(C) 2013 Mehmet Sait Taylan. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
This study focuses on the evolutionary relationships among Turkish species of the cave cricket genus Troglophilus.Fifteen populations were studied for sequence variation in a fragment (543 base pairs) of the mitochondrial DNA (mtDNA) 16S rDNA gene (16S) to reconstruct their phylogenetic relationships and biogeographic history. Genetic data retrieved three main clades and at least three divergent lineages that could not be attributed to any of the taxa known for the area. Molecular time estimates suggest that the diversification of the group took place between the Messinian and the Plio-Pleistocene.
Troglophilus, Rhaphidophoridae, Orthoptera, 16S rDNA, mitochondrial DNA, molecular systematics, cave crickets
Caves are traditionally considered as natural laboratories to understand evolutionary processes related to allopatric divergence because, similarly to remote oceanic islands, by their very nature greatly reduce or hamper gene flow among populations (Poulson and White 1969;
In the peri-Mediterranean area the family is represented by two genera only (Dolichopoda and Troglophilus) with a fairly overlapping Eastern-Mediterranean distribution. Dolichopoda (49 described species) is by far more species-rich than Troglophilus (17 described species). Until now, seven species of Dolichopoda (Dolichopoda aranea Bolivar, 1899, Dolichopoda pusilla Bolivar, 1899, Dolichopoda euxina Semenov, 1901, Dolichopoda sbordonii Di Russo & Rampini, 2006, Dolichopoda lycia (Galvagni, 2006), Dolichopoda noctivaga Di Russo & Rampini, 2007, Dolichopoda sutini Rampini & Taylan, 2012) and five species of Troglophilus (Troglophilus escalerai Bolivar, 1899, Troglophilus gajaci Us, 1974, Troglophilus adamovici Us, 1974, Troglophilus bicakcii Rampini & Di Russo, 2003, Troglophilus tatyanae Di Russo & Rampini, 2007) have been reported from Anatolian caves. As far as Troglophilus is concerned, the first species to be described from the area was Troglophilus escalerai (Jenidje-Kale cave) by Bolivar in
Of these two genera of cave crickets inhabiting the peri-Mediterranean area, Dolichopoda has received comparatively more scientific attention than Troglophilus. Both genera have been the object of a number of studies based on a variety of molecular markers. Nowadays for Dolichopoda we have a very detailed knowledge from the population level (with special emphasis on those species inhabiting the Italian peninsula) up to the phylogenetic relationships among the vast majority of taxa ascribed to the genus (
For this study, we explored 71 caves from the Black Sea, Aegean, Mediterranean and inland areas of Turkey and found and collected cave crickets belonging to the genus Troglophilus from 15 of them (Figure 1; Table 1). We included in the study all the five known Turkish species of Troglophilus, including Troglophilus escalerai that was not analyzed in
Species list and details of the sampling localities of Turkish Troglophilus populations and species. Numbers in the first column match those in Figure 1.
| No | Species | Cave name | Locality | N (north), E (east) | Date | Altitude (m a.s.l.) | |
|---|---|---|---|---|---|---|---|
| Black Sea Region | |||||||
| 1 | Troglophilus tatyanae | Epigian forest | Artvin, Kafkasor | 41.098, 41.475 | 29–30/06/2000 | 1300 | |
| Aegean Region | |||||||
| 2 | Troglophilus sp .4 | Havran cave | Balıkesir, Havran | 39.34499, 27.10336 | 01/11/2008 | 115 | |
| 3 | Troglophilus sp .1 | Gökçeler cave | Muğla, Milas | 37.11378, 27.45982 | 25/11/2008 | 120 | |
| 4 | Troglophilus sp .1 | Güroluk cave | Muğla, Fethiye | 36.47564, 28.58646 | 26/06/2008 | 450 | |
| Mediterranean and Central Anatolia Region | |||||||
| 5 | Troglophilus adamovici | Zindan cave | Isparta, Aksu | 37.48424, 31.05060 | 03/05/2009 | 1286 | |
| 6 | Troglophilus bicakcii | Direkliin cave | Konya, Beyşehir | 37.35548, 31.28549 | 02/07/2008 | 1209 | |
| 7 | Troglophilus bicakcii | Bıçakçı cave | Konya, Derebucak | 37.23648, 31.32166 | 23/08/2009 | 1372 | |
| 8 | Troglophilus bicakcii | Balatini cave | Konya, Derebucak | 37.21706, 31.35060 | 22/08/2009 | 1379 | |
| 9 | Troglophilus bicakcii | Feyzullah cave | Konya, Derebucak | 37.15771, 31.27314 | 22/08/2009 | 1508 | |
| 10 | Troglophilus adamovici | Ferzene cave | Konya, Seydişehir | 37.22854, 31.50071 | 24/08/2009 | 1390 | |
| 11 | Troglophilus sp .2 | Ferzene cave | Konya, Seydişehir | 37.22854, 31.50071 | 24/08/2009 | 1390 | |
| 12 | Troglophilus adamovici | Tınaztepe cave | Konya, Seydişehir | 37.14855, 31.35692 | 24/08/2009 | 1461 | |
| 13 | Troglophilus sp .3 | Dim cave | Antalya, Alanya | 36.32405, 32.06549 | 30/08/2009 | 232 | |
| 14 | Troglophilus gajaci | Cennet cave | Içel, Silifke | 36.27120, 34.06383 | 05/06/2009 | 135 | |
| 15 | Troglophilus escalerai | Döngel cave | Maraş, Narliseki | 37.51557, 36.38476 | 06/06/2009 | 647 | |
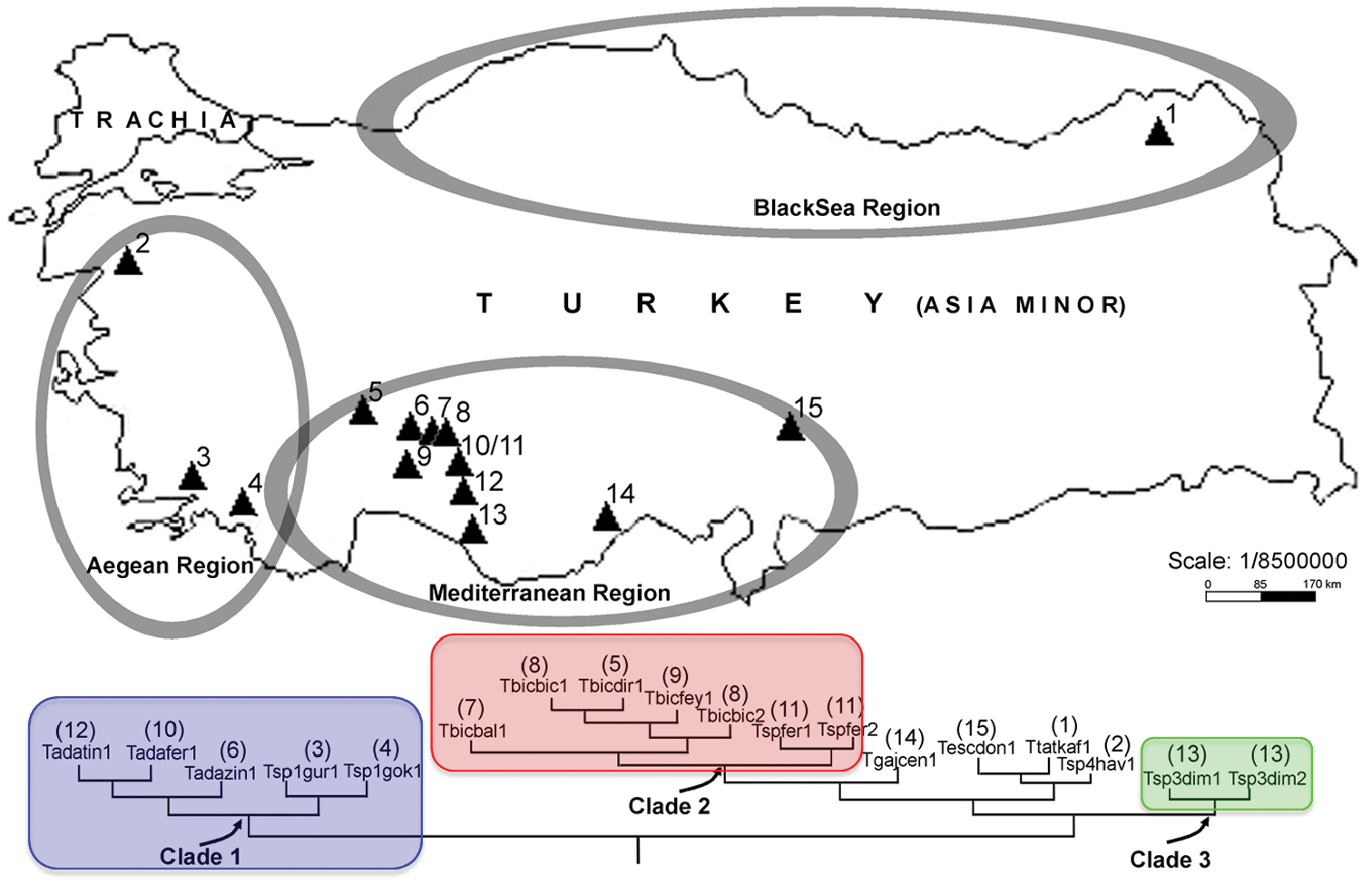
Geographic position of the fifteen caves were we sampled the Troglophilus populations analyzed in the study. Numbers correspond to those in Table 1. The lower half of the figure depicts the phylogeography of Troglophilus in Turkey (for details see Discussion); colors of clades match those in Figure 2.
Ten caves have been checked for each region in Turkey (Mediterranean, Central Anatolian, Aegean and Black Sea region) to collect cave crickets and fifteen sampled populations belonged to the genus Troglophilus; of these eleven were in the Mediterranean and Anatolian region, three in the Aegean region and one in the Black Sea region (Figure 1). All the known five Turkish species (Troglophilus escalerai, Troglophilus gajaci, Troglophilus adamovici, Troglophilus bicakcii, Troglophilus tatyanae) and four new taxa/populations from Muğla, Alanya, Seydişehir and Balıkesir provinces (see Table 1, Figure 1) were included in this study. The latter four taxa are hereto considered as non-described species because it was not possible to attribute them on morphological grounds to any of the Troglophilus species known for the area. Specimens were collected between 2008 and 2009 by hands searching on walls and grounds of caves through the day. Morphological identification of specimens was performed using a stereomicroscope Leica MZ 12.5 equipped with a “camera lucida” and photo camera. Specimens were preserved in absolute ethyl alcohol at AUZM (Akdeniz University Zoology Museum, Antalya, Turkey).
Genomic DNA was extracted from the hind femoral muscle using I-genomic CTB DNA Extraction Mini Kit (type G protocol for Insect, Cat. No 17341, Macrogen Inc.). A 532-535 base pair (bp) fragment of the mitochondrial 16S rDNA gene was amplified through the Polymerase Chain Reaction (PCR) from each individual samples. The primers used were ER232 (5’-CGCCTGTTTAACAAAAACAT-3’) and ER233 (5’-CCGGTCTGAACTCAG ATGACTG-3’) (
Sequences were aligned in ClustalX (
Divergence times were calculated in a Bayesian MCMC framework by using Beast 1.4.6 (
The 16S alignment consisted of 543 nucleotidic positions. Sequences were obtained for each individual and a total of 38 samples belonging to 15 populations were analyzed and 18 different haplotypes found. Sequences of these unique haplotypes have been deposited in GenBank under the Accession N. JX968473-JX968490. Table 2 shows the absolute frequency of these 18 haplotypes in the different populations included in the study. In the final alignment 123 sites were variable and 53 were parsimony informative. The transition/transversion (ti/tv) ratio ranged from 1.7 to 2.2. Ti values accounted for about 62% or 69% of all substitutions when the outgroup was alternatively included or excluded. Divergence in the 16S rDNA gene ranged from 1.1% to 13.1% at the ingroup level (16.1% with the outgroup included).
Troglophilus species included in this study, the names of the sampling locations, their sample size per locality (N), number of haplotypes, the codes of the haplotypes as they appear in Figure 1 and 2.
| Species | Population | Locality | N | Haplotype number | Haplotype code |
|---|---|---|---|---|---|
| Black Sea Region | |||||
| Troglophilus tatyanae | Kafkasor | Artvin | 1 | 1 | Ttat-kaf1 |
| Aegean Region | |||||
| Troglophilus sp .1 | Güroluk cave | Muğla, Fethiye | 3 | 1 | Tsp1-gur1 |
| Gökçeler cave | Muğla, Milas | 1 | 1 | Tsp1-gok1 | |
| Troglophilus sp .4 | Havran Cave | Balıkesir, Havran | 3 | 1 | Tsp4-hav1 |
| Mediterranean and Central Anatolia region | |||||
| Troglophilus escalerai | Döngel cave | K.Maraş, Döngel | 3 | 1 | Tesc-don1 |
| Troglophilus gajaci | Cennet cave | İçel, Silifke | 5 | 1 | Tgaj-cen1 |
| Troglophilus adamovici | Zindan cave | Isparta, Aksu | 4 | 1 | Tada-zin1 |
| Tınaztepe cave | Konya, Seydişehir | 2 | 1 | Tada-tin1 | |
| Ferzene cave | Konya, Seydişehir | 1 | 1 | Tada-fer1 | |
| Troglophilus bicakcii | Bıçakçı cave | Konya, Derebucak | 2 | 2 | Tbic-bic1, Tbic-bic2 |
| Direkliin cave | Konya, Beyşehir | 2 | 1 | Tbic-dir1 | |
| Feyzullah cave | Konya, Derebucak | 2 | 1 | Tbic-fey1 | |
| Balatini cave | Konya, Derebucak | 2 | 1 | Tbic-bal1 | |
| Troglophilus sp .2 | Ferzene cave | Konya, Seydişehir | 2 | 2 | Tsp2-fer1, Tsp2-fer2 |
| Troglophilus sp .3 | Dim Cave | Antalya, Alanya | 5 | 2 | Tsp3-dim1, Tsp3-dim2 |
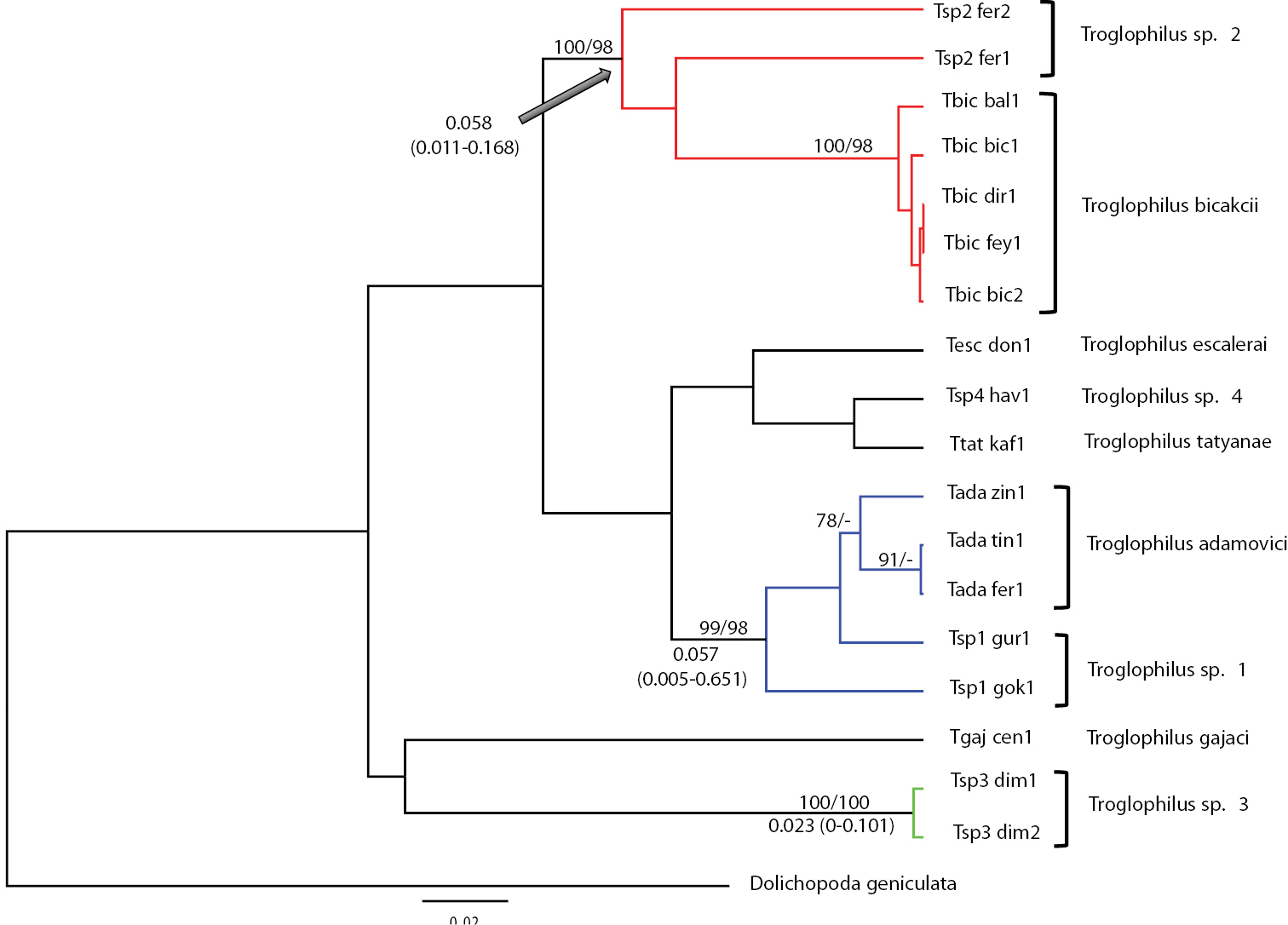
Figure 2 shows the Bayesian phylogram based on the GTR + G (gamma distribution shape parameter a = 0.188) model chosen by JMODELTEST as the one best fitting our data and summarizes the results of the other phylogenetic methods employed in the study. Bayesian and MP searches were all largely congruent with one another. MP searches yielded three equally parsimonious trees with length (L) = 193 steps, homoplasy index (HI) = 0.249, consistency index (CI) = 0.751, retention index (RI) = 0.780. All analyses consistently recovered three well-supported clades, whose geographic distribution is shown in Figure 1.
Clade 1 includes Troglophilus adamovici and Troglophilus sp. 1 populations, which are distributed in the Northern Mediterranean region (Isparta) through the western Taurus Mountain, Southern Central Anatolian regions with a Mediterranean climate and Southern Aegean region (Muğla, Fethiye, Milas). Clade 2 contains Troglophilus bicakci and Troglophilus sp. 2 populations, which are distributed in the Southern Central Anatolian region through Kembos Polye and Konya, Seydişehir, Derebucak and Beyşehir Provinces. This clade overlaps with Clade 1 in the Seydişehir Province (Ferzene cave). Clade 3 comprises Troglophilus sp. 3 population only and it is geographically restricted to the Antalya area (Alanya, Dim cave). The cave is located near the Dim River in the Southern Mediterranean Region.
Average GTR + G distance between Clade 1 and 2 is 0.063 ± 0.025, between Clade 2 and 3 is 0.058 ± 0.021 and between Clade 1 and 3 is 0.050 ± 0.005. Time estimates retrieved from the Bayesian MCMC analyses for the three main clades are illustrated in Figure 2. In all cases 95% credible intervals for node age estimates overlapped. The data did not conform to a clock-like behavior, the coefficient of variation being 0.87 (95% High Posterior Density, HPD: 0.393-1.435; ESS: 1214.24). Parent and daughter branches showed no co-variation, the mean covariance being -5.83-2 (HPD: -0.321-0.237; ESS: 7191.33). The 95% High Posterior Density spans zero; this implies that branches with fast and slow rates are next to each other in the phylogenetic tree. There is thus no evidence of autocorrelation of rates in the tree. Ages of Clades 1, 2, and 3 ranges between 5.8 and 2.3 million years; the lack of a clear calibration point resulted in a chronogram with relatively ample confidence intervals (Figure 2).
Results of the Mantel test (
Bayesian phylogram among Troglophilus haplotypes from Turkey. Haplotype codes match those in Table 2. Numbers at nodes are statistical supports for the Bayesian and MP searches (first and second value, respectively); only values ≥ 75% are reported. The three supported clusters are described in the text are highlighted here in blue (clade 1), red (clade 2) and green (clade 3). Bold values are node ages (in Myr %) as obtained by the BEAST analyses; 95% HPD intervals are shown in parentheses.
The genetic data confirmed the validity of the already described species, with conspecific populations firmly forming monophyletic clusters. On the other hand, four deeply genetically divergent lineages (Troglophilus sp. 1, 2, 3 and 4) could not be attributed to any of the previously described species and could hence represent new taxa. The mean GTR + G genetic distance between the described Troglophilus species included in our study (
Overall, we could distinguish three main clades; all received strong support in our phylogenetic analyses (Figures 1 and 2). Clade 1 includes Troglophilus adamovici and the new species Troglophilus sp. 1 distributed in the Isparta, Konya and Izmir provinces. Clade 2 comprises Troglophilus bicakcii and the new species Troglophilus sp. 2 (from Ferzene cave) both from the Konya province, while Clade 3 includes Troglophilus sp. 3 population distributed in the Dim Cave in Antalya. The phylogenetic placement of Troglophilus gajaci, Troglophilus escalerai, Troglophilus tatyanae, and Troglophilus sp. 4 could not be resolved by the data and remains controversial.
The Mantel test (
An additional point of interest of this study is the confirmation of the results of
The estimated divergence times range from the Messinian to the Plio-Pleistocene (Figure 2). The oldest estimated divergence times are around 5.8 Ma (Messinian) and coincide with the last period of the uplifting the Anatolian Plateau, which arose 5-10 Ma as a consequence of the northward movement of the Arabian Plate (
The estimated divergence time for Troglophilus sp. 3 is more recent (2.3 Ma), dating to the Plio-Pleistocene, which was characterized by alternating dry/cold and warm/humid phases. The climatic fluctuations during the Plio-Pleistocene likely led to ecological fragmentation with subsequent genetic isolation and speciation in the area. This hypothesis is also supported by the results from the Dolichopoda species, whose radiation also appears to have followed the climatic changes of the Plio-Pleistocene (
Since the syntopic Troglophilus adamovici and Troglophilus sp. 2 in the Ferzene cave do not interbreed, their secondary contact must have taken place after the diversification within Clades 1 and 2, certainly more recently than the Messinian. Even though we are not in the position to date when the secondary contact actually happened, we suspected that this was favored by one of the many warm and humid climatic phases of the Quaternary, which allegedly promoted epigean dispersal among lineages that had been previously confined to caves.
Our time estimates for the splitting events within the Anatolian representatives of Troglophilus are in agreement with those reported in
Finally, it should not be overlooked that this study is limited to the Turkish area and is based on a single mitochondrial marker. To place these results in a broader perspective and to understand in details the evolutionary trajectories followed by the genus, we need to expand our sampling by covering its whole distribution range and by combining multiple mitochondrial and nuclear loci. To these aims our ongoing research activity is currently devoted.
We thank Dr. Mehmet Öz for his advices and Dr. Deniz Şirin (Namık Kemal University, Biology Department), Emrah Özel (Hacettepe University, Biology Department), Yusuf Dikmentepe for their help in collecting cave samples. For the revision of English we thank Metin Albükrek and Duygu Yaşar Şirin. Two reviewers provided useful comments. This study includes data of the Project no° 2008.03.0121.002 (Prof. Dr. M. Öz), which was supported by Akdeniz University Scientific Research Found (Turkey, Antalya).