Citation: Baldwin CC, Robertson RD (2014) A new Liopropoma sea bass (Serranidae, Epinephelinae, Liopropomini) from deep reefs off Curaçao, southern Caribbean, with comments on depth distributions of western Atlantic liopropomins. ZooKeys 409: 71–92. doi: 10.3897/zookeys.409.7249
Collecting reef-fish specimens using a manned submersible diving to 300 m off Curaçao, southern Caribbean, is resulting in the discovery of numerous new fish species. The new Liopropoma sea bass described here differs from other western Atlantic members of the genus in having VIII, 13 dorsal-fin rays; a moderately indented dorsal-fin margin; a yellow-orange stripe along the entire upper lip; a series of approximately 13 white, chevron-shaped markings on the ventral portion of the trunk; and a reddish-black blotch on the tip of the lower caudal-fin lobe. The new species, with predominantly yellow body and fins, closely resembles the other two “golden basses” found together with it at Curaçao: L. aberrans and L. olneyi. It also shares morphological features with the other western Atlantic liopropomin genus, Bathyanthias. Preliminary phylogenetic data suggest that western Atlantic liopropomins, including Bathyanthias, are monophyletic with respect to Indo-Pacific Liopropoma, and that Bathyanthias is nested within Liopropoma, indicating a need for further study of the generic limits of Liopropoma. The phylogenetic data also suggest that western Atlantic liopropomins comprise three monophyletic clades that have overlapping depth distributions but different depth maxima (3–135 m, 30–150 m, 133–411 m). The new species has the deepest depth range (182–241 m) of any known western Atlantic Liopropoma species. Both allopatric and depth-mediated ecological speciation may have contributed to the evolution of western Atlantic Liopropomini.
Liopropoma aberrans, Liopropoma olneyi, submersible, Substation Curaçao, Deep Reef Observation Project (DROP), DNA barcoding, phylogeny, modes of speciation
Submersible diving to 300 m off Curaçao in the southern Caribbean as part of the Smithsonian Institution’s Deep Reef Observation Project (DROP) is expanding our knowledge of the deep-reef Caribbean fish fauna (
Liopropoma (Atlantic and Pacific), Bathyanthias (western Atlantic), and the monotypic Rainfordia (Indo-Pacific) form the monophyletic epinepheline serranid tribe Liopropomini (
The manned submersible Curasub (http://www.substation-Curacao.com) was employed to collect fishes and invertebrates during various field periods between 2011 and 2013. Fish specimens were collected using the fish anesthetic quinaldine pumped from a reservoir through a tube attached to one hydraulic arm of the sub and a suction hose (that uses the same pump as the anesthetic-delivery apparatus) attached to the other arm. The latter empties into a vented plexiglass cylinder attached to the outside of the sub. At the surface, the specimens were measured, photographed, tissue sampled (muscle biopsy from right side) and preserved. They were later x-rayed with a digital radiography system. Counts and measurements included in the description follow
Tissue samples for DNA Barcoding were stored in saturated salt-DMSO (dimethyl sulfoxide) buffer (
The label for each entry on the neighbor-joining tree is an assigned DNA number, and we include that number in the designation of type specimens and in some figure captions. Abbreviations used in DNA numbers are as follows: BAH–Bahamas, BLZ–Belize, CUR–Curacao, FLST–Florida Straits, FWRI–Florida Wildlife Research Institute, MBIO–Moorea Biocode Project, MCgroup–Matthew Craig, MOC–Miguel Oliver Caribbean Cruise, MOOP–Moorea Deep Reef, TOB - Tobago. GenSeq nomenclature for DNA sequences (
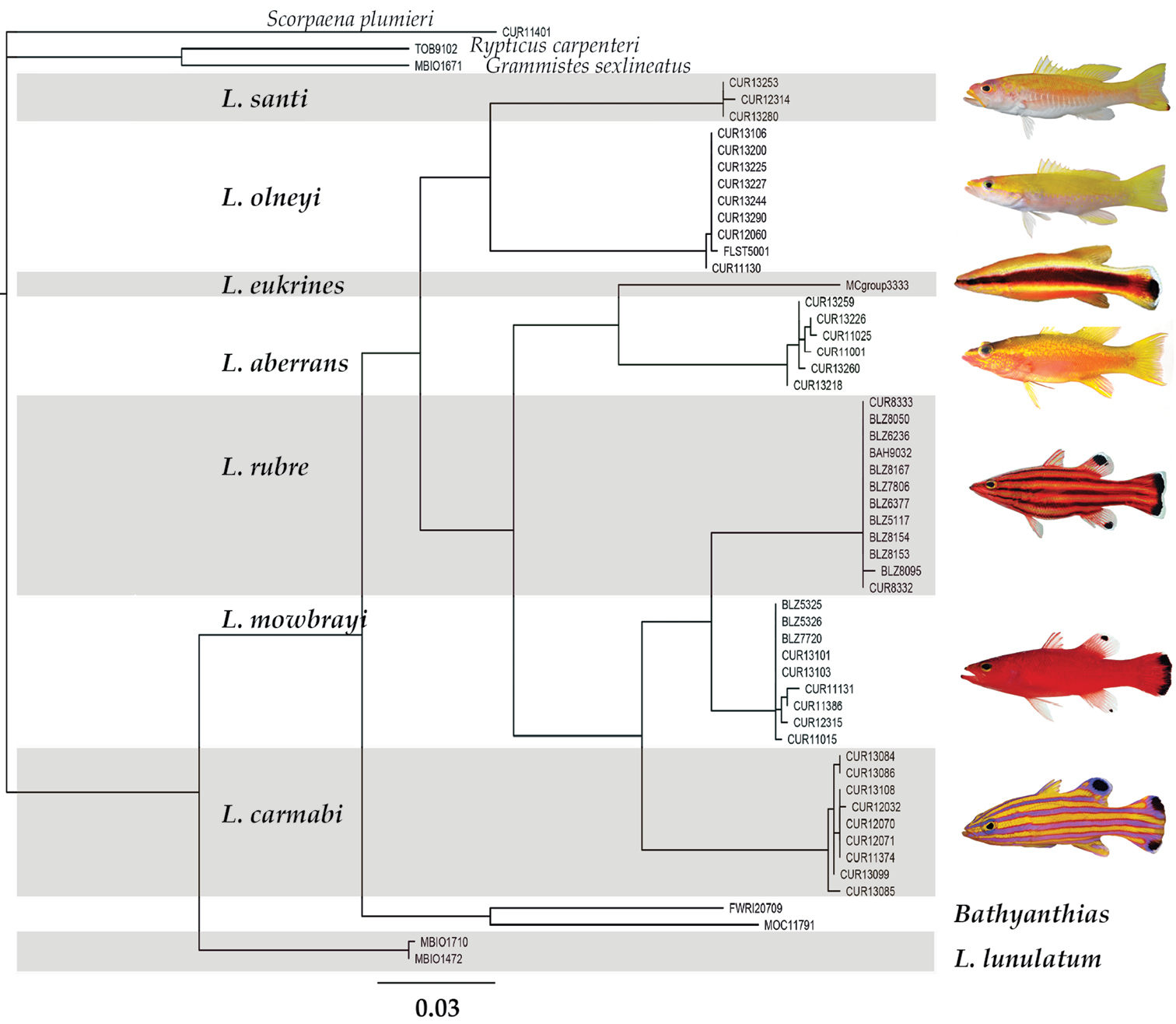
The neighbor-joining tree (Fig. 1) shows how individual specimens of western Atlantic Liopropoma sort into genetic lineages based on similarity in COI sequences. Lineages correlate well with currently recognized species. Genetic distance in COI between pairs of species of western Atlantic Liopropoma ranges from 5–18%, and distance between Liopropoma santi sp. n., and other western Atlantic Liopropoma species is 13–18% (Table 1). Average intraspecific variation for western Atlantic Liopropoma is 0–0.3%, 0.2% for Liopropoma santi.
Neighbor-joining tree derived from COI sequences for western Atlantic Liopropoma, the Indo-Pacific Liopropoma lunulatum, and related taxa. The tree was rooted on Scorpaena plumieri. Divergence represented by scale bar = 3%. Photographs of Liopropoma rubre and Liopropoma mowbrayi by James Van Tassell and Ross Robertson.
Average (and range) Kimura two–parameter distance summary for species of western Atlantic Liopropoma (7), Indo–Pacific Liopropoma (1), western Atlantic Bathyanthias (2), and outgroups Grammistes, Rypticus, and Scorpaena based on cytochrome c oxidase I (COI) sequences of individuals represented in the neighbor–joining tree in Figure 1. Intraspecific averages are shown in bold. “na” = not applicable (n=1).
| Liopropoma aberrans | Liopropoma carmabi | Liopropoma eukrines | Liopropoma lunulatum | Liopropoma mowbrayi | Liopropoma olneyi | Liopropoma rubre | Liopropoma santi sp. n. | Bathyanthias mexicanus | Bathyanthias sp. | Grammistes sexlineatus | Rypticus carpenteri | Scorpaena plumieri | |
| W. Atl. | W. Atl. | W. Atl. | Indo-Pacific | W. Atl. | W. Atl. | W. Atl. | W. Atl. | W. Atl. | W. Atl. | Indo-Pacific | W. Atl. | W. Atl. | |
| (n=6) | (n=9) | (n=1) | (n=2) | (n=9) | (n=9) | (n=12) | (n=3) | (n=1) | (n=1) | (n=1) | (n=1) | (n=1) | |
| Liopropoma aberrans | 0.3 (0–0.6) | ||||||||||||
| Liopropoma carmabi | 14.6 (14.2–15.2) | 0.2 (0–0.6) | |||||||||||
| Liopropoma eukrines | 10.5 (10.2–10.8) | 15.1 (14.8–15.6) | na | ||||||||||
| Liopropoma lunulatum | 14.8 (14.6–15.1) | 16.4 (16.1–16.9) | 14 (14.0–14.1) | 0.1 (0–0.2) | |||||||||
| Liopropoma mowbrayi | 12.2 (11.9–12.5) | 8.6 (8.2–9.1) | 13.5 (13.4–13.9) | 15.5 (15.3–15.8) | 0.2 (0–0.6) | ||||||||
| Liopropoma olneyi | 11.8 (11.5–12.1) | 13.6 (13.4–14.2) | 13 (12.8–13.1) | 14.7 (14.4–14.9) | 13.3 (13.0–13.7) | 0 (0–0.3) | |||||||
| Liopropoma rubre | 11.9 (11.5–12.4) | 10.5 (10.1–10.9) | 12.9 (12.8–13.3) | 15.8 (15.3–16) | 5.7 (5.3–6.0) | 12.5 (12.2–13.2) | 0 (0–0.3) | ||||||
| Liopropoma santi sp. n. | 16.2 (16.0–16.7) | 17.6 (17.1–18.4) | 15.2 (15.0–15.6) | 16.4 (16.0–16.9) | 15.5 (14.8–16.5) | 13.3 (13.0–13.5) | 16.4 (15.9–17.6) | 0.2 (0.0–0.3) | |||||
| Bathyanthias mexicanus | 16.1 (15.8–16.4) | 17.6 (17.1–18.4) | 15.2 (15.0–15.6) | 16.4 (16.0–16.9) | 13.9 (13.8–14.1) | 13.4 (13.4–13.5) | 13.8 (13.7–14.3) | 16.2 (15.9–16.9) | na | ||||
| Bathyanthias sp. | 16.8 (16.5–17) | 15.4 (15.2–15.7) | 15.4 (-) | 16 (15.9–16.1) | 14.8 (14.6–14.9) | 15.2 (15.1–15.4) | 14.6 (14.5–14.8) | 16.4 (16.2–16.7) | 13.7 (-) | na | |||
| Grammistes sexlineatus | 18.6 (18.3–18.8) | 17.9 (17.7–18.4) | 18 (-) | 17 (16.9–17.1) | 18.1 (18.0–18.5) | 18.9 (18.7–18.9) | 18 (17.8–18.3) | 20.8 (20.5–21.5) | 19.8 (-) | 15.9 (-) | na | ||
| Rypticus carpenteri | 17.3 (17.1–17.5) | 17.9 (17.9–18.1) | 14.8 (-) | 15.4 (15.4–15.5) | 16.8 (16.6–17) | 18.6 (18.4–18.6) | 17.4 (17.2–17.5) | 17.5 (17.5–17.6) | 18.9 (-) | 16.8 (-) | 13.2 (-) | na | |
| Scorpaena plumieri | 21.4 (21.2–21.5) | 21.6 (21.4–22) | 20.9 (-) | 19.8 (19.7–19.8) | 19.7 (19.5–20.4) | 20.8 (20.8) | 20.3 (20.2–20.6) | 24.5 (24.4–24.8) | 19.2 (-) | 20.7 (-) | 19.5 (-) | 19.6 (-) | na |
http://zoobank.org/83D20375-39CA-457D-8D54-127ACC3ED0B7
http://species-id.net/wiki/Liopropoma_santi
Figs 2–4, Spot-tail Golden BassCuraçao, southern Caribbean
USNM 426811, 116 mm SL, DNA #CUR 13253, Curasub submersible, sta. 13-14, southern Caribbean, Curaçao, off Substation Curaçao downline, near 12°05.069'N, 68°53.886'W, 241 m, quinaldine, 9 Aug 2013, C. C. Baldwin, D. R. Robertson, A. Driskell, B. van Bebber.
Paratypes. USNM 426813, 76.2 mm SL, DNA #CUR 13280, Curasub submersible, sta. 13–19, southern Caribbean, Curaçao, Playa Forti, Westpoint, 12°22.001'N, 69°9.005 W, 182 m, quinaldine, 15 Aug 2013, A. Schrier, N. Knowlton, R. Sant, B. van Bebber. USNM 414824, 42.0 mm SL, DNA #CUR 12314, Curasub submersible, sta. 12–19, southern Caribbean, Curaçao, east of Substation Curaçao downline, near 12°05.069'N, 68°53.886'W, 209 m, 15 Aug 2012, C. C. Baldwin, B. Brandt, B. van Bebber.
A liopropomin serranid with the following combination of characters: dorsal fin VIII, 13; anal fin III, 8; pectoral fin 15; total gill rakers on first arch (including rudiments) 20–21; lateral-line scales 47–48; length of first dorsal spine 2.9–4.2% SL; margin of spinous dorsal fin moderately indented posteriorly in adults (fourth spine 11–12% SL, fifth and sixth spines only slightly shorter than fourth—6.9–10% SL); depth at origin of dorsal fin 23–26% SL; least depth of caudal peduncle 11–13% SL; orbit diameter 9.4–12% SL; yellow-orange stripe externally on upper lip; series of approximately 13 white, chevron-shaped markings on ventral portion of trunk; reddish-black blotch on distal portion of lower caudal-fin lobe; inhabiting depths of 182–241 m.
Counts and measurements of holotype, if different from those of paratypes, are given in parentheses. Dorsal-fin rays VIII, 13; anal-fin rays III, 8; pectoral-fin rays (both sides) 15; pelvic-fin rays I, 5; principal caudal-fin rays 9+8=17; procurrent caudal-fin rays 9+9=18; pored lateral-line scales 48 (47), two additional pored scales present on base of caudal fin not included in total count; scales from lateral line to dorsal-fin origin 3 or 4 (3); gillrakers on first arch, including rudiments, 6+14-15 (6+14); upper limb with 3 rudiments + 3 rakers, lower limb with 11-13 rakers + 2-3 rudiments, total 20–21 (20); vertebrae 10 + 14.
Body proportions expressed as percentage of SL. Body depth at origin of dorsal fin 23–26 (26); body width just behind gill opening 11–14 (14); head length 37–39 (37); snout length 7.4–9.1 (9.1), relative length increasing with increasing SL; orbit diameter 9.4–12 (9.4) relative diameter decreasing with increasing SL; bony interorbital width 4.5–5.5 (5.5); upper-jaw length 16–18 (18); greatest depth of maxilla 5.0–6.1 (6.1); least caudal-peduncle depth 11–13 (13); caudal-peduncle length 22–24 (23); lengths of dorsal-fin spines: (I) 2.9–4.2 (4.2); (II) 11–12 (12); (III) 13–15 (14); (IV) 11–12 (11); (V) 6.9–10 (10); (VI) 6.9–8.2 (8.2); (VII) 5.0–7.5 (7.5); (VIII) 4.8–6.9 (6.9); longest dorsal soft ray the 11th, length 15–20 (20); length of 3rd anal-fin spine 6.9–9.3 (9.3); longest anal soft ray the 5th, length 15–17 (16); caudal-fin length 23–28 (23), relative length decreasing with increasing SL; pectoral-fin length 27–30 (27), fin reaching vertical between anus and origin of anal fin, falling short of anal fin in all specimens; pelvic-fin length 18–20 (19), fin reaching vertical through base of 6th dorsal-fin spine, well short of anus.
Interorbital region flat to slightly convex; mouth oblique, maxilla reaching vertical beyond posterior border of pupil; prominent bony projection on posteroventral corner of maxilla; lower jaw slightly projecting. Anterior nostril in thin, membranous tube, nostril situated just posterior to groove between tip of snout and premaxilla; posterior nostril a simple opening, nostril situated close to orbit (the distance approximately 1.5 nostril diameters). Lateral line strongly arched above pectoral fin, highest point below fourth and fifth dorsal-fin spines.
Trunk covered with ctenoid scales, scales becoming weakly ctenoid anteriorly and cycloid on head. Head fully scaled except over branchiostegal area. Holotype with short column of scales on dorsal-fin spines III and IV, scales on basal portion of membranes between spines VI and VIII, three rows of scales covering basal portion of soft dorsal fin, and some scales extending distally onto soft dorsal-fin membranes; paratypes with same squamation except no scales present on spinous dorsal fin, and 42.0-mm SL paratype having only basal scale rows on soft dorsal fin. In holotype and larger paratype, anal fin with two or three rows of scales basally and additional scales that extend distally onto fin membranes and cover most of fin. In smaller paratype, scales confined to basal portion of fin. Caudal fin completely scaled in holotype except for distal tips of rays; larger paratype with scales covering only proximal half of fin; smaller paratype with scales confined to basal portion of fin. Scales present on pectoral-fin base, and elongate scales present on proximal portion of fin. Scales present on pelvic-fin base and on proximal portion of fin; pelvic axillary scales present.
Jaw teeth small and depressible; upper and lower jaws with bands of villiform teeth, bands widest anteriorly, largest teeth in innermost row. Vomer with a chevron-shaped patch of small teeth. Palatines with several rows of small teeth in a long, narrow band. Opercle with three flattened spines, only the middle one conspicuous. Margin of upper limb of preopercle and angle with small serrations, lower limb smooth.
Prior to preservation (Figs 2, 3), background color of upper portions of trunk and caudal peduncle yellow, grading to pale pink around midbody, then to white ventrally; no abrupt transitions between those colors; many individual scales on upper half of body marked with orange spots in adults, densely so in holotype; a series of about 13 narrow, bright-white, chevron-shaped bars that point posteriorly present on lower half of trunk, series extending from just behind pectoral-fin base to vertical through center or posterior portion of anal fin; upper arms of white bars more strongly defined; nape yellow from dorsal midline ventrally to about mid-eye level (with some orange spots on scales in adults), grading anteriorly into an irregularly shaped area of purplish-pink over and behind eye, on upper portion of iris, and on snout; a yellow blotch present behind center of eye (in adults) and a smaller one present on dorsal midline of snout just anterior to orbit; iris mostly orange-yellow, grading to fine inner yellow ring; prominent, mostly deep-yellow (adults) or mostly orange (juvenile) stripe along outside of entire upper lip, this pigment spreading slightly above lip along anterior half of jaw in adults and merging with the pink/orange pigment on snout of juvenile; inside of lower lip with small blotch of yellow pigment in adults, inside of upper lip with stripe of yellow (adults) or orange (juvenile); photographic angle did not permit characterization of pigment on inside of lower lip of juvenile; lower jaw and lower two thirds of head white, with pinkish cast in holotype; in adults, dorsal fin with yellow spines and mostly white inter-spinous membranes; soft dorsal-fin rays yellow, membrane between anterior rays yellow, and membrane between rays of remainder of fin with small to large pale area centrally, size of pale area increasing posteriorly such that membrane between posteriormost rays completely pale; some rays and membranes in posterior portion of soft dorsal fin with pale rose pigment in smaller adult; a thin white margin extending along outer edge of entire dorsal fin, this margin appearing blue-white when fish photographed against black background (Fig. 3); in juvenile, inter-spinous membranes of dorsal fin mostly pale and soft dorsal mostly pale except for yellow stripe at the base and yellow stripe near outer margin of fin; caudal fin mostly yellow in holotype, central portion of fin with pale outer margin and with pale to pinkish-orange membranes between rays; thin pinkish-orange stripe present along dorsal and ventral margins of fin; distal tip of lower lobe with reddish-black blotch, a few thin streaks of black extending proximally from this blotch; pigment on caudal fin of smaller adult similar but with less pinkish-orange pigment, and caudal fin of juvenile mostly clear with a large, oval-shaped, oblique yellow blotch on outer half of both upper and lower lobes; dark spot on distal portion of ventral caudal lobe relatively larger in juvenile; anal fin white, with faint pinkish-yellow streak on first through fifth rays in holotype, little or no color in smaller adult and juvenile; pelvic fin white; pectoral fin translucent with pale pinkish-orange cast; general coloration most intense in the holotype and least intense in the juvenile.
Liopropoma santi sp. n., type series: A USNM 426811, holotype, 116 mm SL, DNA #CUR 13253 B USNM 426813, paratype, 76.2 mm SL, DNA #CUR 13280 C USNM 414824, paratype, 42.0 mm SL, DNA #CUR 12314.
Liopropoma santi sp. n., USNM 426811, holotype, 116 mm SL (photographed against a black background).
In alcohol (see Fig. 6A), body pale, the only pigment a dark blotch on distal tip of ventral caudal-fin lobe.
Known only from Curaçao, southern Caribbean.
Off Curaçao, Liopropoma santi is found from 182–241 m inhabiting rocky slopes and ledges. It retreats into small caves and crevices when approached and illuminated by the submersible. Figure 4 shows an in-situ photograph taken from the Curasub submersible at 204 m on a reef slope off Jan Theil Bay, Curaçao.
In-situ photograph of Liopropoma santi sp. n., taken from the Curasub submersible at 204 m on a reef slope off Jan Theil Bay, Curaçao, 5 Nov 2013. Photo courtesy of Substation Curaçao.
The specific name honors Roger Sant, who participated in the Curasub submersible dive at Playa Forti during which the USNM 426813 paratype was collected. Roger and Victoria Sant have provided generous funding to the Smithsonian Institution’s National Museum of Natural History for ocean-related activities.
“Spot-tail golden bass” is in reference to the dark spot on the lower lobe of the caudal fin, which, along with other characters, distinguishes Liopropoma santi from the two other species of western Atlantic Liopropoma that have predominantly golden coloration, Liopropoma aberrans and Liopropoma olneyi.
Counts and measurements of the three western Atlantic “golden basses” collected off Curaçao, Liopropoma santi, Liopropoma aberrans, and Liopropoma olneyi, are given in Table 2, representative images of the three are provided in Figure 5, and a summary of major differences among them appears in Table 3. An image of a freshly collected specimen of a species of the related genus Bathyanthias is also included in Figure 5 for comparative purposes. Liopropoma santi is easily distinguished from the others by color in life, especially by the presence of a yellow or orange stripe externally on the upper lip, a series of white chevron-shaped markings on the ventral portion of the trunk, and the reddish-black blotch on the distal portion of the lower caudal-fin lobe. The last also visually distinguishes Liopropoma santi from Liopropoma aberrans and Liopropoma olneyi in preservative. Liopropoma santi is further distinguished from both of those species by having more dorsal-fin rays, more gill rakers on the first arch, and usually a larger eye (Table 2). From Liopropoma aberrans, Liopropoma santi is further distinguished by having more pectoral-fin rays, a narrower body at the dorsal-fin origin, a narrower caudal peduncle, longer fourth-sixth dorsal-fin spines, and a more shallow indentation in the spinous dorsal fin (Tables 2, 3).
Comparison of the three species of “golden basses” off Curaçao and Bathyanthias sp. from Panama: A Liopropoma santi sp. n., USNM 426811, holotype, 116 mm SL, DNA #CUR 13280 B Liopropoma aberrans, USNM 426807, 102 mm SL, DNA #CUR 12226 C Liopropoma olneyi, USNM 426805, holotype, 84.3 mm SL, DNA #CUR 13200 D Bathyanthias sp., USNM 407791, 110 mm SL, DNA #MOC 11791.
Selected counts and measurements for the type series of Liopropoma santi sp. n., Liopropoma aberrans from Curaçao, and Liopropoma olneyi. Measurements are in percentages of SL. Data for Liopropoma aberrans are from Curaçao specimens examined in this study, those for Liopropoma olneyi are from
| Liopropoma santi | Liopropoma santi | Liopropoma santi | Liopropoma olneyi | Liopropoma aberrans | |
|---|---|---|---|---|---|
| Museum Catalog Numbers | USNM 426811 Holotype | USNM 426813 Paratype | USNM 414824 Paratype | See Appendix | See Appendix |
| SL (mm) | 116 | 76.2 | 42.0 | 53.2-84.3 | 64.8-116 |
| Dorsal Fin | VIII, 13 | VIII, 13 | VIII, 13 | IX, 11 | VIII, 12 |
| Pectoral Fin | 15 | 15 | 15 | 14-15 | 14 |
| Gill Rakers on First Arch | 6+14=20 | 6+14=20 | 6+15=21 | 5-6+12-13=17-19 | 5-6+11-13=17-18 |
| Orbit diameter | 9.4 | 10 | 12 | 7.8–9.4 | 7.4–8.7 |
| Body depth at dorsal-fin origin | 26 | 25 | 23 | 20–24 | 27–29 |
| Least depth of caudal peduncle | 13 | 13 | 11 | 13–15 | 16–17 |
| Length of dorsal-fin spine IV | 11 | 11 | 12 | 9.7–12 | 8.1–9.7 |
| Length of dorsal-fin spine V | 9.5 | 10 | 6.9 | 8.3–9.3 | 3.7–5.6 |
| Length of dorsal-fin spine VI | 8.2 | 7.9 | 6.9 | 7.3–8.9 | 3.6–5.6 |
Summary of differences in morphology and depth ranges among the three golden-colored Liopropoma species off Curaçao.
| Character | Liopropoma santi sp. n. | Liopropoma olneyi | Liopropoma aberrans |
|---|---|---|---|
| Relative body depth | Shallow (23–26% SL) | Shallow (20–24% SL) | Deeper (27–29% SL) |
| Dorsal fin indentation | Moderate (6th spine 7–8% SL) | Weak (6th spine 7–9% SL) | Strong (6th spine 4–6% SL) |
| Dorsal-fin rays | VIII, 13 | IX, 11 | VIII, 12 |
| Gill rakers on first arch | 20–21 | 17–19 | 17–18 |
| Orbit diameter (% SL) | 9.4–12 | 7.8–9.4 | 7.4–8.7 |
| White flank chevrons | yes | no | no |
| Body ground colors | yellow over white | yellow over white | yellow over orange |
| Yellow stripe through eye | no | yes | yes |
| Yellow-orange upper lip | yes | no | no |
| Yellow spots on body | no | adult & juvenile | juvenile only |
| Dark spot on lower caudal-fin lobe | yes | no | no |
| Depth range (m) | 181–241 | 133–193 | 98–149 |
Curaçao and Bahamas Liopropoma aberrans, however, appear to have different depth preferences, with Robins’ Liopropoma aberrans occurring deeper—229 m. At Curaçao, Liopropoma aberrans was collected between 98 and149 m and observed by us only within that depth range during nearly 100 submersible dives over a three-year period. This is unlikely to be due to effects of differences in habitat availability at the two locations, as Liopropoma santi and Liopropoma olneyi occur at deeper depths than Liopropoma aberrans at Curaçao.
Liopropoma santi differs from Poey’s and Robins’ Liopropoma aberrans in number of dorsal-fin rays (VIII, 13 vs. IX, 12 and VIII, 12, respectively) and shape of dorsal fin (with only a moderate indentation in spinous dorsal fin in Liopropoma santi, deep indentation in the others). It further differs from Robins’ Liopropoma aberrans in numbers of pectoral-fin rays (15 vs. 14) and gill rakers on the first arch (20–21 vs. 17–18), and color pattern (presence of diagnostic color features of Liopropoma santi–see Diagnosis–vs. absence). From other western Atlantic Liopropoma (Liopropoma carmabi [
Dorsal-fin counts of western Atlantic Liopropomini fishes. Data for Bathyanthias atlanticus, Bathyanthias cubensis, and Bathyanthias mexicanus are from
| SPINES | SOFT RAYS | ||||||
|---|---|---|---|---|---|---|---|
| VIII | IX | 11 | 12 | 13 | 14 | 15 | |
| Bathyanthias atlanticus | + | + | |||||
| Bathyanthias cubensis | + | + | |||||
| Bathyanthias mexicanus | + | + | + | ||||
| Bathyanthias roseus |
+ | + | |||||
| Liopropoma aberrans (Curaçao) | + | + | |||||
| Liopropoma aberrans (Cuba) | + | + | |||||
| Liopropoma aberrans (Bahamas) | + | + | |||||
| Liopropoma carmabi | + | + | + | ||||
| Liopropoma eukrines | + | + | |||||
| Liopropoma mowbrayi | + | + | |||||
| Liopropoma olneyi | + | + | |||||
| Liopropoma rubre | + | + | |||||
| Liopropoma santi sp. n. | + | + | |||||
1 As noted by
Counts of Liopropoma santi closely match those of Bathyanthias cubensis (
Comparison of Liopropoma and Bathyanthias: A Liopropoma santi sp. n., USNM 426811, holotype, 116 mm SL (photographed after preservation) B Bathyanthias cubensis (
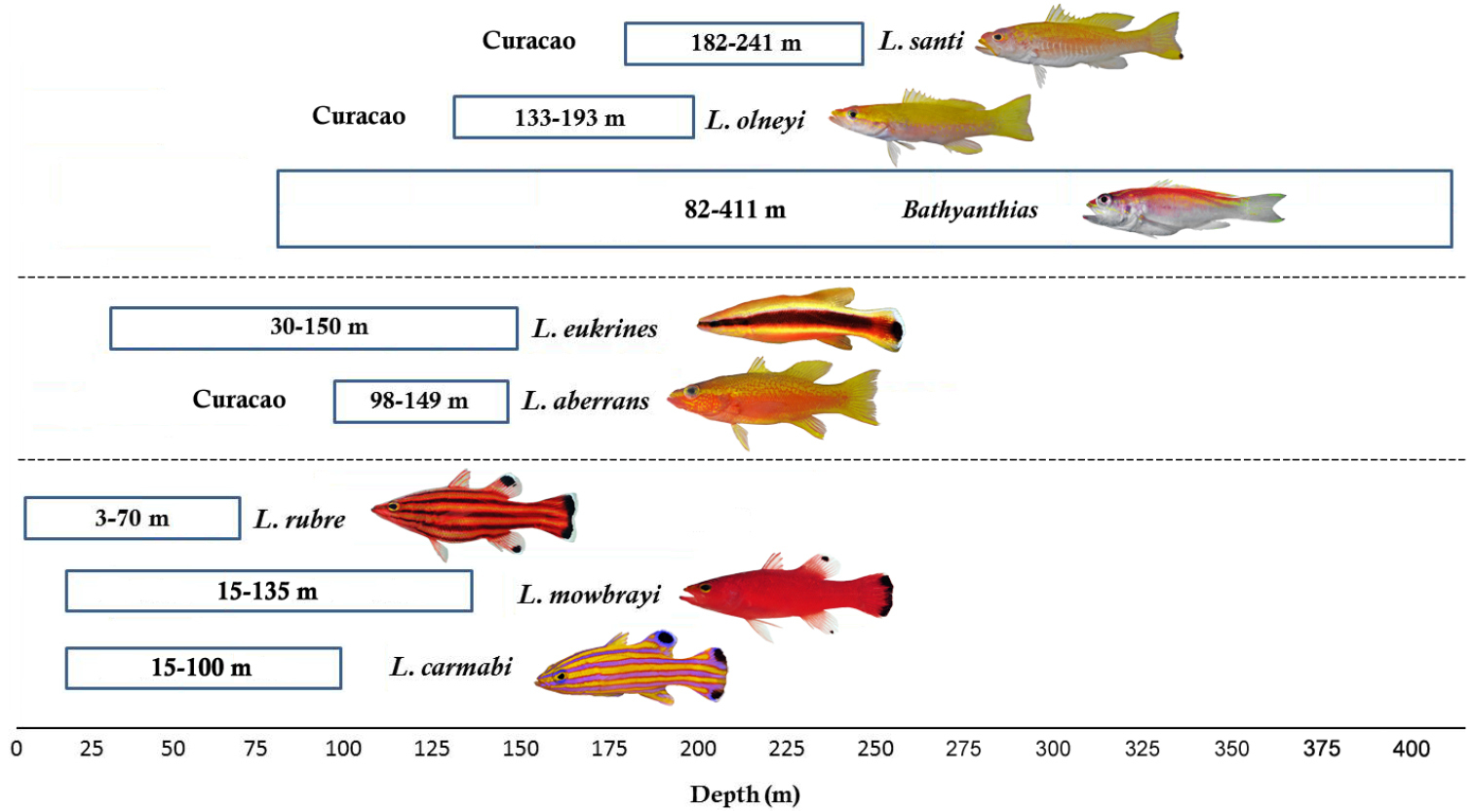
A combination of morphological and genetic differences supports the recognition of Liopropoma santi as a valid new species of Liopropoma. Liopropoma santi inhabits depths of 182-241 m off Curaçao, making it the deepest known Liopropoma species in the western Atlantic (Fig. 7). The shallower portion of its depth range overlaps the deeper portion of the depth range of Liopropoma olneyi (133–193 m), but with the exception of
Depth distributions of western Atlantic Liopropoma and Bathyanthias species that were included in the phylogenetic analysis (see Fig. 8). Photographs of Liopropoma rubre and Liopropoma mowbrayi by James Van Tassell and Ross Robertson.
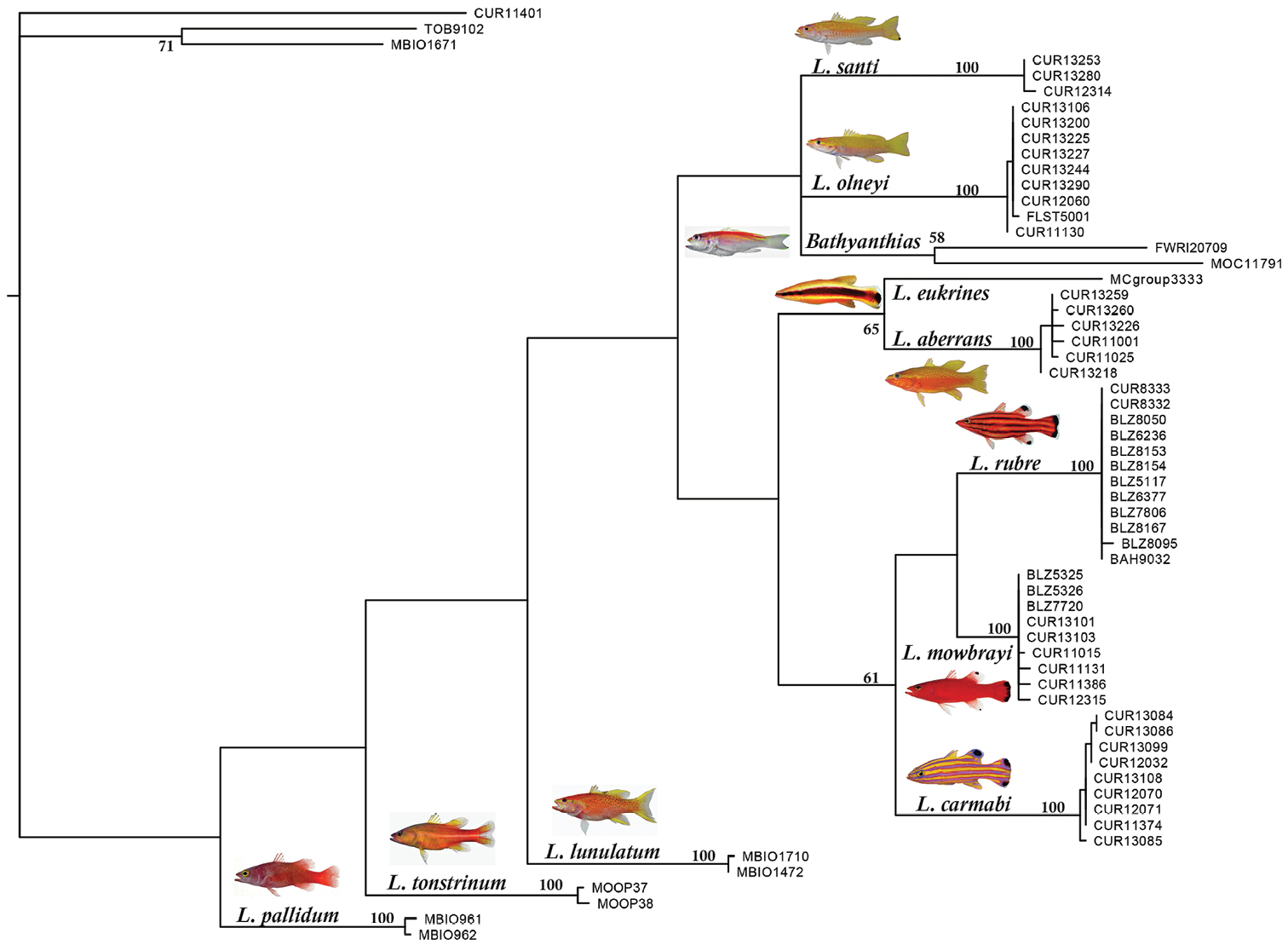
The strict consensus of a maximum parsimony analysis of the COI region among western Atlantic Liopropoma and related taxa. The tree was rooted on Scorpaena plumieri, (CUR11401), and the non-liopropomin serranids Rypticus carpenteri (TOB9102) and Grammistes sexlineatus (MBIO1671) were included as additional outgroups. Photographs of Liopropoma rubre and Liopropoma mowbrayi by James Van Tassell and Ross Robertson; photos of Liopropoma pallidum and Liopropoma lunulatum by Jeffrey Williams (from Encyclopedia of Life); photo of Liopropoma tonstrinum by Richard Winterbottom (from Encyclopedia of Life).
The COI data provide excellent support for the monophyly of species of western Atlantic Liopropoma but poor support for clades within the genus (see bootstrap values in Fig. 8). Nevertheless, the strict consensus (Fig. 8) suggests that western Atlantic liopropomins are monophyletic with respect to Indo-Pacific Liopropoma (Liopropoma lunulatum, Liopropoma tonstrinum, and Liopropoma pallidum in Fig. 8). A more robust phylogenetic hypothesis is needed that is derived from additional genes and more Indo-Pacific species of Liopropoma, but the COI data suggest a relationship between depth and monophyletic clades in western Atlantic Liopropomini that warrants further investigation. Members of the three clades of western Atlantic liopropomins identified in the phylogeny show a tendency to occupy different depth strata (3–135 m, 30–150 m, and 82–411 m). Based on our few specimens, it appears that Liopropoma santi has larger eyes than its sister species, Liopropoma olneyi (Table 3), which may represent an adaptation allowing Liopropoma santi to extend its range to greater depths. Among the three golden basses at Curaçao (Liopropoma aberrans, Liopropoma olneyi, Liopropoma santi), Liopropoma aberrans has the shallowest range and shows a tendency to have the smallest eyes (Table 3). Adaptation to life at different depths may have been involved in the speciation of this co-occuring species group. It may also be involved in the divergence between Liopropoma mowbrayi and Liopropoma rubre, which represent sister species that show only partial overlap in their depth ranges (Fig. 7) but broadly overlapping geographic ranges that incorporate most of the Caribbean and adjacent areas. Adaptation to use of different depth strata may also have been involved in the initial diversification of western Atlantic liopropomins into three clades that now occupy the same geographic area. Such parapatric ecological speciation, in which species diverge along environmental gradients, has been proposed for other marine fishes including Halichoeres (
Conversely, the sister species Liopropoma eukrines and Liopropoma aberrans overlap substantially in depth range but show a significant amount of geographic separation: Liopropoma eukrines is largely restricted to the Gulf of Mexico and southeastern USA, whereas Liopropoma aberrans is primarily Caribbean. However, there is one inconsistency in this pattern of either geographic or depth segregation among members of the same clade: Liopropoma carmabi and both species in its sister group, Liopropoma rubre and Liopropoma mowbrayi, have both geographic- and depth ranges that broadly overlap. Liopropomins have pelagic larvae, and allopatric speciation might be facilitated by larval dispersal to new areas. Possibly both ecological and allopatric speciation have occurred in the group, but, if so, more information on depth and geographic distributions, morphological traits associated with life at different depths, and evolutionary relationships is needed to estimate their relative roles. Depth and morphological information for the three members of the Liopropoma rubre clade collected at the same geographic location would be highly relevant in this regard. At Puerto Rico all three species in that clade occur on the same mesophotic reefs, where they reach the same maximum depth (
The phylogeny (Fig. 8) further suggests the need to reanalyze generic relationships within the Liopropomini, as Bathyanthias is embedded within western Atlantic Liopropoma. Morphologically, Liopropoma santi, Liopropoma olneyi, and Bathyanthias differ from other western Atlantic Liopropoma in having a smaller indentation in the margin of the dorsal fin, and those liopropomins lack body stripes and have similar pale orange/yellow/rose coloration. Four species of Bathyanthias have been described–Bathyanthias atlanticus (Schultz, 1860), Bathyanthias cubensis (Schultz, 1860), Bathyanthias mexicanus (Schultz, 1860), and Bathyanthias roseus
Of the three western Atlantic species of Liopropoma with depth distributions entirely below depths accessible using conventional scuba gear—Liopropoma aberrans, Liopropoma olneyi, and Liopropoma santi—two have been discovered only recently through submersible diving to 300 m off Curaçao in the southern Caribbean (Liopropoma olneyi and Liopropoma santi). More exploration of western Atlantic tropical mesophotic and other deep-reef depths is needed to fully document fish diversity even in well-studied taxonomic groups such as the Serranidae.
For contributing in various ways to this project, we thank the following (in alphabetical order): Bruce Brandt, Barry Brown, Cristina Castillo, Patrick Colin, Matthew Craig, Tico Christiaan, Dave Johnson, Rob Loendersloot, Dan Mulcahy, Diane Pitassy, Sandra Raredon, Rob Robins, Laureen Schenk, Adriaan Schrier, Ian Silver-Gorges, Raymond Simpson, Jennifer Strotman, Laura Tancredi, Barbara van Bebber, Lee Weigt. Victor Springer provided helpful comments on a draft of the manuscript. Funding for the Smithsonian Institution’s Deep Reef Observation Project was provided internally by the Consortium for Understanding and Sustaining a Biodiverse Planet to CCB, the Competitive Grants for the Promotion of Science program to CCB and DRR, the Herbert R. and Evelyn Axelrod Endowment Fund for systematic ichthyology to CCB, and externally by National Geographic Society’s Committee for Research and Exploration to CCB (Grant #9102-12). This is Ocean Heritage Foundation/Curacao Sea Aquarium/Substation Curacao (OHF/SCA/SC) contribution number 4.
Links between DNA voucher specimens, GenBank accession numbers, and cytochrome c oxidase subunit I (COI) sequences of Liopropoma santi sp. nov., related Liopropomini, and outgroup taxa.
| Catalog Number/DNA Number | GenBank No. | GenSeq Designation |
|---|---|---|
| Liopropoma santi sp. n. | ||
| USNM 426811, CUR 13253, Holotype | KJ526147 | Geneseq-1 COI |
| USNM 426813, CUR 13280, Paratype | KJ526148 | genseq-2 COI |
| USNM 414824, CUR 12314, Paratype | KJ526146 | genseq-2 COI |
| Liopropoma olneyi | ||
| USNM 426805, CUR 13200, Holotype | KF770874 | genseq-1 COI |
| USNM 406130, CUR 11130, Paratype | KF770856 | genseq-2 COI |
| USNM 414828, CUR 12060, Paratype | KF770862 | genseq-2 COI |
| USNM 426808, CUR 13225, Paratype | KF770876 | genseq-2 COI |
| USNM 426809, CUR 13227, Paratype | KF770878 | genseq-2 COI |
| USNM 426810, CUR 13244, Paratype | KF770879 | genseq-2 COI |
| USNM 426815, CUR 13290, Paratype | KF770882 | genseq-2 COI |
| USNM 422698, CUR13106, Paratype | KF770872 | genseq-2 COI |
| USNM 426868, FLST 5001, Paratype (larva) | KF770883 | |
| Liopropoma aberrans | ||
| USNM 406001, CUR 11001 | KF770853 | genseq-4 COI |
| USNM 406025, CUR 11025 | KF770855 | genseq-4 COI |
| USNM 426806, CUR 13218 | KF770875 | genseq-4 COI |
| USNM 426807, CUR 13226 | KF770877 | genseq-4 COI |
| USNM 426814, CUR 13259 | KF770880 | genseq-4 COI |
| USNM 426812, CUR 13260 | KF770881 | genseq-4 COI |
| Liopropoma carmabi | ||
| USNM 406374, CUR 11374 | KF770858 | genseq-4 COI |
| USNM 414825, CUR 12032 | KF770861 | genseq-4 COI |
| USNM 414826, CUR 12070 | KF770863 | genseq-4 COI |
| USNM 414827, CUR 12071 | KF770864 | genseq-4 COI |
| USNM 413959, CUR 13084 | KF770866 | genseq-4 COI |
| USNM 413960, CUR 13085 | KF770867 | genseq-4 COI |
| USNM 413961, CUR 13086 | KF770868 | genseq-4 COI |
| USNM 422694, CUR 13099 | KF770869 | genseq-4 COI |
| USNM 422687, CUR 13108 | KF770873 | genseq-4 COI |
| Liopropoma eukrines | ||
| SIO 01-11, MCgroup 3333 | KF770885 | genseq-4 COI |
| Liopropoma mowbrayi | ||
| USNM 420350, BLZ 5325 | JQ840569 | genseq-4 COI |
| USNM 420349, BLZ 5326 | JQ840570 | genseq-4 COI |
| BLZ 7720 (photo voucher only) | JQ841243 | genseq-5 COI |
| USNM 406015, CUR 11015 | KF770854 | genseq-4 COI |
| USNM 406131, CUR 11131 | KF770857 | genseq-4 COI |
| USNM 406386, CUR 11386 | KF770859 | genseq-4 COI |
| USNM 414815, CUR 12315 | KF770865 | genseq-4 COI |
| USNM 422684, CUR 13101 | KF770870 | genseq-4 COI |
| USNM 422675, CUR 13103 | KF770871 | genseq-4 COI |
| Liopropoma rubre | ||
| USNM 414697, BAH 9032 | KF770852 | genseq-4 COI |
| USNM 419340, BLZ 5117 | JQ840571 | genseq-4 COI |
| USNM 416331, BLZ 6236 | JQ840899 | genseq-4 COI |
| USNM 416379, BLZ 6377 | JQ840900 | genseq-4 COI |
| USNM 416009, BLZ 7806 | JQ841244 | genseq-4 COI |
| USNM 415207, BLZ 8050 | JQ841640 | genseq-4 COI |
| USNM 415226, BLZ 8095 | JQ841637 | genseq-4 COI |
| USNM 415180, BLZ 8153 | JQ841638 | genseq-4 COI |
| USNM 415181, BLZ 8154 | JQ841641 | genseq-4 COI |
| USNM 415244, BLZ 8167 | JQ841639 | genseq-4 COI |
| USNM 414498, CUR 8332 | JQ842192 | genseq-4 COI |
| USNM 414499, CUR 8333 | JQ842193 | genseq-4 COI |
| Liopropoma lunulatum (Pacific) | ||
| MBIO 1710 (no photo or specimen voucher) | JQ431889 | no classification |
| MNHN 2008-1023, MBIO 1472 | JQ431888 | genseq-4 COI |
| Liopropoma tonstrinum (Pacific) | ||
| USNM 425632, MOOP37 | KJ526149 | genseq-4 COI |
| USNM 425630, MOOP38 | KJ526150 | genseq-4 COI |
| Liopropoma pallidum (Pacific) | ||
| MNHN 2009-0793, MBIO 961 | JQ431890 | genseq-4 COI |
| MNHN 2009-0794, MBIO 962 | JQ431891 | genseq-4 COI |
| Bathyanthias mexicanus | ||
| FWRI 20709 (DNA number same) | KF770884 | genseq-4 COI |
| Bathyanthias sp. | ||
| USNM 407791, MOC 11791 | KF770886 | genseq-4 COI |
| Outgroup Taxa | ||
| Grammistes sexlineatus | ||
| MNHN 2008-1105, MBIO 1671 | JQ431776 | genseq-4 COI |
| Rypticus carpenteri | ||
| USNM 401296, TOB 9102 | JN828097 | genseq-4 COI |
| Scorpaena plumieri | ||
| USNM 406401, CUR 11401 | KF770860 | genseq-4 COI |