(C) 2013 Vlada K. Peneva. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Peneva VK, Lazarova SS, De Luca F, Brown DJF (2013) Description of Longidorus cholevae sp. n. (Nematoda, Dorylaimida) from a riparian habitat in the Rila Mountains, Bulgaria. ZooKeys 330: 1–26. doi: 10.3897/zookeys.330.5750
A description is provided of Longidorus cholevae sp. n., a bisexual species associated with wild cherry (Prunus avium L.) from the Rila Mountains, Bulgaria. The position of L. cholevae sp. n. among other species of the genus was elucidated by using morphological and molecular data. Phylogenetic analyses were performed of D2-D3 expansion domains of the 28S rRNA and the partial ITS1 containing regions by Neighbor-Joining, Maximum Likelihood and Bayesian Inference methods. The species is characterised by a female body length of 6.1–8.1 mm; long odontostyle (106–129 μm); lip region wide (21.5–24 μm) rounded and continuous with the body profile; amphidial pouches short and wide, funnel-shaped; a posteriorly situated guide ring (30–37 μm); normal arrangement of pharyngeal glands, and short bluntly rounded to hemispherical tail. Four juvenile stages indentified, first stage with elongate conoid tail. Males with 2–4 adanal pairs and a row of 11–13 single ventromedian supplements, spicules 96–120 μm long. Based both on morphological and molecular data the new species appearred to be the most similar witha group of species distributed in Europe sharing common charcters such as amphidial fovea, lip region and tail shapes, and having similar odontostyle and body length: L. poessneckensis, L. caespiticola, L. macrososma, L. helveticus, L. carniolensis and L. pius. An updated list of Longidorus species and a partial polytomous keys to the Longidorus species with long odontostyle (code A45) and short tail (code H1) are provided.
D2D3, ITS, Longidoridae, morphology, phylogeny
Molecular approaches and phylogenetic studies provide additional tools to the routine identification of plant parasitic nematodes. Further, the ribosomal DNA sequences represent a useful diagnostic aproach in the characterisation and phylogenetic reconstruction within Longidoridae, above all, where morphological characters may led to ambiguous identification (
During a study of the longidorid fauna of natural habitats in Bulgaria (2005-2009) several populations of the genus Longidorus were recovered from various locations in the Rila Mountains, one of which represented an undescribed species.
The aim of the present study was to characterise morphologically and molecularly this new species and to infer its phylogenetic relationships with other species of the genus Longidorus by using the D2-D3 expansion domains of the 28S rDNA and the ITS containing region.
Nematodes were isolated from soil samples by a decanting and sieving technique. Longidorus specimens recovered were heat killed at 55°C for two minutes, fixed in a 4% formalin/1% glycerol mixture, processed to anhydrous glycerol (
A partial polytomous keys was prepared for the identification of Longidorus species with long odontostyle (A45) and short tail (H1). This key, based on that by
Specimens for molecular analysis were kept in DESS solution (
PCR products of the ITS region from two individual nematodes were purified for cloning and sequencing using the protocol provided by the manufacturer (High Pure PCR elution kit, Roche, Germany). Purified ITS fragments were cloned in TA cloning vector (Invitrogen) and several clones were sequenced using an ABI Prism 377 sequencer (PE Applied Biosystem, Foster City, CA). Similarly, the D2-D3 regions of rDNA from two individual nematodes were purified and used for direct sequencing. The sequences of the new species have been deposited in GenBank with the accession numbers: FR775757 – FR775760 for the ITS clones; and FR775761, FR775762 for the D2-D3 regions. Additionally, another four sequences (ITS and D2-D3) belonging to a population identified as Longidorus cf. caespiticola Hooper, 1961 were produced and deposited using the same methodology (see Table 1 for accession numbers and locality). The morphometrics of this population and detailed discussion will be presented in another publication.
Species of fam. Longidoridae used in phylogenetic reconstructions.
| Nematode species | Locality | Accession number | Reference |
|---|---|---|---|
| Longidorus caespiticola Hooper, 1961 | Brdo, Slovenia | HM447030 | |
| Longidorus caespiticola | Brussegem, Belgium | AF480079 | |
| Longidorus caespiticola | Gandesbergen, Germany | AF480080 | |
| Longidorus caespiticola | Viermaal, Belgium | AF480081 | |
| Longidorus caespiticola | Scotland, UK | AY601567 | |
| Longidorus cf. caespiticola | Sokolovo, Bulgaria | HG329719–HG329721 | Present study |
| Longidorus carniolensis | Krmačina, Slovenia | JN631811 | |
| Longidorus carniolensis | Drašiči, Slovenia | JN631812 | |
| Longidorus cholevae sp. n. | Bulgaria | FR775757–FR775762 | Present study |
| Longidorus elongatus (de Man, 1876) Thorne & Swanger, 1936 | Scotland, UK | AF511417 | |
| Longidorus helveticus Lamberi, Kunz, Grunder, Molinari, De Luca, Agostinelli & Radicci, 2001 | Trška gora, Slovenia | HM447031 | |
| Longidorus helveticus | Stari Ledinci, Serbia | EF538753, JN627412 | |
| Longidorus helveticus | Camenzuid, Switzerland | AY601566 | |
| Longidorus helveticus | Chodovlice, Czech Republic | JN627410, JN627414 | |
| Longidorus helveticus | Silničná, Czech Republic | JN627411, JN627415 | |
| Longidorus helveticus | Switzerland | AJ549985 | |
| Longidorus macrosoma Hooper, 1961 | Liége, Belgium | AF480082 | |
| Longidorus macrosoma | Austria | EF538752 | |
| Longidorus macrosoma | unknown | AY580055 | unpublished |
| Longidorus macrosoma | Switzerland | AY601565 | |
| Longidorus macrosoma | Switzerland | AJ549978, AJ549979 | |
| Longidorus macrosoma | unknown | AY430184 | unpublished |
| Longidorus pius Barsi & Lamberti, 2000 | Republic of Macedonia | AM743178–AM743184 | |
| Longidorus poessneckensis Altherr, 1974 | Czech Republic | EF538750 | |
| Longidorus poessneckensis | Slovakia | EF538751 | |
| Longidorus raskii Lamberti & Agostinelli, 1993 | Switzerland | AJ549983, AJ549984 | |
| Longidorus diadecturus Eveleigh & Allen, 1982 | Elkins, White river, USA | AY601584 | |
| Xiphinema diversicaudatum (Micoletzky, 1927) Thorne, 1939 | Slovakia | EF538755 | |
| Xiphinema index Thorne & Allen, 1950 | Argentina | AY601628 | |
| Xiphinema insigne Loos, 1949 | Taiwan | AY563427 |
Further, a BLAST (Basic Local Alignment Search Tool) search at NCBI (National Center for Biotechnology Information) was performed using the obtainedITS and D2-D3 sequences as queries to confirm their nematode origins and to identify the most closely related nematode sequences. Different Longidorus species were used in the phylogenetic analyses of ITS1-5.8S-ITS2 and D2-D3 regions due to sequence availability in the GenBank database (Table 1). The multiple sequence alignments (MSA) of both datasets were performed using MAFFT algorithm (
Base compositional differences were evaluated using the c2-test. Sequence divergences (uncorrected p distance) were calculated using MEGA 5.0 (
http://zoobank.org/882B3067-D244-4B8F-9312-B6E0F55B6C90
http://species-id.net/wiki/Longidorus_cholevae
Figures 1–9See Table 2
Measurements of females, males and juvenile stages of Longidorus cholevae sp. n. from Bachevo village. All measurements are given in μm (mean ± standard deviation, with range in parentheses).
| – | Holo- | Females | Males | J1 | J2 | J3 | J4 |
|---|---|---|---|---|---|---|---|
| n | type | 11 | 11 | 9 | 8 | 9 | 11 |
| L | 7199 | 6788 ± 573 (6127–8083) |
6390 ± 594 (5415–7111) |
1209 ± 63 (1135–1289) |
1874 ± 236 (1554–2251) |
3048 ± 406 (2336–3447) |
4798 ± 442 (4148–5666) |
| a | 83.3 | 72.1 ± 7.4 (61.1–83.3) |
70.2 ± 6.2 (63.9–82.0) |
47.0 ± 1.9 (43.8–50.3) |
51.1 ± 2.4 (49.0–55.3) |
56.5 ± 3.8 (50.2–61.3) |
63.8 ± 5.9 (54.8–76.6) |
| b | 13.1 | 14.3 ± 1.5 (12.3–17.9) |
12.7 ± 1.2 (10.7–14.7) |
4.5 ± 0.4 (3.9–5.1) |
5.8 ± 0.9 (4.5–7.2) |
7.9 ± 0.9 (7.2–9.9) |
10.9 ± 1.5 (9.2–14.1) |
| c | 202.4 | 199.7 ± 15.4 (171.2–220.4) |
199.6 ± 18.3 (171.1–227.8) |
29.6 ± 3.8 (26.1–36.6) |
48.2 ± 3.6 (43.2–53.9) |
78.3 ± 7.2 (66.1–91.3) |
136.5 ± 19.9 (115.5–181.1) |
| c’ | 0.6 | 0.6 ± 0.06 (0.5–0.7) |
0.6 ± 0.06 (0.6–0.8) |
2.1 ± 0.19 (1.8–2.4) |
1.4 ± 0.1 (1.2–1.5) |
0.9 ± 0.07 (0.8–1.0) |
0.7 ± 0.06 (0.6–0.8) |
| V (%) | 52.5 | 50.5 ± 2.2 (46.7–53.4) |
- | - | - | - | - |
| G1 (%) | 13.0 | 14.0 ± 2.8 (8.6–17.7) |
- | - | - | - | - |
| G2 (%) | 11.5 | 14.2 ± 1.6 (11.6–17.1) |
- | - | - | - | - |
| d | 1.3 | 1.3 ± 0.04 (1.2–1.4) |
1.3 ± 0.04 (1.3–1.4) |
1.7 ± 0.08 (1.6–1.8) |
1.6 ± 0.09 (1.5–1.7) |
1.6 ± 0.11 (1.4–1.7) |
1.5 ± 0.08 (1.3–1.5) |
| d’ | 1.5 | 1.5 ± 0.06 (1.4–1.6) |
1.5 ± 0.03 (1.4–1.6) |
1.6 ± 0.07 (1.5–1.7) |
1.7 ± 0.1 (1.6–1.8) |
1.7 ± 0.1 (1.5–1.8) |
1.7 ± 0.08 (1.6–1.9) |
| Odontostyle | 121 | 120.1 ± 7.2 (106–129) |
121.2 ± 5.1 (115–131) |
61.1 ± 3.5 (56–66) |
65.9 ± 2.8 (62–71) |
84.7 ± 3.3 (79–90) |
99.1 ± 5.3 (88–105) |
| Replacement odontostyle | - | - | - | 65.0 ± 1.8 (61–67) |
78.1 ± 4.3 (74–86.5) |
101.4 ± 4.5 (96–109) |
117.5 ± 7.9 (105.5–131) |
| Developing gonads |
- | - | - | 19.9 ± 3.2 (16–25) |
28.3 ± 3.2 (24–34) |
53.1 ± 8.7 (41–65) |
135.5 ± 11.4 (114–147) |
| Odontophore | 88 | 76.3 ± 3.3 (74-81) |
73.7 ± 5.0 (69.5-81) |
41.7 ± 5.4 (36-48) |
48.6 ± 3.3 (42-52) |
62.6 ± 2.5 (60-66) |
71.1 ± 3.6 (67-79) |
| Pharynx | 550 | 481.9 ± 47.8 (439–577) |
507.0 ± 45.6 (421–584) |
273.6 ± 19.3 (250–302) |
318.4 ± 23.6 (277–349) |
398.1 ± 47.7 (311.5–450.5) |
445.7 ± 38.9 (362–491) |
| Anterior to guiding ring | 36 | 32.6 ± 2.21 (30–37) |
33.5 ± 1.1 (32–36) |
16.2 ± 0.7 (15–18) |
19.3 ± 0.74 (18–20) |
24.2 ± 1.6 (22.5–27) |
28.4 ± 1.4 (25.5–31) |
| Bulb length | 139 | 128 ± 12.5 (114.5–146.5) |
124 ± 7.2 (115–137) |
60.7 ±5.1 (53–66) |
72.5 ± 8.9 (65–90) |
100.7 ±5.6 (92–108) |
116.8 ± 9.4 (105–128) |
| Bulb width | 34 | 34.1 ± 2.9 (30–38) |
33.0 ± 3.3 (28–38) |
15 ± 1.2 (14–17) |
20.2 ± 2.0 (18–23) |
26.3 ± 1.2 (24–27) |
29.8 ± 2.5 (26–34) |
| Tail | 35.5 | 34.1 ± 2.9 (28.5–38) |
32.2 ± 3.3 (29–39) |
41.3 ± 4.8 (35–48.5) |
38.8 ± 2.8 (34–43) |
38.9 ± 3.3 (34–44.5) |
35.4 ± 2.6 (31–38) |
| Length of hyaline part | 19 | 18.1 ± 1.10 (17–20) |
14.5 ± 2.2 (12–18) |
10.9 ± 1.8 (9–14) |
11.8 ± 2.09 (9–15) |
13.9 ± 1.6 (12–17) |
14.5 ± 1.4 (12–16.5) |
| Body diameter at: - lip region |
22.5 |
22.8 ± 0.8 (21.5–24) |
23.0 ± 0.7 (22–24) |
9.5 ± 0.30 (9–10) |
12.0 ± 0.6 (11–13) |
15.5 ± 1.2 (14–17) |
19.5 ± 1.0 (18–21) |
| - guiding ring | 39 | 37.5 ± 2.5 (35–43) |
37.7 ± 1.7 (34–40) |
15.1 ± 0.32 (14–15.5) |
20.1 ± 1.09 (18–22) |
26.2 ± 1.8 (23.5–28.3) |
33.2 ± 1.7 (30 - 35.5) |
| - base of pharynx | 74.5 | 75.9 ± 8.4 (69–100) |
77.5 ± 8.8 (66–90.5) |
26.3 ± 2.0 (24–30) |
35.2 ± 2.8 (31–38) |
49.0 ± 4.60 (41–53) |
63.4 ± 5.2 (57–77) |
| - mid-body/at vulva | 86 | 93.7 ± 9.1 (83–106) |
91.7 ± 11.5 (73–111) |
25.9 ± 2.1 (23–29.5) |
36.6 ± 3.8 (31–41) |
54.1 ± 7.2 (43.5–68) |
75.6 ± 8.5 (66–94) |
| - anus | 57 | 54.8 ± 4.4 (48–66) |
52.4 ± 4.0 (46–58) |
19.5 ± 1.2 (18–22) |
28.7 ± 2.3 (25–31) |
42.8 ± 4.3 (36–48) |
52.2 ± 2.5 (47–56) |
| - hyaline part | 47 | 42.9 ± 3.8 (37–48) |
36.0 ± 4.0 (27–42) |
10.9 ± 1.77 (9–14) |
17.9 ± 2.4 (14.5–21) |
29.0 ± 3.4 (25–37) |
37.2 ± 3.0 (33–42) |
| Spicules | - | 105.9 ± 6.9 (96–120) |
Female. Body plump, assuming a C to open spiral shape. Lip region continuous, anteriorly rounded. Labial papillae prominent. Cuticle 8–10 μm thick at poslabial region, 5–7 μm along the body and 12–14 μm on tail posterior to anus. Guide ring 6–7 μm wide. One lateral pore anterior to guide ring, 2–4 along odontostyle, 1–2 along odontophore, 4–5 in narrow part of the oesophagus and 3–4 in bulb region as well as 3–5 dorsal and 7–10 ventral; numerous lateral body pores observed. Amphidial fovea pouch like, short, almost as wide as long, funnel shape with code E5 according to
Male. Habitus as in females, posterior part more strongly coiled ventrad. Shape of lip region similar to that in females. Cuticle 5–8 μm thick at poslabial region, 7–9 at guiding ring level, 4–6 μm along the body and 9–13 μm on tail posterior to cloaca. One lateral pore anterior to guide ring, 2–3 along odontostyle, 1–2 along odontophore, 3–5 in narrow part of the oesophagus and 3–4 in pharyngeal bulb region as 4 dorsal and 7–10 ventral; numerous lateral body pores present. Fusus at 52.3±3.7 (47–57) μm, n=7 from anterior end. Nerve ring surrounding odontophore base, at 231.8±12.2 (217.5–259.5) μm from anterior end, a second nerve ring situated at a short distance behind the first one. Pharyngo-intestinal valve, variable in shape (broadly rounded to heart-shape) and size, almost as long as wide: 16.6±3.2 (13–23) × 18±3.1 (13–22) μm, n=6. Lateral chord 20–25 μm wide. Supplements 3–4 adanal pairs followed by 10–14 arranged irregularly in a single row. Spicules massive, slightly curved ventrally, lateral guiding piece 27–28 μm long. Spermatozoids round small (4–6 μm diam.). Tail short, bluntly conoid, dorsally convex, ventrally slightly concave, three pairs of lateral pores.
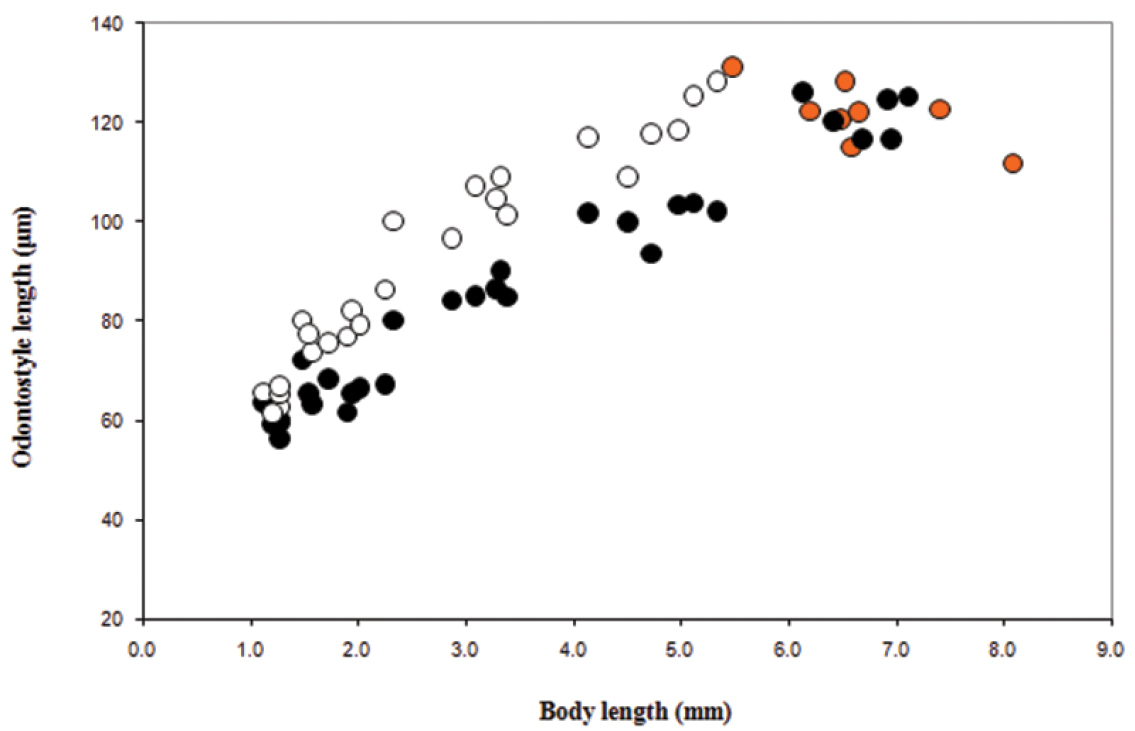
Juveniles. Morphometrics obtained from juvenile specimens, and of the relationship between the lengths of their functional and replacement odontostyles and body lengths, confirmed the presence of four juvenile stages (Figure 9). Habitus in the shape of more or less open C, tail of the first stage juvenile conoid elongated whereas in the subsequent developmental stages the tail is conoid (second stage) to bluntly conoid (third and fourth stage).
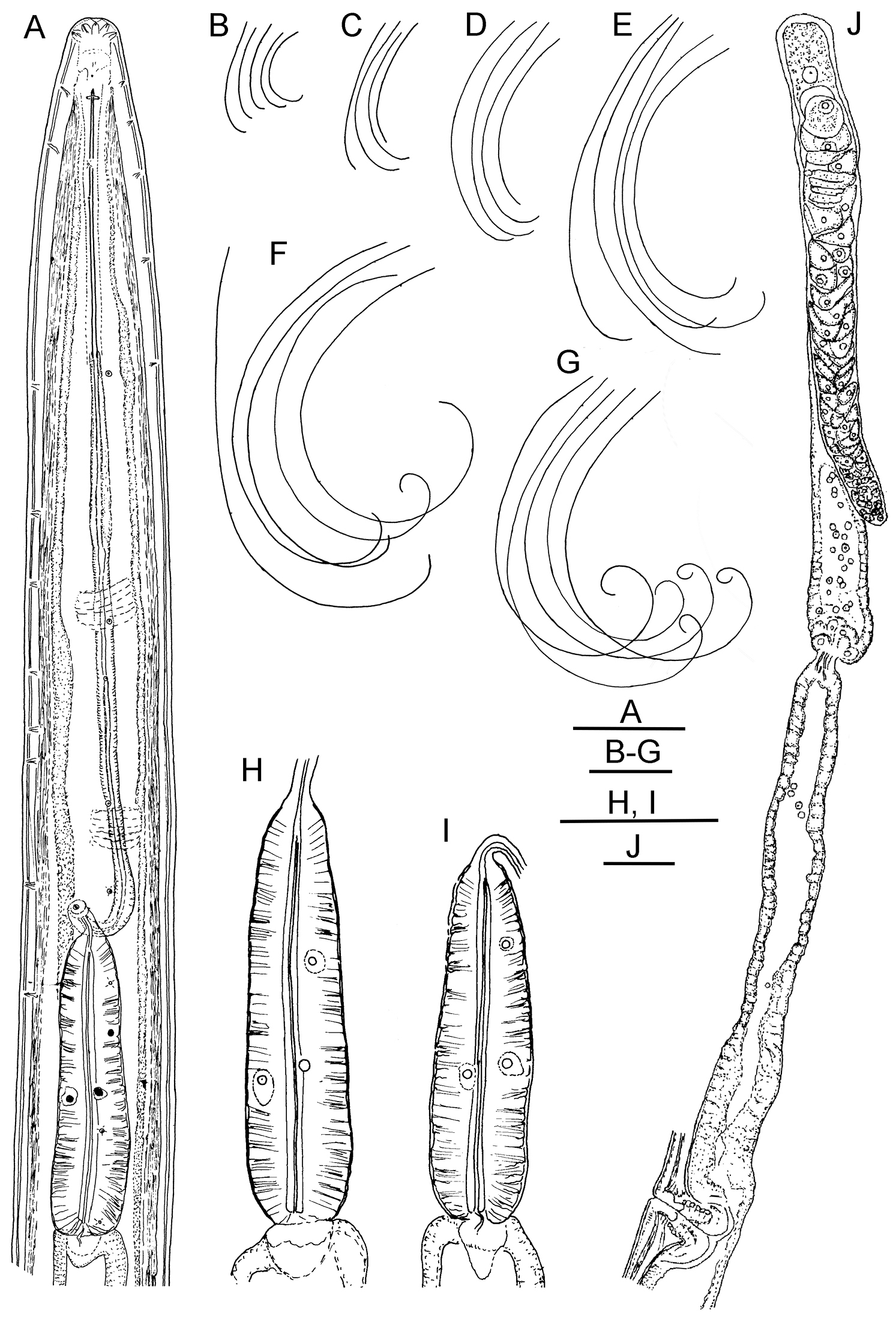
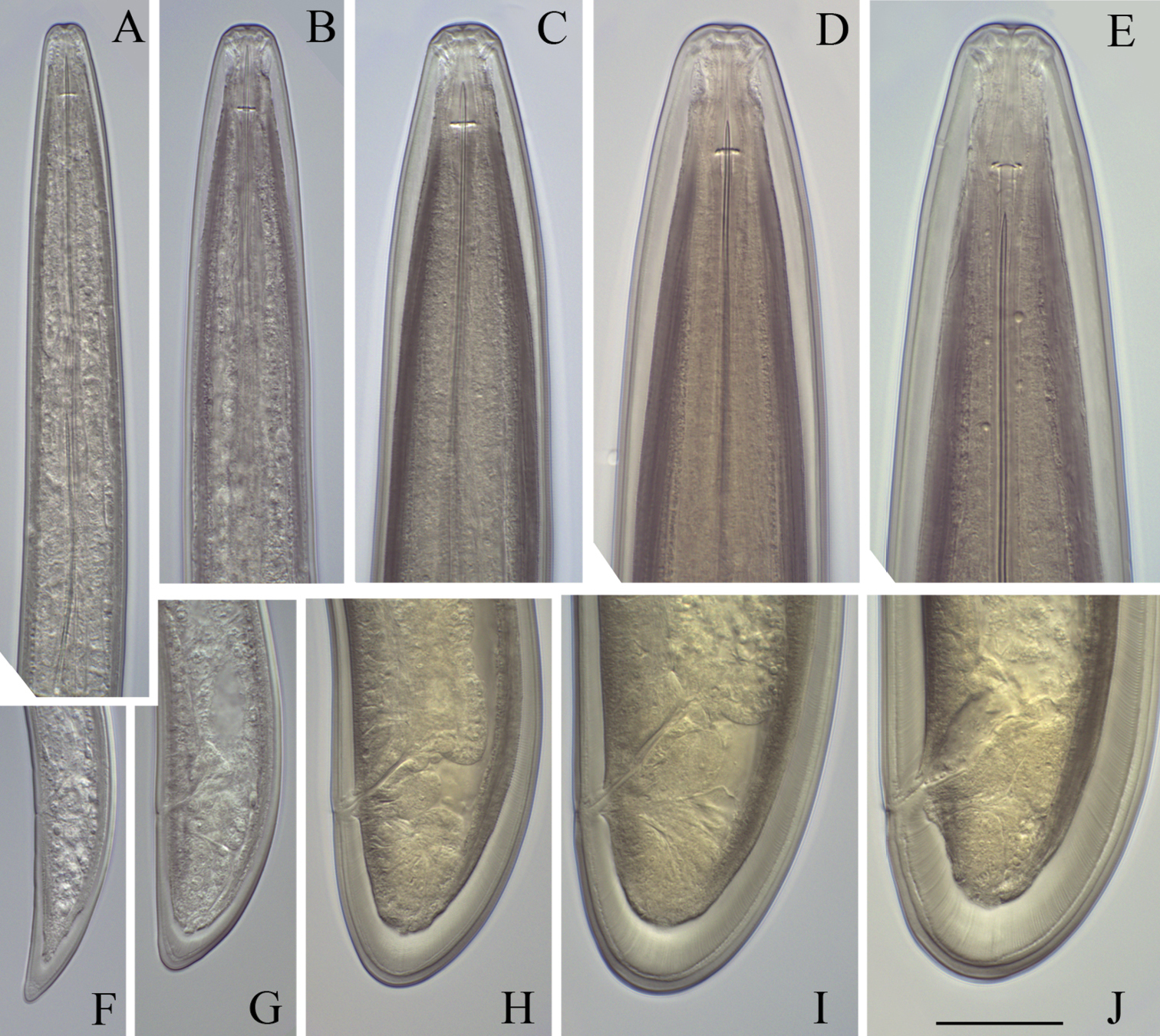
Longidorus cholevae sp. n. Female: A Anterior end F Habitus I Pharyngeal bulb J Anterior genital branch Male: G Habitus H Pharyngeal bulb Juveniles: B–E Habitus of first-, second-, third- and fourth-stage juveniles. Scale-bars: A, H, I, J 50 μm; B–G 1 mm.
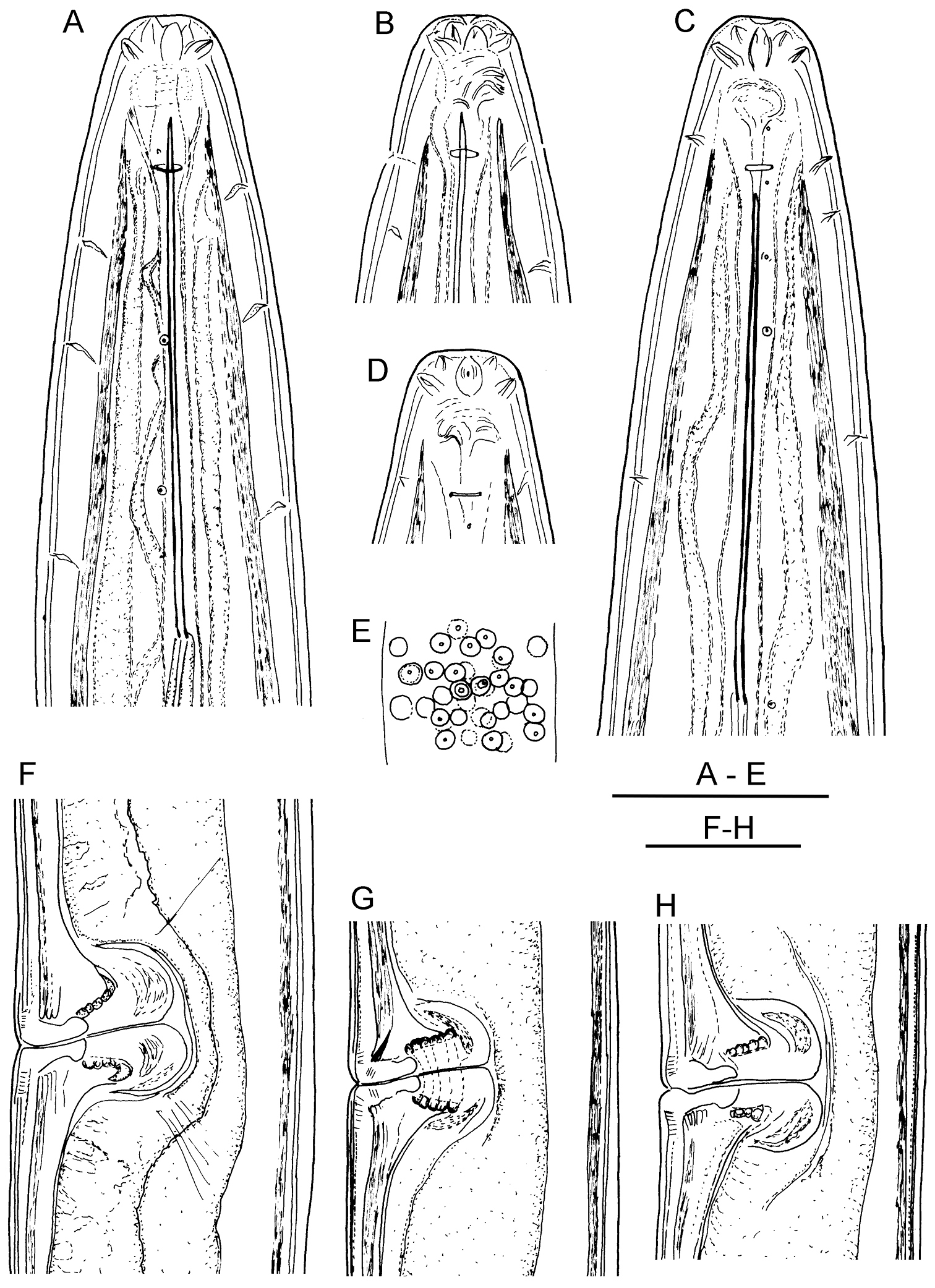
Longidorus cholevae sp. n. Female: A Anterior end B Lip region/amphidial fovea F–H Variations in vagina shape; Male: C Anterior end D Lip region/amphidial fovea E Sperms. Scale-bars: A–H 50 μm.
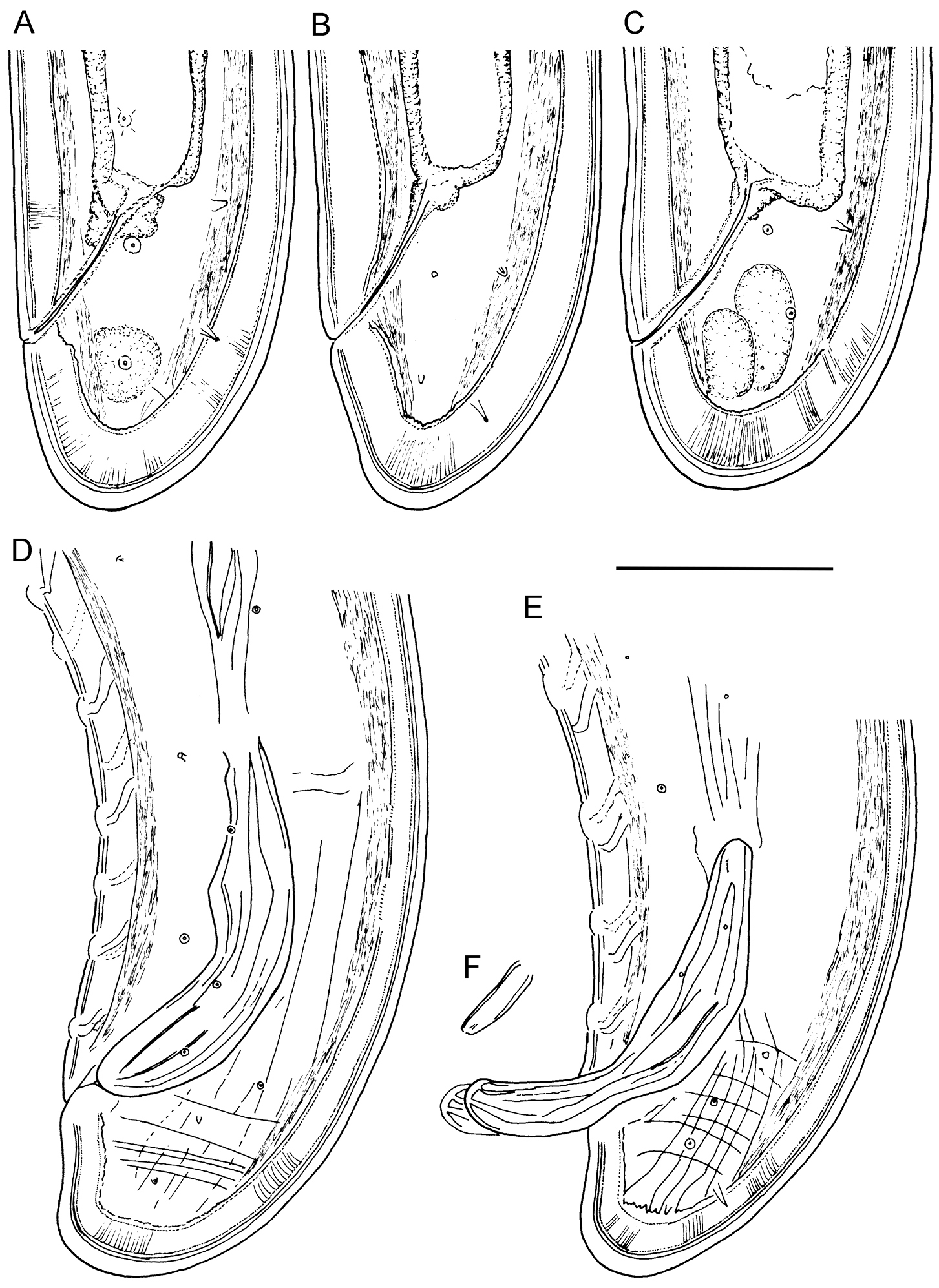
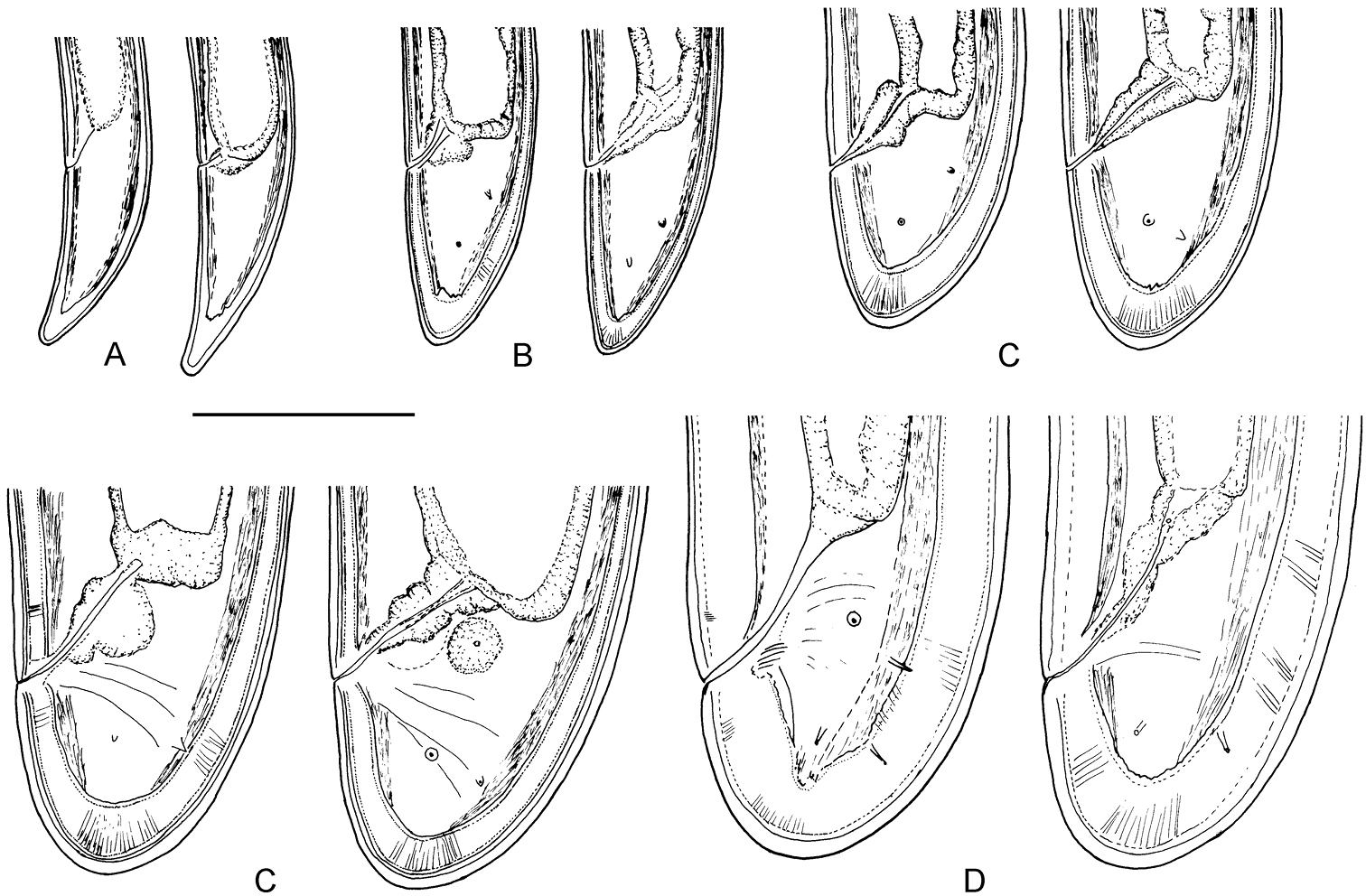
Longidorus cholevae sp. n. Female: A–C Variations in tail shape; Male: D, E Tail ends with spicules F Lateral piece. Scale-bars: 50 μm.
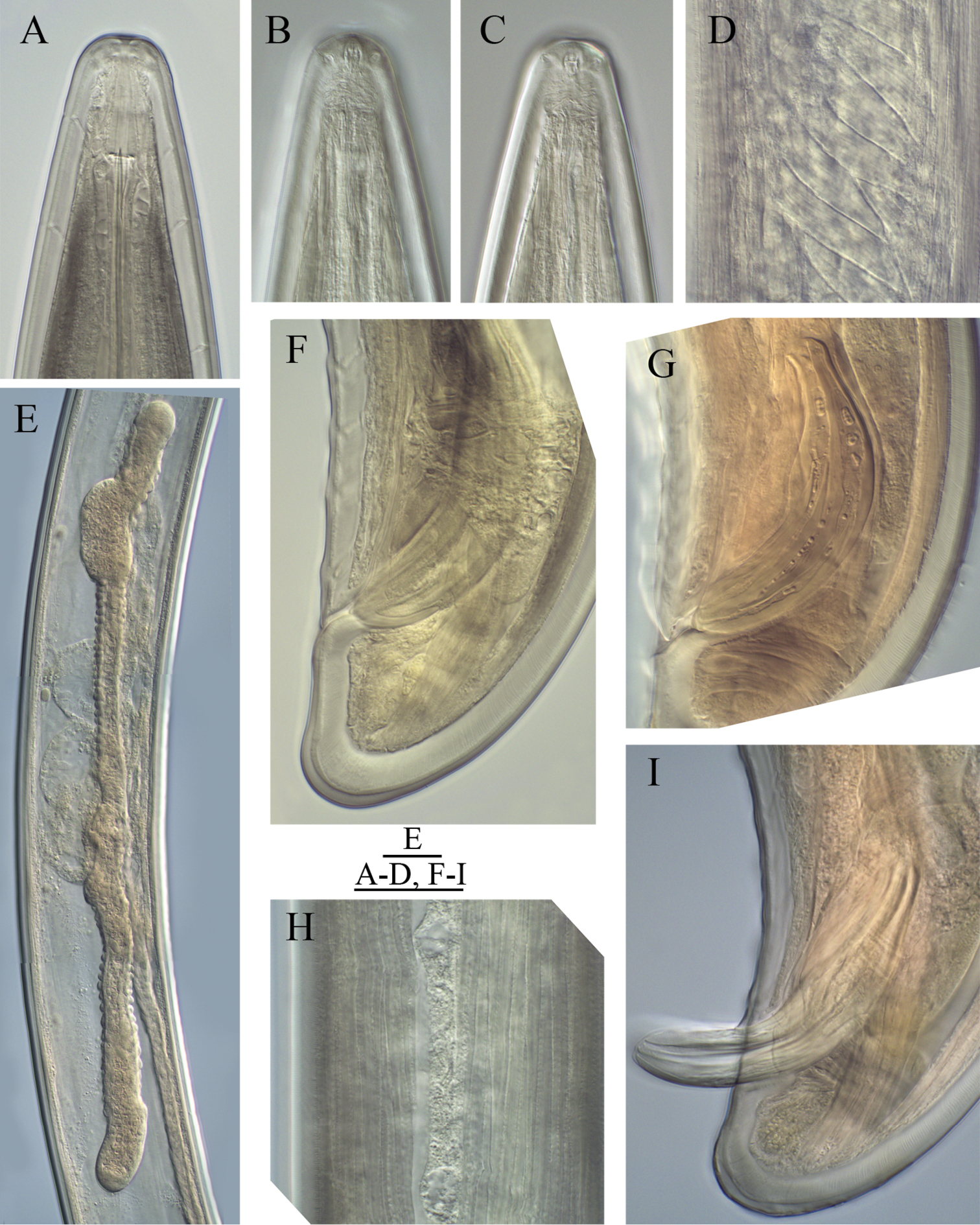
Longidorus cholevae sp. n. Female: A Anterior region of holotype B Head end C Amphidial fovea of holotype D Sphincter between uterus and pars dilatata oviductus E Caudal pores F Pharyngeal bulb G, H Variations in vagina shape; I–K Variations in tail shape. Scale bars: A, F 40 μm; E 20 μm; B–D, G–K 30 μm.
Longidorus cholevae sp. n. Male: A Lip region B, C Amphidial fovea, B upper view C, lower view D Part of testis with radial muscles E Genital system of a young male F Tail, G Spicule H Lateral field I Protracted spicule. Scale bars: A–D, F–I 30 µm; E 40 µm.
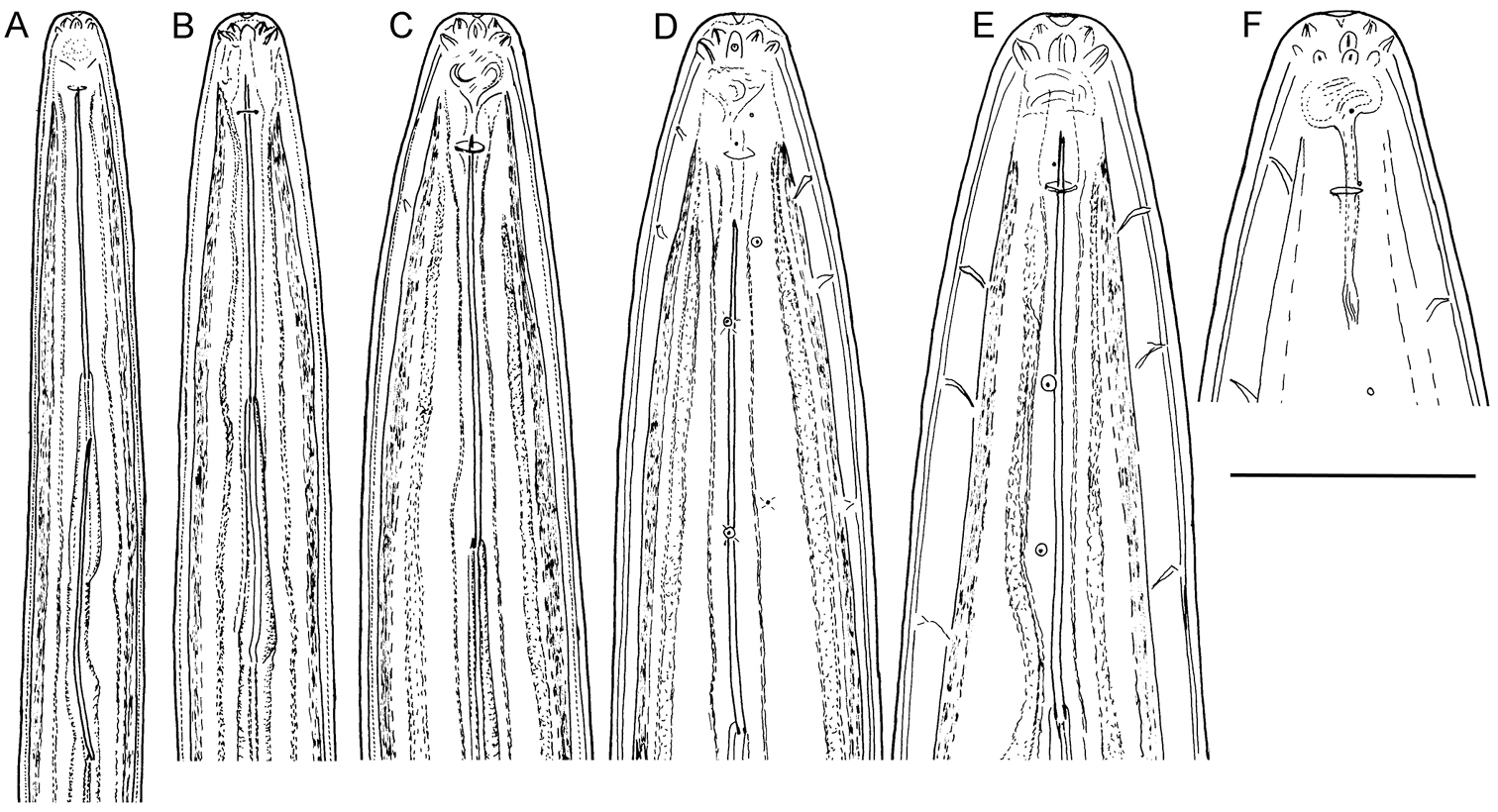
Longidorus cholevae sp. n. Anterior region of: A–D First-fourth juvenile stage F Female G Male. Scale-bar: 50 μm.
Longidorus cholevae sp. n. Variations in tail shape: A–D Tail of first-fourth juvenile stageE Female Scale-bar: 50 μm.
Longidorus cholevae sp. n. Juveniles: A–D Head ends of first- to fourth-stages F–I Tail end of first- to fourth-stages Female: E Anterior end J Tail. Scale-bar: 30 μm.
Longidorus cholevae sp. n. Scatter plot of the functional (•, juveniles and adults, females in orange) and replacement (◦, juveniles) odontostyle in relation to body length of the juvenile developmental stages and adults.
Bachevo village, Rila Mountains, co-ordinates 41°56'14.97"N, 23°25'15.02"E, 1032 m asl, riparian vegetation; soil around the roots of wild cherry (Prunus avium L.), Juniperus communis L., Urtica dioica L. and grasses.
Holotype and 1 paratype females, 2 males, and 23 juveniles deposited in the nematode collection of the Institute of Biodiversity and Ecosystem Research, Sofia, Bulgaria. Other paratypes deposited as follows: two females, one male and 8 juveniles in the Nematode collection of the Foodand Environment Research Agency, Sand Hutton, UK (former Rothamsted Nematode Collection); one female, one male and 6 juveniles in the USDA Nematode Collection, Beltsville, Maryland, USA; one female, one male and 8 juveniles in the Riverside Nematode Collection, University of California, Riverside, USA; one female, one male and 5 juveniles in the Nematode Collection of the Institute of Plant Protection, Bari, Italy; one female, one male and 12 juveniles in the Wageningen Nematode Collection (WANECO), Wageningen, the Netherlands.
Longidorus cholevae sp. n. is a comparatively large bisexual species (6.1–8.1 mm) with odontostyle over 100 μm (106–129 μm) long, lip region wide (21.5–24 μm), continuous, anteriorly rounded, amphidial fovea pouch like, almost as wide as long, posteriorly situated guide ring, short, bluntly rounded to hemispherical tail and normal arrangement of pharyngeal glands.
The alpha-numeric codes for Longidorus cholevae sp. n. to be applied to the polytomic identification key for Longidorus species by
Longidorus caespiticola by females having wider (21.5–24 vs 16–18 μm) and differently shaped lip region (rounded vs smoothly rounded, almost conical), shorter tail
(28.5–38 vs 39–47 μm) and longer spicules (96–120vs 88.5–93 μm), and tail in first stage juveniles (elongate conoid vs bluntly conoid) (
Longidorus macrosoma by female specimens having a somewhat shorter body (L= ave. 6.8 mm (6.1–8.1) vs ave. 9.1 mm (6.8–12), diferently shaped lip region (rounded vs slightly concave) and differently
shaped tail of the first stage juvenile (elongated conoid vs digitate) (
Longidorus helveticus by females having different shape of amphidial fovea (almost rounded vs elongated), shorter odontostyle (ave. 120.1 (106–129) μm vs ave. 135. 4 (127–145.5) in the type
population and reported range for other populations 127–142 μm, differently shaped tail in first stage juvenile (elongated conoid vs mucronated) and shorter hyaline portion of tail (J=10–14 vs J=17.5–33 μm) (
Longidorus carniolensis – by having a shorter odontostyle (106–129 vs 136–157 μm); males with shorter spicules (96–120 vs 122–145 μm); different tail shape in first stage juvenile
(elongate conoid vs rounded, conoidal, c’=1.8–2.2 vs c’=1.2–1.5) (
Longidorus macroteromucronatus – by females having wider lip region (21.5–24 vs 17.5 μm (calculated from the drawing by
Longidorus pius – by different d and d’ values (following
Longidorus pseudoelongatus – by having a longer body (L=6.1–8.1 vs L=5.1–5.6 mm), differently shaped (continious vs separated by constriction) and wider
lip region (21.5–24 vs 12 μm), higher c (c=171.2–220.4 vs c=100–150) and lower c’ values (c’=0.5–0.7 vs c’=0.93) (
Further, Longidorus cholevae sp. n. is similar in body and odontostyle lengths (codes F34 and A45), and shape of anterior region and tail (codes D1 and H1) with a group of several other species from which it differs in amphidial fovea shape (see Appendix 1: a partial polytomous key): Longidorus kheirii, Longidorus raskii Lamberi & Agostinelli, 1993, Longidorus arthensis Brown, Grunder, Hooper, Klingler & Kunz, 1994, Longidorus fasciatus Roca & Lamberti, 1981, Longidorus uroshis Krnjaić, Lamberti, Krnjaić, Agostinelli & Radicci, 2000, Longidorus silvae Roca, 1993, Longidorus iuglandis Roca, Lamberti & Agostinelli, 1984, Longidorus saginus Khan, Seshardi, Weischer & Mathen, 1971, Longidorus picenus Roca, Lamberti & Agostinelli, 1984, Longidorus baeticus). The new species can be distinguished from Longidorus raskii, Longidorus arthensis, Longidorus fasciatus, Longidorus uroshis, Longidorus silvae, Longidorus picenus and Longidorus baeticus by its wider lip region (21.5–24 μm vs 15–19 μm; 14–19 μm; 12–14 μm; 14–20.5; 14–17 μm; 14–16 μm; 19–22 μm; 12–14.5 μm); from Longidorus raskii, Longidorus uroshis and Longidorus saginus by having different odontostyle length (106–129 μm vs 76–103 μm; 120–152 μm and 135–155 μm); from Longidorus kheirii, Longidorus raskii, Longidorus arthensis and Longidorus uroshis by the shorter tail (28.5–38 vs 47–72 μm; 36–50 μm; 36–46.5 μm and 38–57 μm); from Longidorus kheirii, Longidorus silvae and Longidorus picenus by having more anteriorly situated guide ring (30–37 vs 36.5–45 μm, 36–48 μm and 37–42 μm). Additionally, it can be differentiated from:
Longidorus kheirii by females having differently shaped lip region (rounded vs slightly concave), higher c value (171.2–220.4 vs 119–167.8), smaller pharyngeal bulb (114.5–146.5
× 30–38 vs 149.5–193.5 × 39.5–48 μm), males abundant, functional vs rare and not functional, differently shaped tail of the first stage juvenile as well as different morphometrics concerning the main characters such as body and tail length, functional and replacement odontostyle length (Table 1; Table 2 in
Longidorus raskii by females having different tail shape in first stage juveniles (elongate conoid vs bluntly conoid); (
Longidorus arthensis by females having lower c’ value (c’=0.5–0.7 vs c’=0.8–1.1 and 0.9–1.1); males with longer spicules (96–120 vs 60–66 μm); different tail shape in first stage juveniles
(elongate conoid vs digitate) (
Longidorus fasciatus by females having a more plump body (a=61.1–83.3 vs a=121–143) (
Longidorus uroshis by males with longer spicules (96–120 vs 59–72 and 64–78 μm) and different tail shape in first stage juveniles (elongate conoid vs digitated) (
Longidorus silvae by female specimens having differently shaped lip region (rounded vs sub-acute and flattened anteriorly) and tail of the first and second stage juvenile (elongated conoid vs mucronated; conoid
vs bluntly rounded, respectively), and males abundant vs males rare (
Longidorus iuglandis by having longer uteri (357.5–662.5 vs 140–160 μm) and differently shaped tail in the first stage juvenile (elongate conoid vs bluntly rounded) (
Longidorus saginus by having a longer body (L=6.1–8.1 vs 4.8–6.4 mm); lower c’ value (c’’=0.5–07 vs c’=0.8); more posteriorly situated vulva (V=46.7–53.4 vs V=40–45) (Khan et al. 1971);
Longidorus picenus by having, differently shaped tail in the first stage juvenile (elongate conoid vs mucronated) (
Longidorus baeticus by males having longer spicules (96–120 vs 80–95 μm) and differently shaped tail in the first stage juvenile (elongate conoid vs bluntly rounded to cylindrical).
The species is named after Dr Boryana Choleva, Faculty of Biology, University of Sofia, retired, for her substantial contribution to the knowledge of the fam. Longidoridae in Bulgaria.
The amplification of D2-D3 expansion domains of the 28S rDNA and the ITS containing region yielded single fragments of 800 bp and 1384 bp, respectively, based on sequencing. The ITS1 and ITS2 sizes were 579 bp and 338 bp, respectively that resulted in the shortest ITS recorded for Longidorus so far. Intra-individual and intra-population sequence variability in ITS and no variability in D2D3 domains have been observed.
A BLAST search for D2-D3 region showed a 80-93% degree of similarity among Longidorus spp. suggesting that Longidorus cholevae can be easily identified from other species by using this ribosomal region. The closest species were Longidorus poessneckensis (93% similarity), Longidorus caespiticola, Longidorus macrosoma and Longidorus helveticus (92% similarity). Pairwise BLAST comparisons of the ITS sequence of Longidorus cholevae with those of Longidorus spp. from the database displayed high nucleotide dissimilarity and considerable variation in length.
Our preliminary phylogenetic analyses based on all the D2-D3 Longidorus sequences deposited in NCBI revealed that the new species clusters into a well-supported group of Longidorus species having a European distribution: Longidorus caespiticola, Longidorus macrososma, Longidorus poessneckensis, Longidorus helveticus and Longidorus carniolensis (trees not presented). The monophyly of this group has been highly supported also in other studies, including SSU phylogenetic analyses (
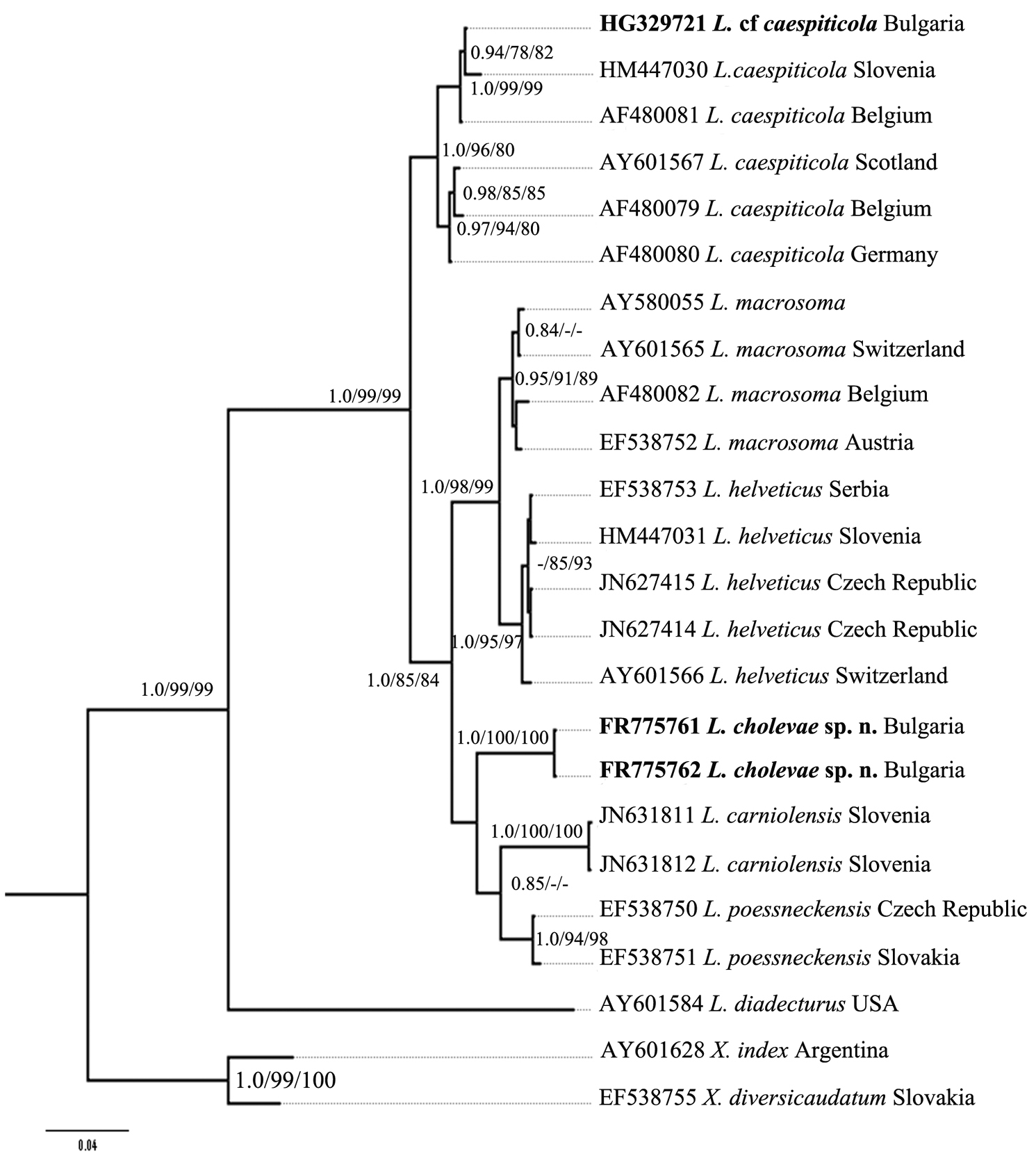
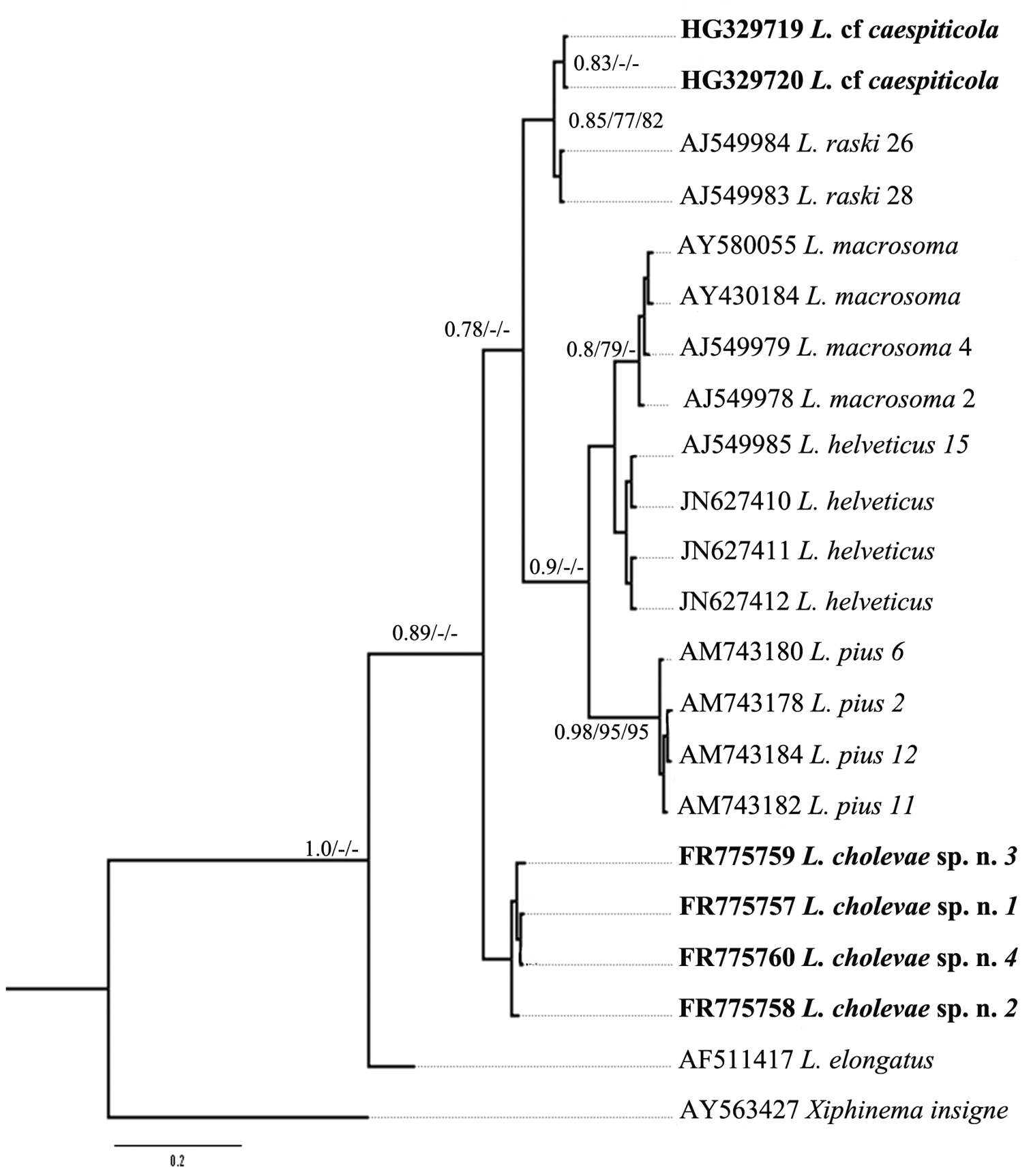
Further, for phylogenetic analysis Longidorus species from GenBank with the highest match of BLAST search were aligned along with Longidorus cholevae D2-D3 and partial 18S-ITS1 sequences and these alignments included sequences from various populations (Table 1). The trees obtained by NJ, ML and BI methods showed similar topology and differed in the position of poorly supported clades, and thus only the BI trees with posterior probabilities higher than 0.8 and bootstrap values above 70% (NJ and ML) are presented (Figs 10–11).
Phylogenetic relationships of Longidorus cholevae sp. n. and its closest species for the D2-D3 rDNA. Bayesian Inference strict consensus tree acquired under GTR+G model. Numbers at the nodes indicating posterior probabilities higher that 0.8 and bootstrap values more that 70% for ML and NJ are presented.
Phylogenetic relationships of Longidorus cholevae sp. n. and its closest species for the partial 18S-ITS1 rDNA regions. Bayesian Inference strict consensus tree acquired under K2+G model. Numbers at the nodes indicating posterior probabilities higher that 0.8 and bootstrap values more that 70% for ML and NJ are presented.
The phylogenetic tree of the D2-D3 region (Fig. 10) showed two well-supported clades: Clade I consists of three subclades: two highly supported subclades containing various populations of I1) Longidorus helveticus and I2) Longidorus macrosoma, and one subclade having lower values for ML bootstrap support (52%) and BI posterior probabilities (0.72) I3) that includes the new species Longidorus cholevae, two populations of Longidorus carniolensis from Slovenia and two populations of Longidorus poessneckensis from the Czech Republic and Slovakia. The second clade (II) consists of two well-supported subclades: II1) consisted of Longidorus caespiticola from Slovenia and Belgium and one Longidorus cf. caespiticola from Bulgaria and subclade II2) consisted of three populations of Longidorus caespiticola from Scotland, Belgium and Germany. It is possible that these populations represent two different species that requires further investigation.
The phylogenetic reconstructions of the partial 18S-ITS1 region revealed more unstable groups due to the shorter sequence length and higher sequence variability. Three of the Longidorus spp. belonging to the above mentioned group (Longidorus cf. caespiticola, Longidorus helveticus and Longidorus macrosoma)and two additional species(Longidorus pius and Longidorus raskii) originating from Macedonia and Switzerland have been separated from other ITS1 Longidorus sequences (the tree not presented) and further analysed (Fig. 11). Three clades were distinguished, two well supported clades consisting of: 1) Longidorus macrosoma, Longidorus helveticus and Longidorus pius and 2) Longidorus cf. caespiticola and Longidorus raskii, and one not well resolved 3) containing only Longidorus cholevae sp. n. The species forming these clades have similar tail shape in first stage juveniles: digitate in clade 1, bluntly conoidal in clade 2, elongate conoidal in clade 3.
The study was supported by the National Science Fund grants: No B-1405/04, CEBDER (DO 02-15) and BioCORE (INI 03/01.08.2005) projects and the project ANIDIV, funded by the Bulgarian Academy of Sciences. Mutual visits of authors have been supported by a mobility program between the Bulgarian Academy of Sciences and the Consiglio Nazionale delle Ricerche (Bilateral Collaboration Project between BAS and CNR).
List of the species of the genus Longidorus (doi: 10.3897/zookeys.330.5750.app1) File format: Microsoft Word Document (doc).
Explanation note: List of the species of the genus Longidorus Micoletzky, 1922.
A partial polytomous key to the species of Longidorus (doi: 10.3897/zookeys.330.5750.app2) File format: Microsoft Word Document (doc).
Explanation note: A partial polytomous key to the species of Longidorus with long odontostyle (A45) and short tail (H1) based on the key by