(C) 2013 Mohammed Ahmed. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Ahmed M, van de Vossenberg BTLH, Cornelisse C, Karssen G (2013) On the species status of the root-knot nematode Meloidogyne ulmi Palmisano & Ambrogioni, 2000 (Nematoda, Meloidogynidae). ZooKeys 362: 1–27. doi: 10.3897/zookeys.362.6352
The root-knot nematode Meloidogyne ulmi is synonymised with Meloidogyne mali based on morphological and morphometric similarities, common hosts, as well as biochemical similarities at both protein and DNA levels. M. mali was first described in Japan on Malus prunifolia Borkh.; and M. ulmi in Italy on Ulmus chenmoui W.C. Cheng. Morphological and morphometric studies of their holo- and paratypes revealed important similarities in the major characters as well as some general variability in a few others. Host test also showed that besides the two species being able to parasitize the type hosts of the other, they share some other common hosts. Our study of the esterase and malate dehydrogenase isozyme phenotypes of some M. ulmi populations gave a perfectly comparable result to that already known for M. mali. Finally, phylogenetic studies of their SSU and LSU rDNA sequence data revealed that the two are not distinguishable at DNA level. All these put together, leave strong evidences to support the fact that M. ulmi is not a valid species, but a junior synonym of M. mali. Brief discussion on the biology and life cycle of M. mali is given. An overview of all known hosts and the possible distribution of M. mali in Europe are also presented.
Morphological, morphometrics, esterase, malate dehydrogenase, Japan, Italy, Mierenbos, Malus prunifolia, Ulmus chenmoui, SSU rDNA, LSU rDNA
The genus Meloidogyne comprises all root-knot nematodes. It contains over 100 described species (
In 2000, Palmisano and Ambrogioni described from Italy the root-knot nematode, Meloidogyne ulmi, from Ulmus chenmoui W.C. Chengon which it was found to induce large galls. For many years, elm remained the only known host of Meloidogyne ulmi. According to the authors, the tree at the type locality was introduced from the Netherlands as part of a breeding programme focussed on resistance to the Dutch elm disease. The Netherlands, like many other countries in Europe and North America, as well as New Zealand, has for years been battling against the notorious Dutch elm disease. It was for this reason that the former Dutch phytopathological laboratory “Willie Commelin Scholten” (WCS), based in Baarn, was mandated with the research on Dutch elm disease. This breeding programme was later on moved to Wageningen at the former institute the Dorschkamp Research Institute for Forestry & Landscape Planning; and in this programme, elm trees from all over the world were tested. Trial field “Mierenbos”, a part of the Dorschkamp Research Institute for Forestry & Landscape Planning, was used for growing and improving resistant elm cultivars. It was from this trial field that resistant elm seedlings were sent to ten other European countries at the end of the breeding programme, among others, Italy in 1992 (
The first observation of galls on elm trees was already in 1960 at Baarn, and the associating nematode was diagnosed as Meloidogyne arenaria (Neal, 1889) Chitwood, 1949 by the National Plant Protection Organization (
In 2006, root samples of the dying type host apple containing Meloidogyne mali from the type locality in Japan sent by Dr. Takayuki Mizukubo were received at the Dutch National Plant Protection organization. About the same time, galled root samples of the type host of Meloidogyne ulmi were obtained from Italy. They were propagated and maintained on the Ulmus × hollandica variety “Wredei”. Juveniles isolated from the Japanese apple root samples were used for sequencing and the resulting SSU rRNA sequence was discovered to be almost identical to that of Meloidogyne ulmi (
The original description of Meloidogyne ulmi differentiates it from Meloidogyne mali on the basis of characters that generally show high intraspecific variations. With the original description being the only paper written about Meloidogyne ulmi, all the known features so far are ones from the original description. On Meloidogyne mali, however, there have been quite a lot of research on the hosts, life cycle, ecology, detailed morphology, as well as their variations within species (
The objectives of this current research, therefore, are:
i) to evaluate the morphological similarities between Meloidogyne mali and Meloidogyne ulmi.
ii) to search for other host plants, than Ulmus sp. present at the trail field “Mierenbos”.
iii) to test Meloidogyne ulmi on selected host plants on which Meloidogyne mali is already known to reproduce.
iv) analyze their biochemical similarities, at the protein and DNA levels.
Paratype slides of Meloidogyne ulmi used for morphological and morphometric studies were obtained from Dr. Z.A. Handoo of the USDA Nematode Collection. In addition to these, we obtained Meloidogyne mali specimens on slides taken to the USDA by Dr. Ichinohe during his visit in 1958 as well as additional specimens of males, second-stage juveniles and females stored in formalin that were only recently isolated from root samples sent to USDA by Ichinohe during that same visit. Also, by courtesy of Dr. Hiromichi Sakai and Shigeyuki Sekimoto, paratypes that were deposited at the National Agriculture and Food Research Organization, Agricultural Research Center (Kannondai, Tsukuba, Ibaraki, Japan), the then Central Agricultural Experiment Station were also obtained. All slides were observed using compound light microscope (DM 2500, LEICA) equipped with differential interference contrast (DIC), and camera (DC 300F, LEICA) for taking images. Comparisons of morphological and morphometric characters were based on the most differential characters, previously used by
This is a combination of sampling undertaken in 2011 and 2012 at the former trial field “Mierenbos” on several plant species and a subsequent greenhouse experiment involving some important plant species already associated with Meloidogyne mali in previous studies (
Esterase and malate dehydrogenase isozymes were analysed for Meloidogyne ulmi sampled at “Mierenbos”, following the method described by
Electrophoresis was run using the PhastSystem (Pharmacia Ltd, Uppsala, Sweden) and the gels were subsequently stained in a Petri dish and placed in an incubator at 37 °C. Staining for non-specific esterase (EST; EC 3.1.1.1) was allowed to stand for 60 minutes whiles that for malate dehydrogenase (MDH; EC 1.1.1.37) stayed for 5 minutes.
Following staining, the gels were rinsed with distilled water and fixed for 5 minutes in a 10% acetic acid / 10 % glycerol/ 80 % distilled water solution. Pictures of the gels were taken by placing them on a glass surface illuminated from below.
Already published sequences of both Meloidogyne mali and Meloidogyne ulmi (
Nucleic acids were isolated from single male or second-stage juveniles of Meloidogyne ulmi populations taken from “Mierenbos” and type populations kept in culture at the Dutch National Plant Protection Organization on an elm tree (Ulmus × hollandica Mill “Wredei”). Genomic DNAs were isolated from these samples using High Pure PCR Template Preparation Kit (www.roche-applied-science.com, Cat. No. 11796828001, Version 16.0) protocol for isolation of nucleic acids from Mammalian Tissue with slight modification in the first step to suit nematode DNA isolation (150 µl tissue lysis buffer added to 50 µl sterile water containing nematodes, minimum protease incubation time of 16 hours and elution volume of 50 µl).
Amplification of 1000 base pairs (bp) of the large ribosomal subunit (LSU) (28S) was performed using primer set 28–81for (forward) 5’-TTAAGCATATCATTTAGCGGAGGAA-3’ and 28–1006rev (reverse) 5’-GTTCGATTAGTCTTTCGCCCCT-3’ described by
To amplify the nearly full length sequence of the small ribosomal subunit (SSU) (18S), two partially overlapping fragments were generated using three universal primers and one nematode-specific primer (1912R) described by
PCR products were purified after amplification using QIAquick PCR Purification Kit (Qiagen), and the genomic DNA concentration measured using a ND1000 spectrophotometer (NanoDrop). This was followed by a cycle sequencing reaction in a final volume of 20 µl (molecular grade water (Sigma-Aldrich, Saint Louis, USA), BigDye Terminator v1.1, 1x sequencing buffer, purified PCR product and 0.5 µM template-specific forward or reverse primers). Cycling reactions were carried out separately for each of forward and reverse primers. The reaction programme was set for 1 min at 96°C, 25x (10 sec 96°C, 5 sec 50°C, 2.5 min 60°C), 1 min 20°C. The cycle sequence products were cleaned up using DyeEx 2.0 Spin Kit (Qiagen) and run on a multi-capillary 3500 Genetic Analyzer DNA sequencer (Applied Biosystems, Carlsbad, USA).
Trace files of D2–D3 expansion segments of 28S and 18S-rRNA genes were assembled into contigs and amplification primer sequences trimmed using Geneouis 6.1.6 (Biomatters, New Zealand). Additional trimming was performed when needed, to obtain high quality consensus sequence data. Conflicts in the consensus sequence were assessed visually and corrected where possible. The sequences were aligned with selected sequences of other species from GeneBank using MAFFT alignment (
The following are the observations made on selected features considered to be the most differential for species discrimination among members of the genus Meloidogyne (
The various forms of type specimens of Meloidogyne mali and Meloidogyne ulmi studied and their sources.
| Form | Meloidogyne mali | Meloidogyne ulmi | ||
|---|---|---|---|---|
| Sex/stage | Source | Sex/stage | Source | |
| Holotype | 1 female | Shigeyuki Sekimoto | – | – |
| Allotype |
1 male | Shigeyuki Sekimoto | – | – |
| Paratype | 17 perineal patterns | Dr. Zafar A. Handoo | 2 PP & 1 anterior part | Dr. Zafar A. Handoo |
| Paratype | 3 males | Dr. Zafar A. Handoo | 2 males | Dr. Zafar A. Handoo |
| Paratype | 4 juveniles | Dr. Zafar A. Handoo | 3 juveniles | Dr. Zafar A. Handoo |
| Paratype | 1 male | Dr. Hiromichi Sakai | – | – |
| Paratype | 1 juvenile | Dr. Hiromichi Sakai | – | – |
*Slide marked as allotype, however at present not recognized by the ICZN, i.e. it is a paratype.
Plant species included in the host plant test with Meloidogyne ulmi in the greenhouse.
| Family | Plant species |
|---|---|
| Brassicaceae | Brassica oleracea L. var. Gemmifera (cabbage) |
| Brassica pekinensis (Lour.) Rupr. (celery cabbage) | |
| Rosaceae | Malus pumila “M9” (apple) |
| Rosa hybrida L. (rose) | |
| Fabaceae | Trifolium repens L. (white clover) |
| Solanaceae | Solanum lycopersicum L. (tomato) |
| Ulmaceae | Ulmus glabra Huds. (wych elm) |
Observations of the differential characteristics of female, male and second-stage juveniles of Meloidogyne mali and Meloidogyne ulmi types in comparison with their interpretation in the original description.
| Species | Meloidogyne mali | Meloidogyne ulmi | ||
|---|---|---|---|---|
| Character | Described | Observed | Described | Observed |
| Female | ||||
| Stylet knobs | Well developed knobs that tend to slope backward or forward in the ratio of 16 to 8 | Rounded to pear-shaped knobs, set off and slightly anteriorly concave to backwardly sloping | Knobs rounded to transversely ovoid, slightly concave anteriorly | Rounded knobs that are slightly anteriorly concave and offset |
| Perineal pattern | Oval, made up of smooth striae, finely spaced, dorsal arch low and flat. Phasmids large, lateral field clearly marked with single or double incisures | Oval, dorsal arch low to slightly high, rounded to square shaped. Phasmids distinct. Lateral field marked by breaks in the striae or showing indistinct lateral lines |
Oval, dorsal arch flattened to medium high, rounded or somewhat square, phasmids conspicuous, lateral field indistinct or marked by folds, sometimes by lateral lines on one or both sides | Oval, dorsal arch low to slightly high, rounded to square shaped. Phasmids distinct. Lateral field marked by breaks in the striae or showing indistinct lateral lines |
| Male | ||||
| Head shape | _ | Head weakly offset, head cap low and slightly narrower than the postlabial region. Postlabial incisures absent | Head slightly set off, labial cap shallowly rounded, one-fifth to one-fourth as high as postlabial region | Head weakly offset, head cap low and slightly narrower than the postlabial region. Postlabial incisures absent |
| Stylet knobs | Knobs rounded | Backwardly sloping with rounded to pear shaped knobs | Knobs rounded to pear shaped more or less backwardly sloping | Backwardly sloping with rounded to pear shaped knobs |
| Second-stage juvenile | ||||
| Stylet knobs | Knobs backwardly sloped | Small rounded knobs, slightly backwardly sloping | Knobs rounded and set off from shaft | Small rounded knobs, backwardly sloping |
| Tail shape | Short | Conical with a broad to finely pointed tip | Conical, tapering to a finely rounded almost pointed terminus or broader and rounded at the tip | Conical and tapers to a broadly or finely pointed tip |
| Hyaline tail part | _ | Constrictions present along hyaline part, length short or long. Anterior part clearly delimitated | Cuticular constrictions present along hyaline tail terminus, variable in length | Constrictions present along hyaline part, anterior part clearly delimitated |
The general shape of the perineal pattern in both species studied ranged from low rounded to oval. The dorsal arch of Meloidogyne mali and Meloidogyne ulmi was mostly low rounded with very few instances where some specimens showed relatively high square patterns. Lateral field was marked by change in direction or breaks in striae resulting in what would appear as weak lateral lines. The double lateral lines mentioned in the description of Meloidogyne mali were not observed in the studied specimens. The interphasmidial distance in both species was about the same as their corresponding vulva slit lengths. As mentioned in the description, the phasmids were distinct but did not appear large when observed at the correct focus. However, attempting to observe them at the same (relatively deeper) focus as the vulva slit makes them look larger and even farther apart than they really are, due to the diagonally sloping phasmid canals.
Same variations in stylet knobs shape as described in Meloidogyne mali were observed for Meloidogyne mali paratypes i.e. slightly backwardly sloping to anteriorly concave, with the former being the more frequent. Such variations, however, cannot be mentioned about the Meloidogyne ulmi paratype since there was only a single anterior part of the female on the slides we obtained. We therefore supplemented it with specimens taken from samples from the “Mierenbos”, where the type host originated from. This population showed similar variation as described for Meloidogyne mali, but not reported in Table 3. Our observation of the shape of the stylet itself was typical of the genus, i.e. straight shaft with a slightly dorsally curved cone.
S-E pore position measured from the anterior end showed quite some variations. Nevertheless, all measurements taken for both species fell within the range described for Meloidogyne ulmi. This character in Meloidogyne mali description was measured on the basis of the number of annuli counted from the anterior end to the one bearing the S-E pore.
Under light microscope, both species have the same head outline. This was already illustrated in the descriptions of the two species (
The stylet moderately slender. Conus with bluntly pointed tip. The shaft width the same along its entire length, although in some specimens it appeared to be broader close to the junction with the knobs. Individual knobs rounded to pear shaped. Knobs backwardly sloping in both species.
The lateral field marked by four incisures. In most of the specimens studied, the outer lines appeared areolated along most part of the body. No difference in the number of lateral incisures was observed along the body, except at the anterior part where it reduces to two and gradually fades out further anterior.
Although not considered to be of any diagnostic significance, this character remained fairly consistent in all specimens studied. The hemizonid always occurred anterior to the S-E pore, at slightly varying distances.
Examination of the second-stage juvenile characters was based on six Meloidogyne mali type specimens and two of Meloidogyne ulmi.
Head slightly set off from the rest of the body, with a low lip. Post-labial region lacking any annule.
Stylet somewhat slender, with conus terminating in a fine tip, in both species. Stylet knobs small and rounded; slightly backwardly sloping.
Almost all our average measurements were within the range of those in the original descriptions (Tables 4–6). In the case of Meloidogyne ulmi, measured values of the female anterior part are based only on a single paratype specimen. Useful differential characters like the stylet length, stylet knob widths and stylet knob heights showed great similarities. From the perineal patterns, measurements of all the known important features also gave comparable values with those in the descriptions. Interphasmidial distance and the vulva slit were in most cases similar, rarely significantly different. In Meloidogyne mali, these two measurements were almost identical. There was however, a slight difference in these two measurements from Meloidogyne ulmi types (Table 4), probably because only two perineal patterns were studied.
Morphometrics of Meloidogyne mali and Meloidogyne ulmi females in comparison with the original descriptions. All measurements are in µm and in the form: mean ± sd. (range).
| Species | Meloidogyne mali | Meloidogyne ulmi | ||
|---|---|---|---|---|
| Character | Described | Observed | Described | Observed |
| N | 25 | 17 | 30 | 2 |
| Body length | 847 (684–1044) |
762 ± 115 (608–890) |
771 ± 140 (568–1043) |
_ |
| Body width | 660 (540–864) |
570 ± 122 (372–700) |
618 ± 152 (357–1007) |
_ |
| Neck length | 166 ± 43.7 (90–252) |
165 ± 62 (60–265) |
165 ± 67 (58–382) |
205 |
| Neck diameter | _ | 100 ± 34.3 (48–160) |
_ | 152 |
| Stylet length | 15 (13–17) |
11.9 ± 1.8 (7.7–15.4) |
14.2 ± 1.0 (12.0–15.7) |
13.4 |
| Stylet knob height | _ | 1.6 ± 0.3 (1–2.2) |
1.8 ± 0.6 (1.1–3.9) |
1.6 |
| Stylet knob width | _ | 3.2 ± 0.4 (2.6–3.8) |
3.5 ± 0.7 (2.6–5.2) |
3.2 |
| DGO | 5.5 (4–7) |
4.3 ± 1.5 (2.2–6.7) |
4.6 ± 0.8 (3.3–6.5) |
3.9 |
| S-E pore | _ | 32.8 ± 5.5 (25–43.5) |
32.3 ± 7.8 (15.7–45.1) |
36.5 |
| Metacorpus | 110 (90–147) |
103 ± 7.9 (90–117) |
_ | 92.8 |
| Metacorpus length | 39 (32–44) |
40.4 ± 4.5 (32–50) |
42.6 ± 6.5 (32.7–58.8) |
39.7 |
| Metacorpus diameter | 49 (40–73) |
39.7 ± 6.5 (29–47) |
40.9 ± 7.0 (31.3–59.0) |
36.5 |
| Metacorpus valve length | 12 (11–13) |
13.0 ± 1.1 (11.5–15.4) |
12.4 ± 1.0 (11.1–14.4) |
17.9 |
| Metacorpus valve width | 10 (9–11) |
9.5 ± 1.3 (7–11.2) |
9.7 ± 1.4 (7.2–12.4) |
10.2 |
| Vulva – anus distance | 17 ± 1.8 (14–22) |
19.1 ± 2.6 (12.8–22.4) |
19.0 ± 1.9 (15.0–22.2) |
19.2 ± 0.8 (18.8–19.8) |
| Interphasmidial distance | 22 ± 3.5 (17–29) |
24.8 ± 4.7 (17.6–35.2) |
19.2 ± 3.8 (13.7–28.9) |
22.4 ± 4.5 (19.2–25.6) |
| Level of phasmids to vulva | 25 ± 2.4 (19–31) |
27.4 ± 2.8 (24–33.9) |
25.1 ± 4.2 (15.7–39.2) |
27.6 ± 0.9 (26.9–28.2) |
| Level of phasmids to anus | _ | 8.1 ± 2.5 (5.1–12.8) |
6.9 ± 2.2 (2.6–15.7) |
8.7 ± 1.3 (7.7–9.6) |
| Vulva slit length | 18 ± 2.5 (12–24) |
24.4 ± 3.3 (16–28.2) |
22.0 ± 2.9 (17–28.7) |
24.5 ± 1.1 (23.7–25.3) |
*Two perineal patterns and a single anterior part.
Morphometrics of Meloidogyne mali and Meloidogyne ulmi males in comparison with the original descriptions. All measurements are in µm and in the form: mean ± sd. (range).
| Species | Meloidogyne mali | Meloidogyne ulmi | ||
|---|---|---|---|---|
| Character | Described | Observed | Described | Observed |
| N | 25 | 3 | 30 | 2 |
| Body length | 1447 (1270–1630) |
1428 ± 41.0 (1380–1452) |
1462 ± 190 (1053–1776) |
1455 ± 64 (1410–15.0) |
| Body width | 38 (30–47) |
34.8 ± 9.6 (28.0–41.8) |
36.9 ± 4.3 (26.6–48.4) |
40.5 ± 0.7 (40.0–41.0) |
| Body width at stylet knobs | _ |
15.7 ± 0.4 (15.4–16.0) |
16.7 ± 1.3 (14.5–19.4) |
17.6 ± 1.1 (16.8–18.4) |
| Body width at S.E pore | _ |
24.5 ± 6.1 (20.2–28.8) |
27.6 ± 2.6 (23.0–31.5 |
28.4 ± 0.6 (28.0–28.8) |
| Stylet length | 20 18–22 |
19.9 ± 1.8 (18.6–21.1) |
19.4 ± 1.2 (17.5–22.9) |
19.9 ± 0.9 (19.2–20.5) |
| Stylet knob height | _ |
_ | 2.4 ± 0.1 (2.0–3.0) |
_ |
| Stylet knob width | _ |
_ |
3.9 ± 0.4 (3.0–4.8) |
_ |
| DGO | 8 (6–13) |
9 |
6.3 ± 0.8 (4.8–8.5) |
7.4 ± 0.5 (7.0–7.7) |
| S–E pore |
_ |
135.2 |
147 ± 18.8 (97–187) |
139 ± 7.1 134–144 |
| Metacorpus |
_ |
98 ± 21.5 (83–114) |
99 ± 9.8 (76–119) |
83 ± 12.7 (134–144) |
| Spicule | 32 (28–35) |
28.1 ± 10.3 (20.8–35.3) |
33.8 ± 1.9 (30.0–37.5) |
29.8 |
| Gubernaculum | 8.5 (7–10) |
10.1 |
9.0 ± 0.9 (7.3–9.8) |
8.4 ± 0.4 (8.1–8.7) |
| Testis length | 788 (540–970) |
803 ± 125.9 (714–892) |
716 ± 167 (324–977) |
752 ± 79 (696–808) |
| T | 55 (34–65) |
55.3 ± 8.6 (49.2–61.4) |
48.7 ± 9.7 (27.9–71) |
52 ± 3.2 (49–54) |
* Distance from anterior end to S-E pore.
** Distance from anterior end to valve plate of median bulb.
Morphometrics of Meloidogyne mali and Meloidogyne ulmi second-stage juveniles in comparison with the original descriptions. All measurements are in µm and in the form: mean ± sd. (range).
| Species | Meloidogyne mali | Meloidogyne ulmi | ||
|---|---|---|---|---|
| Character | Described | Observed | Described | Observed |
| N | 25 | 5 | 30 | 3 |
| Body length | 418 (390–450) |
420 ± 21.7 (390–446) |
413 ± 20.6 (373–460) |
384 ± 9.5 (374–394) |
| Body width | 14.5 (14–16) |
14.0 ± 1.1 (12.2–15.2) |
14.2 ± 1.8 (12.1–18.2) |
12.7 ± 3.8 (8.6–16.9) |
| Body diameter at anus | 8.5 (7–9) |
9.4 ± 1.8 (8.3–11.5) |
8.4 ± 1.0 (7.3–10.9) |
6.5 ± 0.7 (6.0–7.0) |
| Stylet length | 14 (12–15) |
12.1 ± 1.5 (10.9–13.8) |
10.0 ± 0.8 (8.5–11.1) |
11.1 ± 0.6 10.6–11.5 |
| Tail length | 31 (30–34) |
30.2 ± 4.3 (24.3–33.9) |
31.3 ± 3.1 (24.2–37.5) |
24.2 ± 0.8 (23.4–25.0) |
| Tail terminus length | _ | 7.0 ± 2.1 (5.1–9.8) |
8.2 ± 1.8 (4.8–12.7) |
5.7 ± 1.1 (4.5–6.7) |
| Anus–primordium | _ | 139 ± 11.4 (125–152) |
_ | 126 ± 21.8 (111–151) |
| a | 28.5 (27–31) |
30.2 ± 3.2 (27.1–34.8) |
29.5 ± 3.4 (22.3–35.5) |
32.5 ± 10.9 (23.4–44.7) |
| c | 13.3 (12–15) |
14.4 ± 2.3 (12.5–17.4) |
13.3 ± 1.2 (11.5–16.6) |
16.3 ± 0.7 (15.8–16.8) |
| c’ | 3.7 (3–5) |
3.3 ± 0.7 (2.5–3.9) |
3.7 ± 0.5 (2.5–4.7) |
3.7 ± 0.5 (3.3–4.1) |
Three male paratypes of Meloidogyne mali and two of Meloidogyne ulmi were measured. Some of the studied characters were only visible enough for measurement on single specimens, and therefore for such characters absolute values were taken rather than their averages. The stylet knobs widths and heights were examples of characters for which measurements were not taken on either species (Table 5) due to the fact that they appeared slightly degenerated on all slides, and so may give false measurements. Nevertheless there were still some outstanding similarities in the stylet length, spicule length and DGO between the observed and the described values.
Similar to the observations made in the females and the males, the second-stage juvenile morphometrics was very comparable in many features between the two species studied. There was, however an unaccountable difference between stylet length as described for Meloidogyne mali (14µm (12–15µm)) and that which was measured (12.1 ± 1.5µm (10.9–13.8µm)). Values of body width at anus level between the two descriptions were very similar. Somemeasurements taken from Meloidogyne ulmi, likewise were quite similar to those in the original descriptions, particularly, the Demanian ratios a and c’, while others such as stylet lengths showed slight differences (Table 6).
The ability of Meloidogyne ulmi to reproduce on various plant species was examined under greenhouse conditions. Host statuses of the various plants used in the greenhouse test are presented in Table 7. Meloidogyne ulmi population from “Mierenbos” used as inoculum was able to induce galls and reproduce on both Ulmus glabra and Ulmus hollandica ‘belgica’. The apple ‘M9’ also had galls which contained egg-laying females. Although galls were induced by Meloidogyne ulmi on Brassica oleracea var. gemmifera, most of these galls contained small non-gravid females whose development seemed to have ceased at some point. Therefore, it is herein not considered as a host. There were no galls on Rosa hybrida and the other cabbage species, Brassica pekinensis.
Plants species identified as host of Meloidogyne ulmi from the green house experiments and field survey at “Mierenbos”.
| Family | Plant species |
|---|---|
| Greenhouse test | |
| Ulmaceae | Ulmus glabra Huds. |
| Ulmus hollandica ‘belgica’ | |
| Rosaceae | Malus pumila ‘M9’ |
| Solanaceae | Solanum lycopersicum L. |
| Field hosts | |
| Sapindaceae | Acer pseudoplatanus L. |
| Balsaminaceae | Impatiens parviflora DC. |
| Asteraceae | Taraxacum officinale F.H. Wigg. |
| Dyopteridaceae | Dryopteris filix-mas (L.) Schott |
| Dryopteris carthusiana (Vill.) H.P. Fuchs | |
| Fagaceae | Fagus sylvatica L. |
| Quercus robur L. | |
| Geraniaceae | Geranium robertianum L. |
| Rosaceae | Geum coccineum Lindl. |
| Rubus idaeus L. | |
| Sorbus aucuparia L. | |
| Taxaceae | Taxus baccata L. |
| Ulmaceae | Ulmus davidiana var. japonica Rehder |
| Urticaceae | Urtica dioica L. |
A compilation of all known host plants of Meloidogyne mali to date.
| Family | Plant species | Reference |
|---|---|---|
| Rosaceae | Malus pumila Mill. | |
| Malus prunifolia Borkh. | ||
| Malus sieboldii Rehd. | ||
| Malus pumila “M9” | Current work | |
| Prunus yedoensis Matsum | ||
| Rosa hybrida Hort. | ||
| Geum coccineum Lindl. | Current work | |
| Vitis vinifera L. | ||
| Rubus idaeus L. | Current work | |
| Sorbus aucuparia L. | Current work | |
| Moraceae | Morus bombycis Koidz. | |
| Ficus carica L. | ||
| Maclura tricuspidata (Carriere) Bureau | ||
| Broussonetia papyrifera (L.) Vent | ||
| Broussonetia kazinoki Seibold. | ||
| Fagaceae | Castanea crenata Seib. Et Zucc | |
| Fagus sylvatica L. | Current work | |
| Quercus robur L. | Current work | |
| Ulmaceae | Ulmus davidiana var. japonica | |
| Ulmus chenmoui W.C. Cheng | ||
| Ulmus glabra Hud. | ||
| Ulmus × hollandica “belgica” | Current work | |
| Sapindaceae | Acer palmatum Thunb. | |
| Acer pseudoplatanus L. | Current work | |
| Trifolium repens L. | ||
| Taxaceae | Taxus baccata L. | Current work |
| Fabaceae | Impatiens parviflora DC. | Current work |
| Solanaceae | Solanum lycopersicum L. | |
| Solanum melongena L. | ||
| Capsicum annuum L. | ||
| Cucurbitaceae | Cucumis sativus L. | |
| Cucurbita spp. | ||
| Citrillus vulgaris Schrad. Ex Eckl. & Zeyh. | ||
| Cruciferae | Brassica pekinensis Rupy. | |
| Brassica oleracea var. capitata L. | ||
| Brassica napus var. oleifera L. | ||
| Compositae | Arcutium lappa L. | |
| Taraxacum officinale F.H. Wigg. | Current work | |
| Umbelliferae | Daucus carota var. sativa L. | |
| Leguminaceae | Glycine max (L.) Merr. | |
| Urticaceae | Urtica dioica L. | Current work |
| Dryopteridaceae | Dryopteris filix-mas (L.) Schott | Current work |
| Dryopteris carthusiana (Vill.) H.P. Fuchs | Current work | |
| Geraniaceae | Geranium robertianum L. | Current work |
Additionally, samples collected during 2011 and 2012 revealed that Meloidogyne ulmi is able to parasitize one or more species of Acer (Aceraceae), Impatiens (Balsaminaceae), Taraxacum (Compositae), Dryopteris (Dryopteridaceae), Fagus (Fagaceae), Quercus (Fagaceae), Geranium (Geraniaceae), Geum (Rosaceae), Rubus (Rosaceae), Sorbus (Rosaceae), Taxus (Taxaceae), Urtica (Urticaceae), as shown in Table 7.
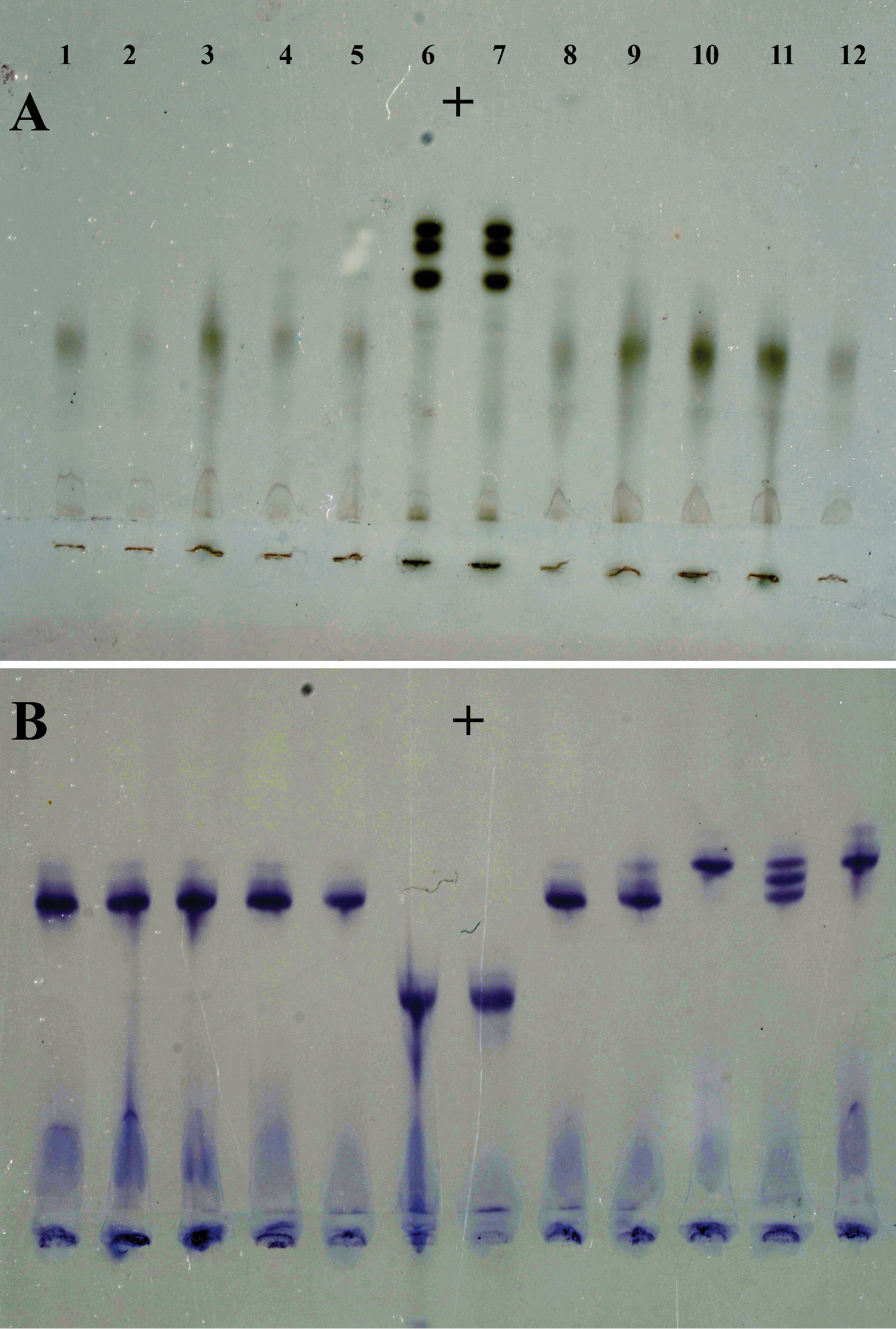
Samples taken from the trial field “Mierenbos” all gave the same type of esterase isozyme pattern of weak single bands, corresponding to the VS1 type (
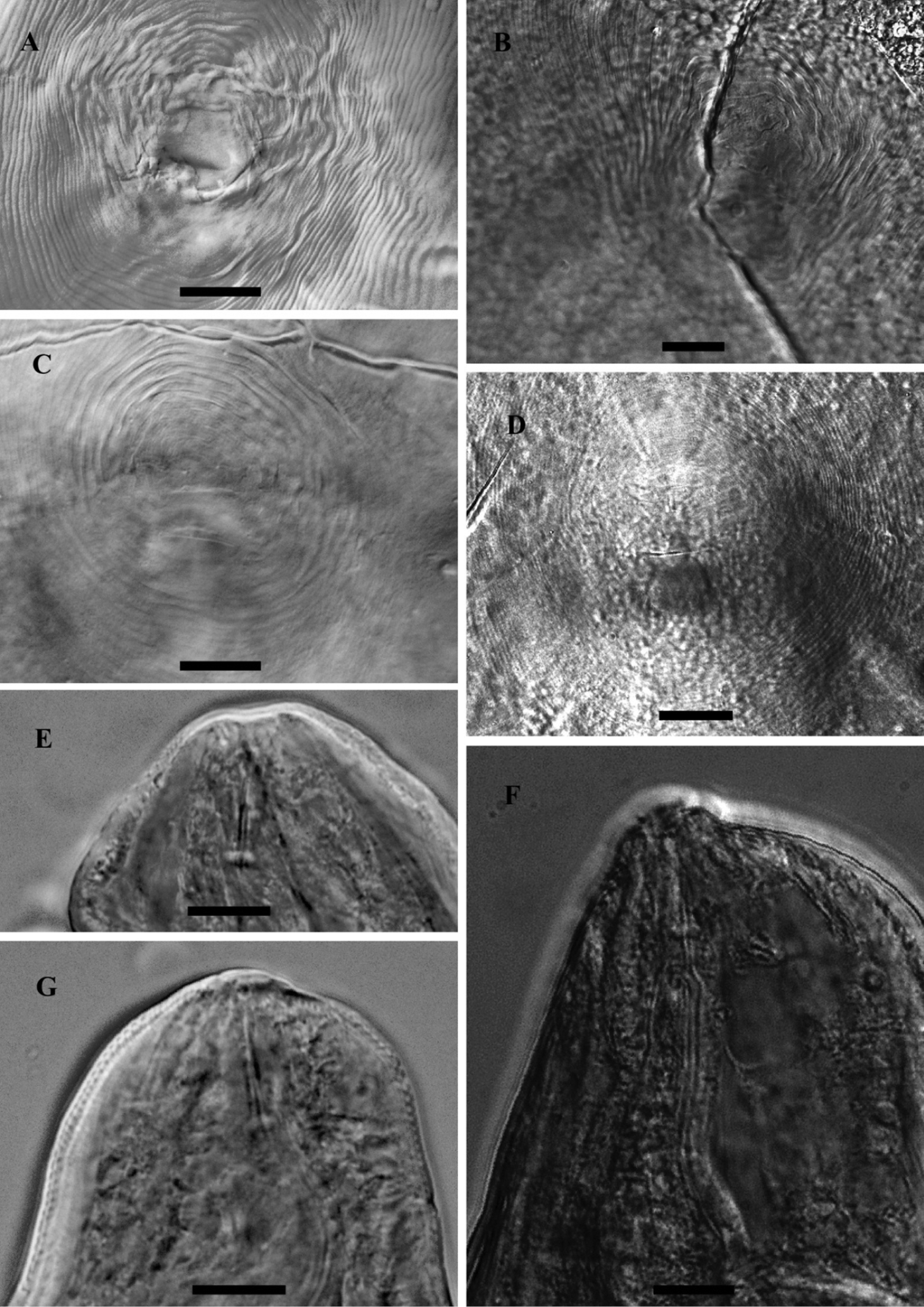
LM photograph of perineal patterns and anterior parts of female Meloidogyne mali (A, C, E, G) and Meloidogyne ulmi (B, D, F), bar = 10 µm.
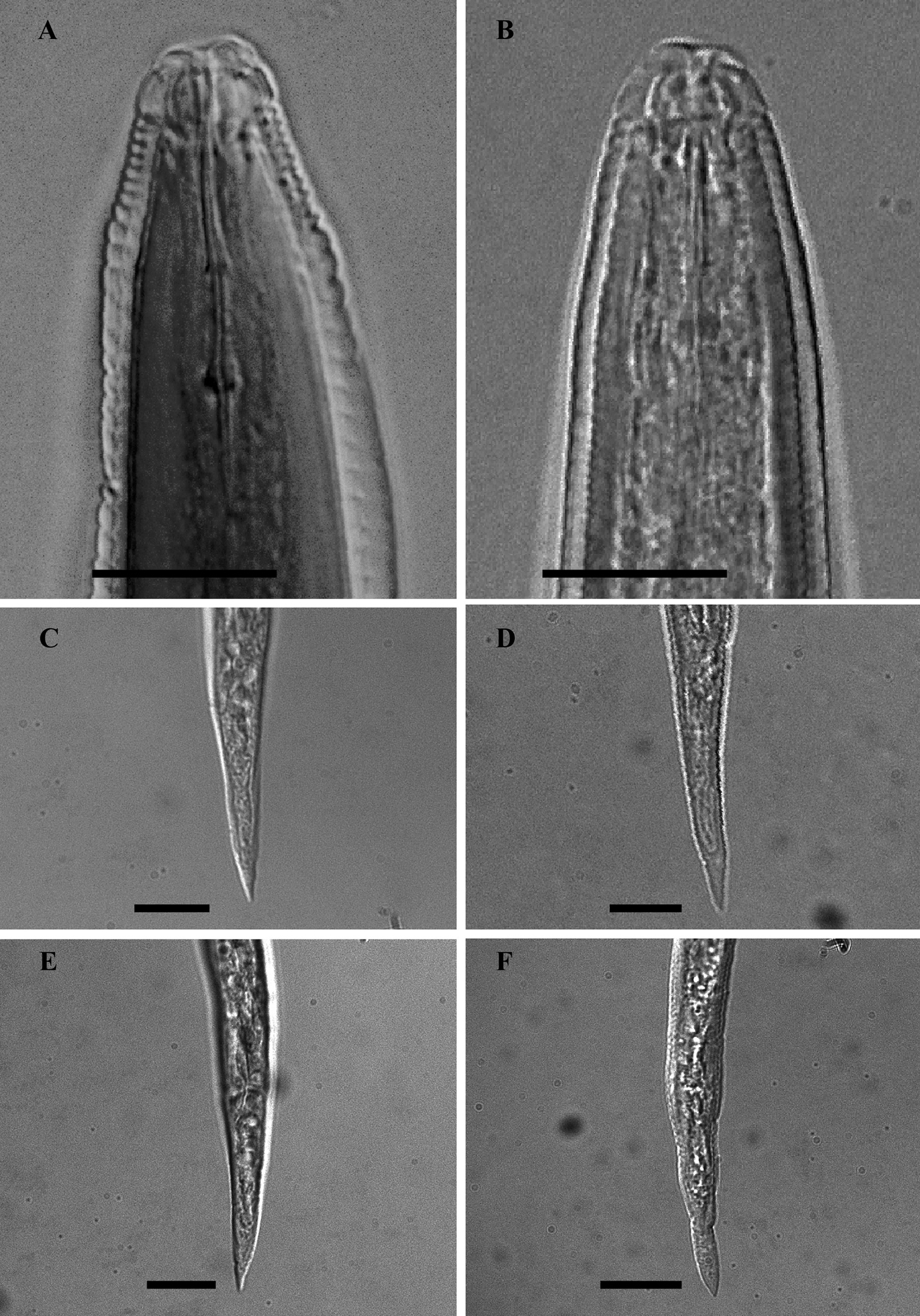
LM photographs of males anterior part and second- stage juvenile tails of Meloidogyne mali (A, C, E) and Meloidogyne ulmi (B, D, F), bar = 10 µm.
Isozyme phenotypes from ten individual females of Meloidogyne ulmi from “Mierenbos”. A Esterase B Malate dehydrogenase. Meloidogyne ulmi (1–5 and 8–12); Meloidogyne javanica (6 and 7) as reference marker.
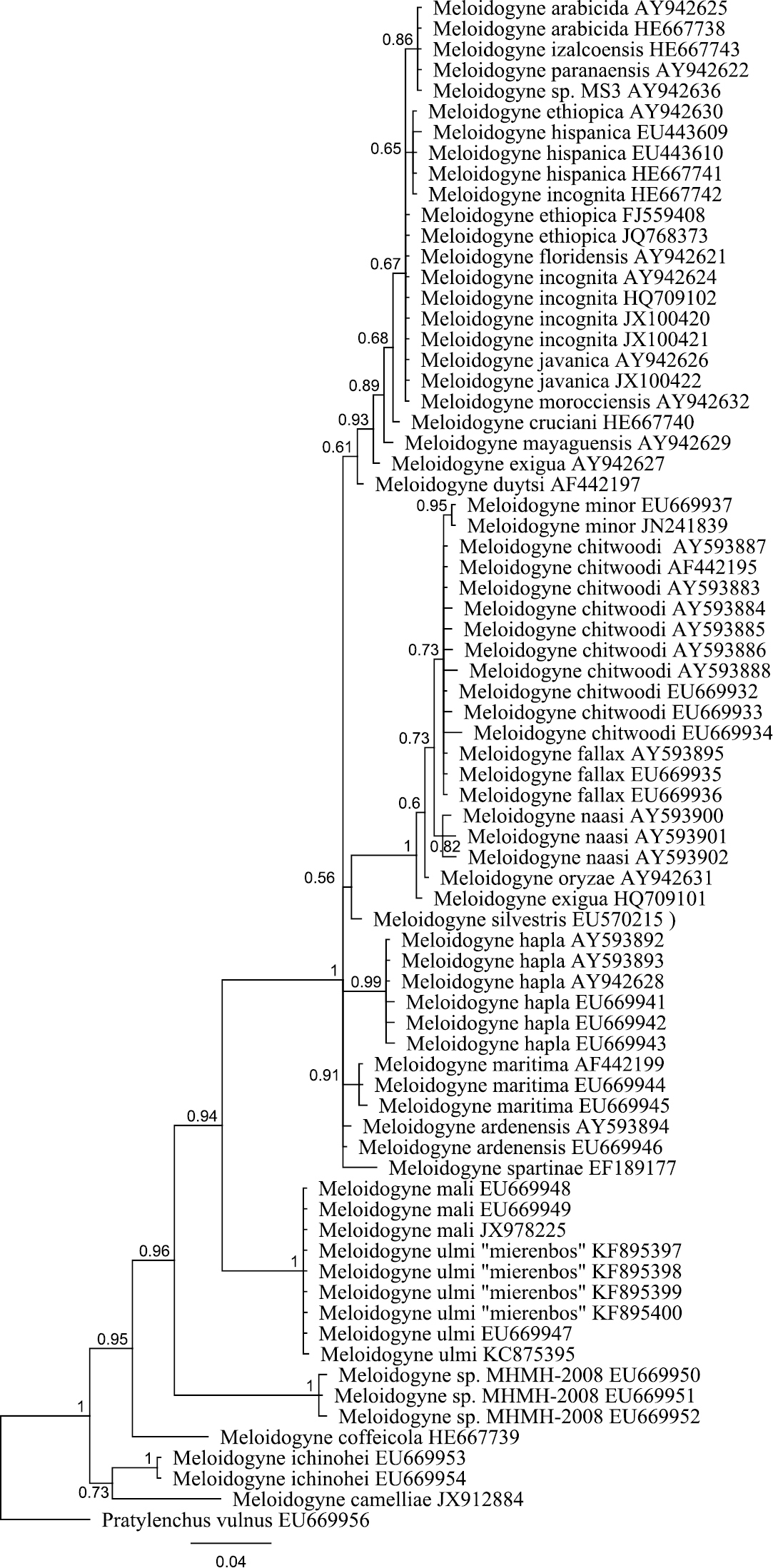
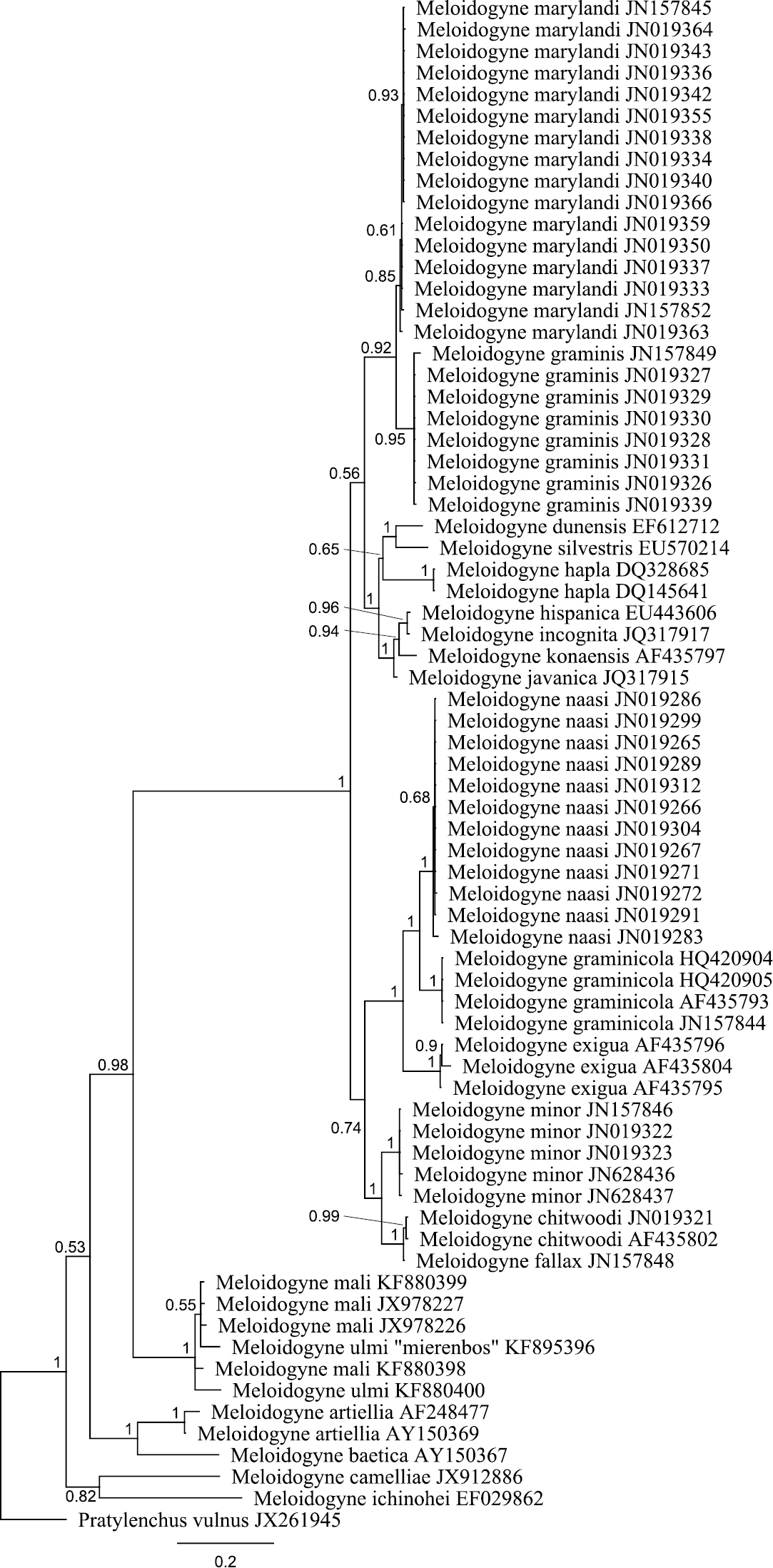
The obtained SSU rDNA and LSU rDNA sequence lengths for Meloidogyne ulmi were 781bp (including gaps) and 698bp (including gaps) respectively. In addition to our four SSU rDNA sequences of Meloidogyne ulmi “Mierenbos” (KF895397, KF895398, KF895399 and KF895400), 69 accessions belonging to other species of Meloidogyne from GeneBank were included in the local alignment (781 aligned positions, including gaps). For LSU rDNA, we had only one sequence of Meloidogyne ulmi “Mierenbos” (KF895396) due to poor data, resulting in lack of consensus sequence. Therefore the multiple sequence alignment included this sequence and 69 GeneBank accessions from other species of Meloidogyne. Pratylenchus vulnus Allen & Jensen, 1951 was selected as outgroup for constructing gene trees using Bayesian inference from both SSU rDNA and LSU rDNA sequences. SSU rDNA-based phylogenetic analysis put all sequences of Meloidogyne ulmi obtained together with those of Meloidogyne mali and Meloidogyne ulmi from GeneBank in one strongly supported polytomous branch. Despite the relatively short sequence length of SSU rDNA, the tree was able to resolve relationship between certain species in a way comparable with that of
Type specimens representing holotypes and paratypes of the two Meloidogyne species were analysed in order to demonstrate the morphological similarities that existed between them. Most of the slides we received were in good conditions except for some few individuals that showed some signs of deterioration, either due to long period of storage or poor conditions at the time they were prepared. Nevertheless, the general states of the important characters were still maintained.
On the morphology, an important character like the occurrence of double lateral lines mentioned in the description and by
Already in the original description of Meloidogyne ulmi, only a few differences could be found to separate it from Meloidogyne mali. And in some cases, the differences emanated from some apparent mistakes in the original description of Meloidogyne mali. An example is the use of male tail length of the two species to draw differences. Tail length in males of Meloidogyne mali was given as ranging between 28 and 44 µm, making it extremely longer than that of Meloidogyne ulmi 10.9 µm. However, it is important to mention that tail length values as long as the range of between 28 and 44 µm never exists among males of species of the genus Meloidogyne.
The DGO position in males with reference to the stylet knobs according to
Both apple and elm trees supported Meloidogyne ulmi reproduction. This does not only provide an additional support for the synonymization of Meloidogyne ulmi with Meloidogyne mali, but represents the first and only test involving the former on an apple plant. In principle, however, the first actual report was the description of Meloidogyne mali on apple in Japan (
It is interesting to mention that the observed variability of the MDH isozyme phenotypes among the different specimens was similar to the findings of
Trimming the SSU and LSU datasets to high quality sequence data may have caused a loss in phylogenetic signal. For SSU rDNA, over half of the target sequence length was trimmed out because of the poor quality of the dataset obtained. Although not ideal for reconstruction of phylogeny, it was still sufficient to resolve the taxa on a species level. Moreover, it has to be emphasized that the purpose here is not to reconstruct any formal phylogeny of Meloidogyne, a subject which is well covered already in previous studies (
Bayesian tree inferred from part of 18S rRNA using TYMef + I model. Sequences were aligned with MAFFT alignment. Numbers near the nodes indicate posterior probabilities. NCBI accession numbers are listed with the species names.
In conclusion, the evidence from morphological and morphometrical studies of holo– and paratype materials of Meloidogyne mali and Meloidogyne ulmi as well as host plant studies, isozyme analysis and DNA analysis all confirm the status of Meloidogyne ulmi as a junior synonym of Meloidogyne mali.
The life cycle of Meloidogyne mali is in many respects typical of the genus. Meloidogyne mali requires 18–22 weeks to complete one full generation on apple and does so only once in a year (
Meloidogyne mali induces a similar type of galls as do Meloidogyne arenaria on tomatoes, a type of gall commonly referred to as bead-like galls (Fig. 6). Concerning the current distribution of the nematode in Europe, no study has yet been done to investigate this. However, it would be rational to speculate that Meloidogyne mali may be found in all the ten European countries to which rooted seedlings were sent after the breeding programme. These countries include Belgium, England, France, Ireland, Italy, Spain, Denmark, Germany, Slovakia and Romania (
Bayesian tree inferred from part of D2 D3 of 28S rRNA using TYM + G model. Sequences were aligned with MAFFT alignment. Numbers near the nodes indicate posterior probabilities. NCBI accession numbers are listed with the species names.
Root gall symptoms of Meloidogyne mali infection on (A, B) Maluspumila “M9” (C, D) Ulmus davidiana var. japonica and (E, F) Solanum lycopersicum.
The authors are grateful to the following people for their help with providing type slides for study: Dr. Z.A. Handoo, Shigeyuki Sekimoto and Dr. Hiromichi Sakai. They also wish to thank Takayuki Mizukubo for providing root sample from the type apple tree, and to Tiziana Irdani and Beatrice Carletti for Elm root materials from the type tree. Evelyn van Heese for helping with slides preparation and electrophoresis, Anne-Sophie van Bruggen and Johan van Woggelum for help with field sampling and host plant tests. They are also grateful to Dr. M. Heybroek, formally of the Dorschkamp Research Institute for Forestry & Landscape Planning, for providing essential information about the Ulmus breeding programme. Very special thanks also to Eveline Metz-Verschure and Dr. Marcel Westenberg for their help with molecular analysis.