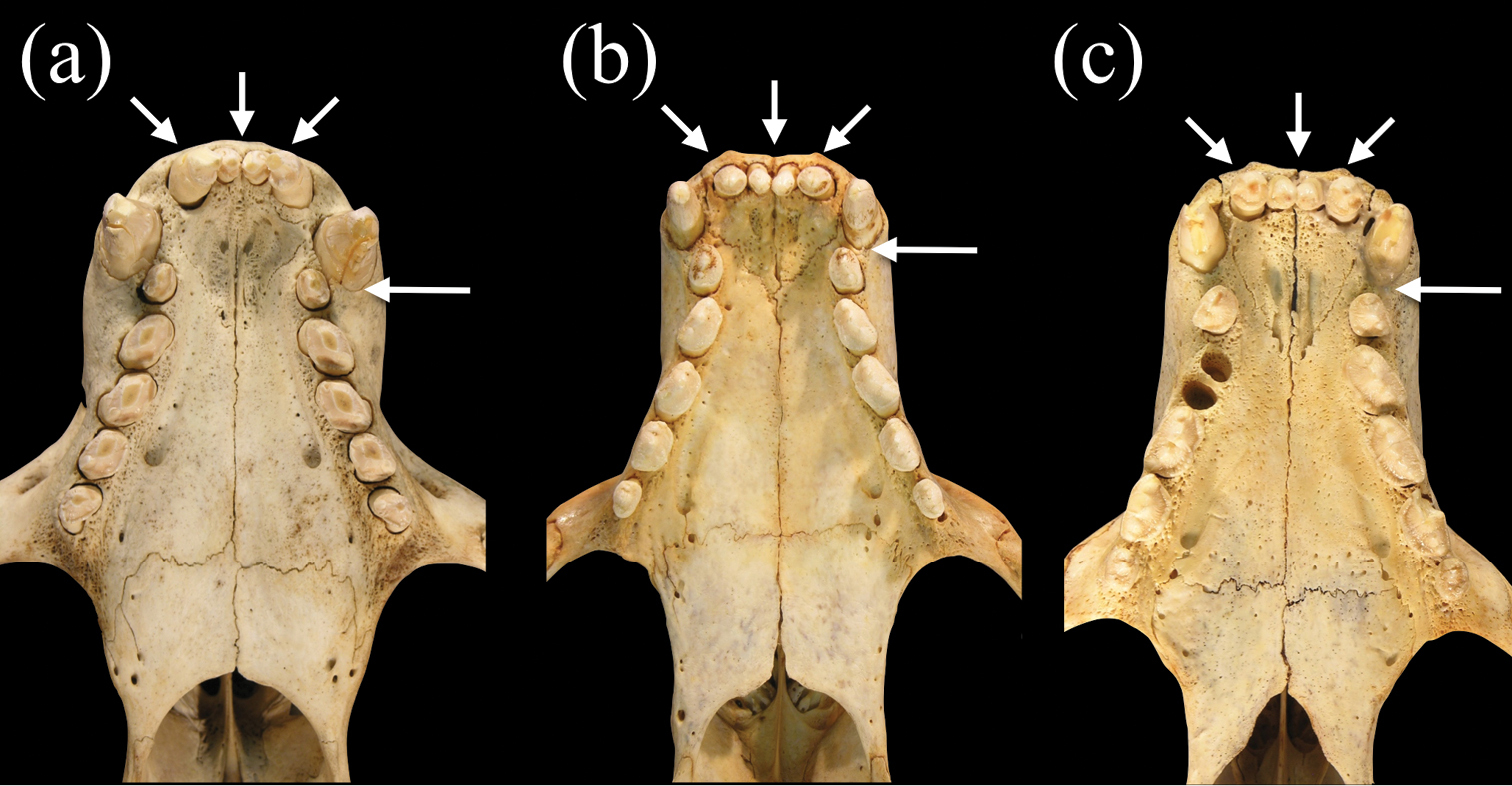
Ventral views of palates of a Monachus monachus b Neomonachus schauinslandi, and c Neomonachus tropicalis. The tooth row of Monachus is more crowded, likely as a result of the shorter rostrum, and this results in a more obliquely oriented set of post-canine teeth and the lack of a diastema between the upper canine and the first premolar. In Neomonachus, there is a distinct diastema between C1 and P1, and the post-canine teeth are arranged more linearly. The upper incisor arcade of Monachus is slightly parabolic due to the posterior placement of the lateral incisors, and the anterior premaxilla appears slightly curved. In Neomonachus, the incisor arcade is linear and the anterior premaxilla is straight.
Dorsal view of rostra of a Monachus monachus b Neomonachus schauinslandi, and c Neomonachus tropicalis. Monachus exhibits a well-developed antorbital process on the maxilla, immediately inferior to the fronto-maxillary suture. The process is reduced or absent in Neomonachus. The nasals of Monachus are short and triangular, tapering smoothly posteriorly to produce a point at their union. The nasals of Neomonachus are longer and do not taper smoothly.
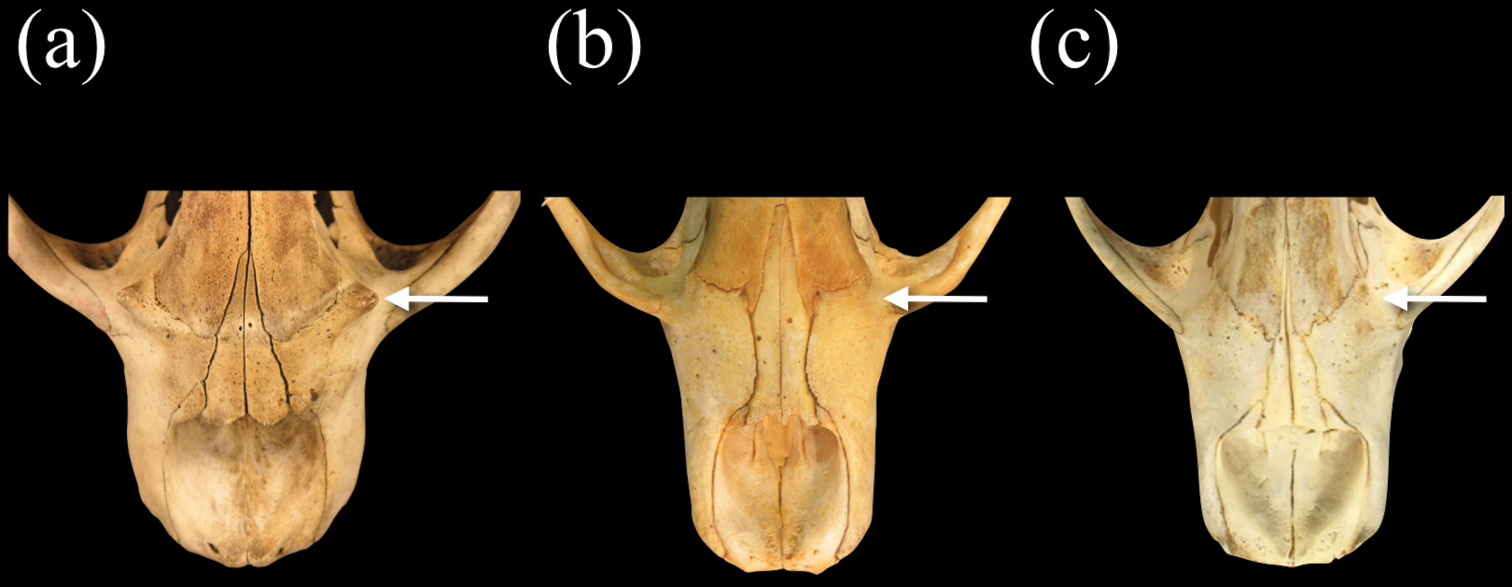
Ventral views of crania of a Monachus monachus b Neomonachus schauinslandi, and c Neomonachus tropicalis, showing the pterygoid region. Neomonachus exhibits a well-developed, laterally flared pterygoid hamulus that is visible in dorsal view. The hamulus may be spatulate (Neomonachus schauinslandi) or hook-like (Neomonachus tropicalis). The hamular process is absent or medially flared in Monachus, and is not visible in dorsal view.
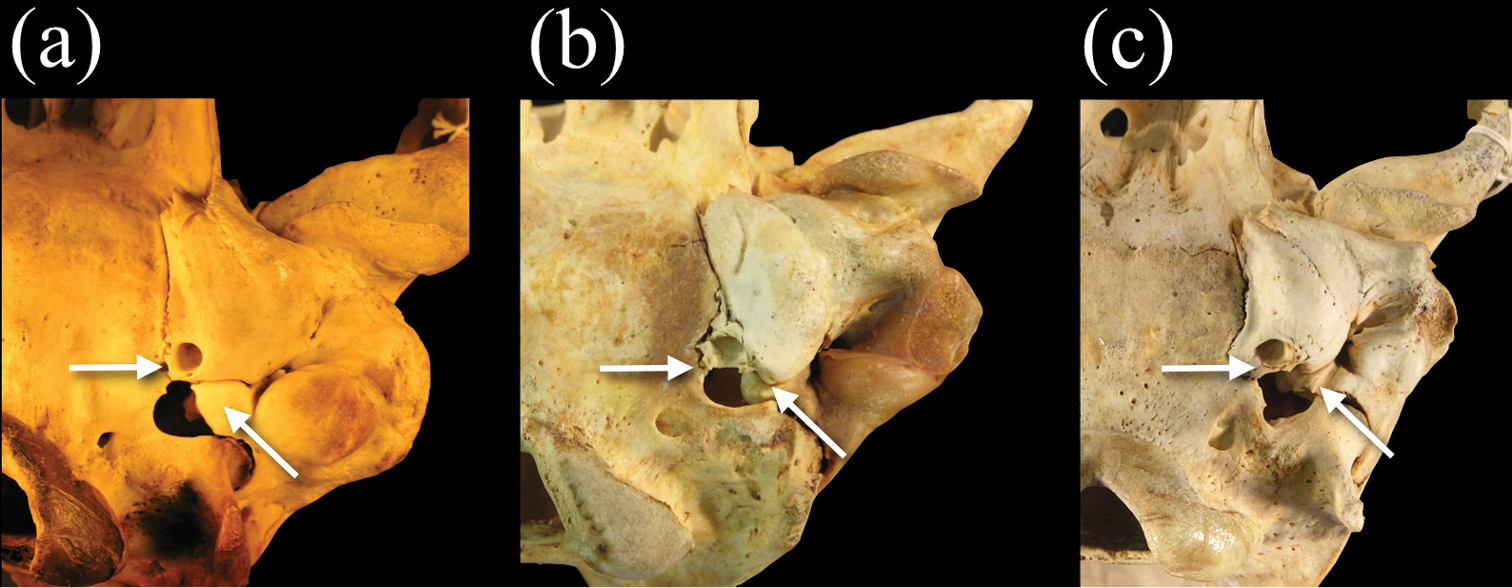
In ventral view, the morphology of the petromastoid (petrosal-mastoid) complex in relation to the auditory bulla in Neomonachus is diagnostic in comparison to Monachus.
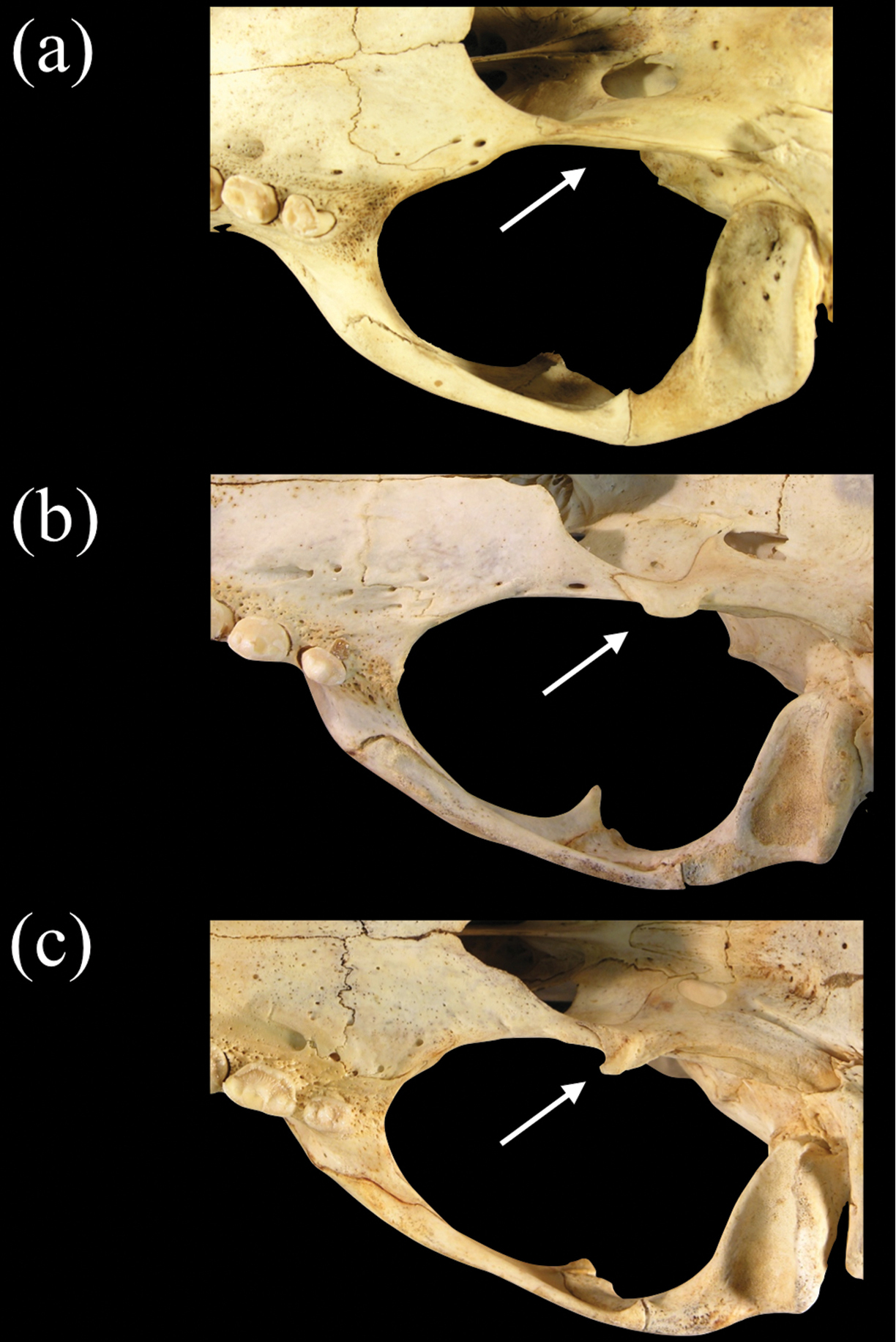
Posteroventral view of the basicranium and left bulla in a Monachus monachus b Neomonachus schauinslandi, and c Neomonachus tropicalis. The bulla of Monachus is bordered posteriorly by a ventrally expanded posterior portion of the petro-mastoid complex. The petrosal abuts the bulla’s posterior wall and in ventral view forms the entire lateral and anterolateral border of the posterior lacerate foramen. In Neomonachus, the posterior part of the petrosal is visible in the posterior lacerate foramen but remains superior to the bulla. In ventral view, this gives the impression that the anterior border of the posterior lacerate foramen is formed entirely by the bulla. The posterior carotid canal opens posteroventrally in Monachus. This apparently results from a relatively complete “ring-like” opening, formed by the bulla. This form of opening is apparent in subadult and juvenile Monachus, suggesting that it is not dependent on ontogenetic development or the robusticity of the Monachus cranium relative to Neomonachus. In contrast, the posterior carotid canal of Neomonachus opens directly posteriorly, the opening being an incomplete ring and the dorsal border formed by a flattening of the bulla, perhaps resulting from the bulla’s extension over the petrosal.
The upper incisor arcade of Neomonachus is sublinear, while that of Monachus appears slightly parabolic due to a more posteriorly set I3 (
Plots of mean upper (a) and lower (b) relative post-canine tooth size. Relative tooth size is computed by dividing the mesio-distal length of each tooth by the length of the 3rd premolar (which is typically largest) in the same row.
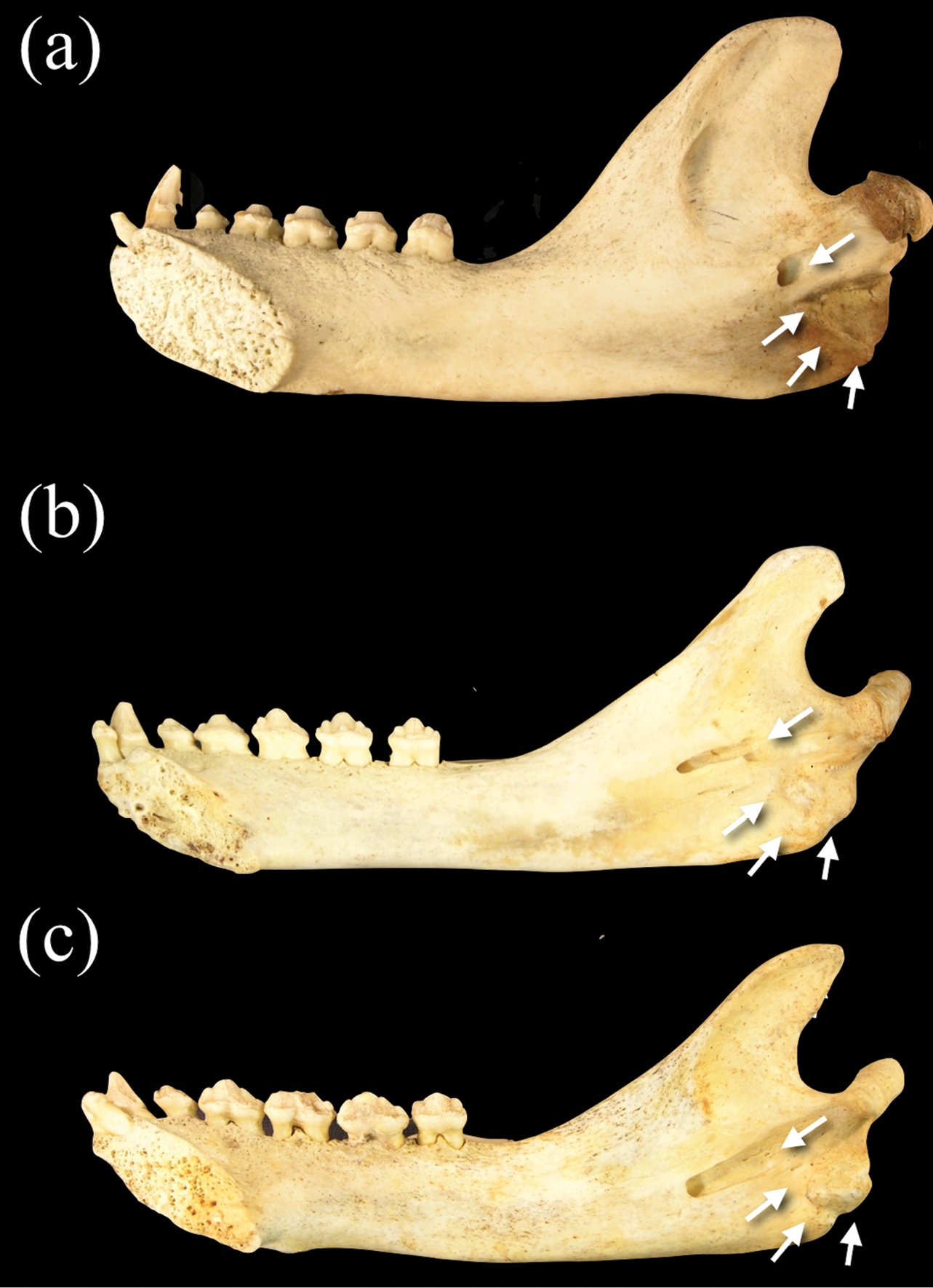
The mandible of Neomonachus is long and slender compared with that of Monachus, and the coronoid process is lower and less broad antero-posteriorly (
Medial view of right dentaries of a Monachus monachus b Neomonachus schauinslandi, and c Neomonachus tropicalis. The mandibular foramen is situated inferior to the mandibular notch in Monachus, and opens immediately to the medial surface of the ramus. In Neomonachus, the foramen is anteriorly displaced and is set in a groove or sulcus that extends from inferior to the mandibular notch. Also note the expanded rugose area for insertion of the pterygoid muscles in Monachus. This region is poorly developed in Neomonachus.
We obtained the first DNA sequence data from the recently extinct Caribbean monk seal. Based on phylogenetic analysis and divergence time estimation, we revealed that the Caribbean and Hawaiian monk seals form a well-supported monophyletic New World clade that diverged from the Mediterranean monk seal lineage ~ 6.3 Mya. Our results further implicate the closure of the Central American Seaway in the Late Pliocene as a driver of divergence between the Caribbean and Hawaiian monk seals, supporting a classical hypothesis in pinniped evolutionary biology. In combination, our morphological examinations of museum specimens and our phylogenetic analyses indicate that the substantial evolutionary divergence and trenchant morphological distinctions between the Mediterranean monk seal and the New World monk seals are similar to or greater than levels of molecular and morphological divergence between other sister phocid genera. Because no genus-level name has previously been proposed for the New World monk seals, we name and diagnose a new genus, Neomonachus, to accommodate the endangered Hawaiian and extinct Caribbean monk seals, leaving the Mediterranean monk seal as the sole species of Monachus.
Our findings and conclusions have broad significance for the two surviving species of monk seal. Because the Caribbean monk seal is already extinct, the elevation of the New World species to a new genus means that both extant monk seals (Monachus monachus and Neomonachus schauinslandi) are the sole remaining representatives of their respective genera—extremely distinctive seal lineages representing deep, independent evolutionary histories. Both species are critically endangered, with an extant populations of about 1000 individuals for Neomonachus schauinslandi and a heavily fragmented and widely distributed population of fewer than 500 individuals for Monachus monachus. Formal recognition of two genera for the living monk seals better indicates their true evolutionary, ecomorphological, and taxonomic uniqueness within the context of pinniped evolution, and this taxonomic change grants even greater poignancy to all efforts to conserve these endangered species.
We dedicate this paper to Judith E. King and André R. Wyss. We thank Karin Hönig for excellent technical support throughout the project, and Tara Fulton, Erich Fitzgerald, Annalisa Berta, Klaus-Peter Koepfli, Jennifer Schultz, Lauren Helgen, and Robert Fleischer for insightful discussion. We thank Darrin Lunde, John Ososky, Esther Langan, and Nicole Edmison (USNM), Eileen Westwig, Neil Duncan, Nancy Simmons, Rob Voss, and Ross MacPhee (AMNH), and William Stanley and Bruce Patterson (FMNH) for access to specimens and assistance with loans. We thank Lauren Helgen and Paige Engelbrektsson for assistance with figures, and Wieslaw Bogdanowicz and two anonymous reviewers for constructive criticisms on a previous version of the manuscript. GJS is supported by a Peter Buck Postdoctoral Fellowship at the National Museum of Natural History, KMH also thanks the staff of the Bishop Museum, Honolulu, Hawaii.
Amplicons covering cytb in this study.
Authors: Dirk-Martin Scheel, Graham J. Slater, Sergios-Orestis Kolokotronis, Charles W. Potter, David S. Rotstein, Kyriakos Tsangaras, Alex D. Greenwood, Kristofer M. Helgen
Data type: Picture
Explanation note: Sequence of the cytb gene and the resulting PCR amplicons with length in number of base pairs (bp). Asterisks indicate that the sequence extends over the 3’ or 5’ border of the target sequence.
Copyright notice: This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Alignment of Neomonachus tropicalis cytb with extant monk seal cytb sequences.
Authors: Dirk-Martin Scheel, Graham J. Slater, Sergios-Orestis Kolokotronis, Charles W. Potter, David S. Rotstein, Kyriakos Tsangaras, Alex D. Greenwood, Kristofer M. Helgen
Data type: Picture
Explanation note: The extinct Caribbean monk seal sequence was used as a reference. Dots indicate identity to the reference. Differences are shown as the base change relative to the reference. Numbering starts from the first base of the ATG start codon.
Copyright notice: This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Alignment of three Neomonachus tropicalis D-loop hypervariable region sequences (from USNM 100358, 102527, and 102534).
Authors: Dirk-Martin Scheel, Graham J. Slater, Sergios-Orestis Kolokotronis, Charles W. Potter, David S. Rotstein, Kyriakos Tsangaras, Alex D. Greenwood, Kristofer M. Helgen
Data type: Picture
Explanation note: The Neomonachus schauinslandi sequence was used as a reference. Dots indicate identity to the reference. Differences are shown as the base change relative to the reference. Numbering starts at the first base after the primer closest to the 5’ end. The X symbols indicate the break between the two amplicons that are approximately 200 bp apart. Sequences were generated from consensus sequences of 3–5 individual PCR product clones. Several products were cloned and sequenced from more than one amplicon to confirm that the differences observed were not DNA damage related or due to sequencing errors.
Copyright notice: This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Fifty percent majority-rule consensus tree based on 1000 bootstrap pseudoreplicates generated using the maximum parsimony phylogenetic optimality criterion.
Authors: Dirk-Martin Scheel, Graham J. Slater, Sergios-Orestis Kolokotronis, Charles W. Potter, David S. Rotstein, Kyriakos Tsangaras, Alex D. Greenwood, Kristofer M. Helgen
Data type: Picture
Explanation note: Values at nodes indicate the proportion of bootstrap trees (>50%) for which a particular bipartition was recovered.
Copyright notice: This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.