(C) 2013 David K-B Cheung. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Cheung DK-B, Brunke AJ, Akkari N, Souza CM, Pape T (2013) Rotational Scanning Electron Micrographs (rSEM): A novel and accessible tool to visualize and communicate complex morphology. ZooKeys 328: 47–57. doi: 10.3897/zookeys.328.5768
An accessible workflow is presented to create interactive, rotational scanning electron micrographs (rSEM). These information-rich animations facilitate the study and communication of complex morphological structures exemplified here by male arthropod genitalia. Methods are outlined for the publication of rSEMs on the web or in journal articles as SWF files. Image components of rSEMs were archived in MorphBank to ensure future data access. rSEM represents a promising new addition to the toolkit of a new generation of digital taxonomy.
Scanning electron microscopy, digital taxonomy, interactive animation
In the effort to discover, describe and organize the
planet’s biodiversity, taxonomists are faced with the
challenge of providing clear and concise diagnostic
characters in species descriptions, often involving
structures with complex morphology. To address this
challenge, a variety of imaging methods have emerged over
the past ten years to complement more traditional line
drawings, which continue to play an important role in
species descriptions. Traditionally, diagnostic characters
are illustrated in one or a set of standard views to
facilitate the comparison of taxa within and between
publications by taxonomists and other users. In this
current paradigm, characters not or suboptimally visible
in these conventional views are difficult for taxonomists
to convey and for users to interpret, placing a constraint
on the range of published morphological data. These issues
are further exacerbated in the case of complex or
asymmetrical male genitalia, which are challenging to
clearly and accurately describe in words. Multifocal,
‘stacked’ images of external structures or habitus are now
standard in most taxonomic descriptions and recent
advances in micro-computed tomography (µCT) and magnetic
resonance imaging (MRI) promise to yield exciting new
character data from both external and internal morphology
(i.e.,
Since the early 1970s (for an early example, see
Recently, we have sought for an accessible method to integrate and present diagnostic differences of taxa existing at non-standard and standard angles, using SEM micrographs. Here we describe an SEM image workflow that results in information-rich, rotatable animations (hereafter referred to as rSEMs – rotational scanning electron micrographs), which can be either web published or embedded in PDF articles for publication in journals accepting such media (e.g., ZooKeys). Examples of rSEM from different arthropod groups (Coleoptera, Diptera, Myriapoda) are provided.
Larger, dry specimens were mounted (see below) without
special cleaning or dehydration. Specimens preserved in
70% ethanol or glycerin were first cleaned of debris and
then dehydrated in 96% ethanol, then acetone, and air
dried before mounting. Fragile specimens (e.g. small
flies, nymphs, larvae) should be dried using critical
point drying to avoid shriveling or collapsing (
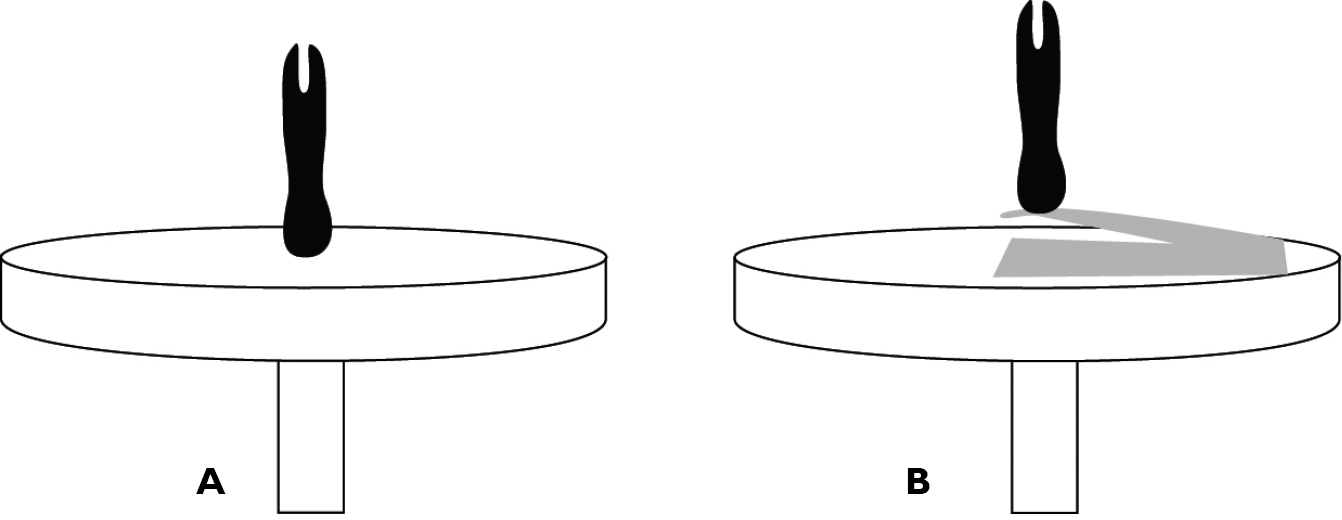
Specimens directly (A) and secondarily (B) mounted on an aluminum SEM stub.
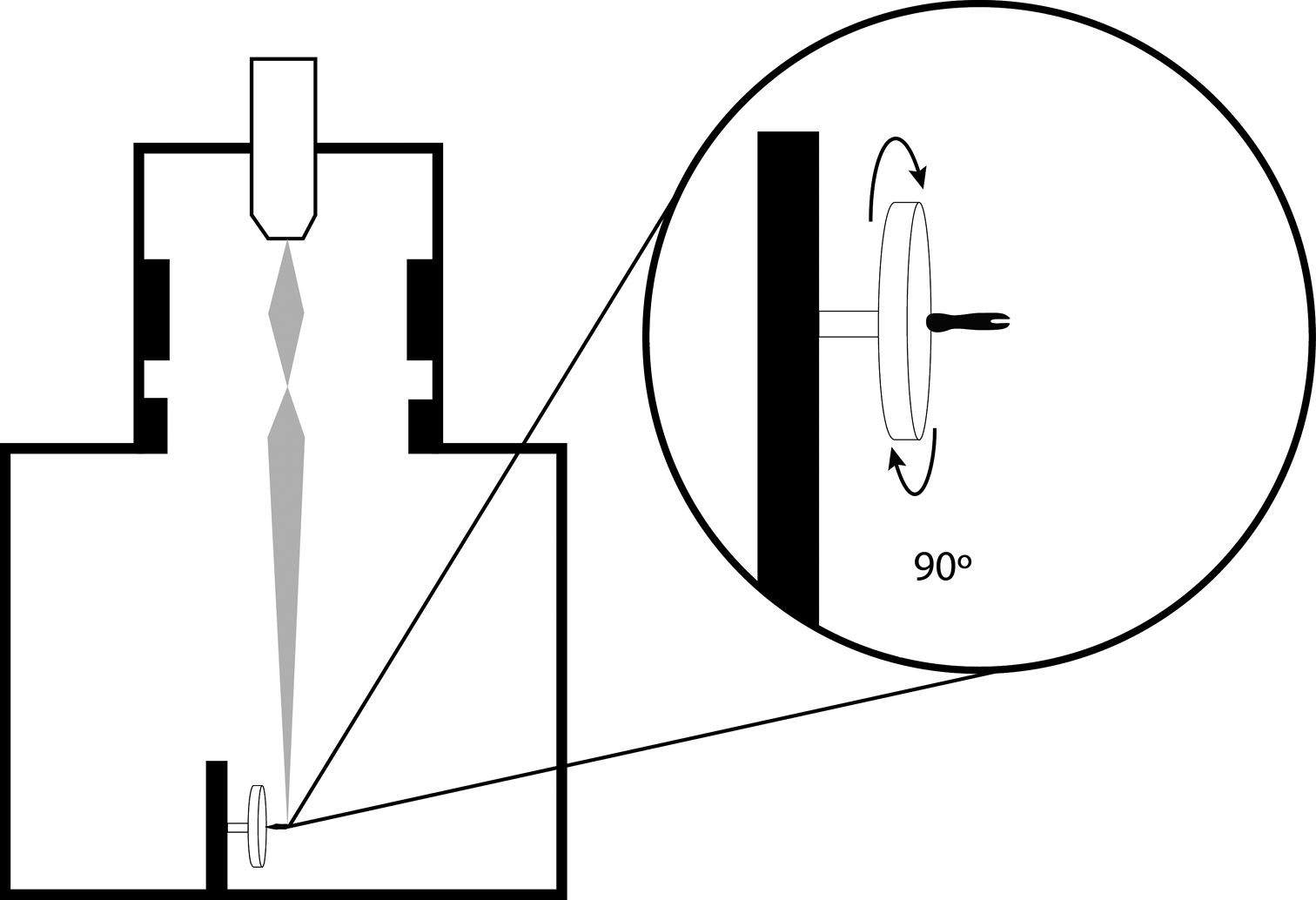
For users unfamiliar with the elements of this workflow, a step-by-step set of instructions is provided in provided in Appendix 1. Specimens were sputter-coated with platinum/palladium and studied with a JEOL JSM-6335F scanning electron microscope. Note that the microscope stage MUST be able to tilt to 90° in order to produce a usable animation (Fig. 2). Images were taken at fixed rotational intervals and named sequentially.
Images were then processed (adjusting the exposure, contrast, highlights, shadows, whites, blacks) and cropped to improve image quality and detail, and to ensure that all images were ‘aligned’, such that they form a smooth transition when browsed in sequence. Images were exported as JPG. Image processing can be accomplished using subscription based software such as Adobe® Lightroom® or Photoshop®, or open-source software such as ImageJ (http://rsbweb.nih.gov/ij/). The images can now be integrated into an animation for submission to a scientific journal or published on a website. The individual images generated for this study were archived in MorphBank under object #830913, 830920-830935, 830953-830970 and 831016-831034; they can be found together by searching for collection ‘rSEM’.
Schematic representation of SEM specimen and stage positioning for creation of rSEM image frames.
rSEMs in the present article and in
import flash.events.Event;
instancename.stop();
var frameTo:Number=0;
addEventListener(Event.ENTER_FRAME, goTo);
function goTo(e:Event):void {
frameTo=int(mouseX/stage.stageWidth*instancename.totalFrames)+1;
instancename.gotoAndStop(frameTo);
}
To publish an rSEM on the web, a program or web plug-in
is needed to integrate the images into an animation.
There is a multitude of these available online of varying
quality and functionality (search ‘object viewer’ or ‘360
panorama’) ranging from subscription-based (Magic 360™ (http://www.magictoolbox.com/magic360/))
to free and open-source (Reel© 1.2.1 (http://jquery.vostrel.cz/reel#dict)).
A set of scripts needs to be downloaded and a folder
containing the sequentially numbered images should be
created. We have tested the two previously mentioned
options and both result in web animations that are
compatible with all operating systems, including mobile
devices. For full instructions, readers are directed to
the tutorials provided by the publishers (see links
above). The rSEMs included in the html version of
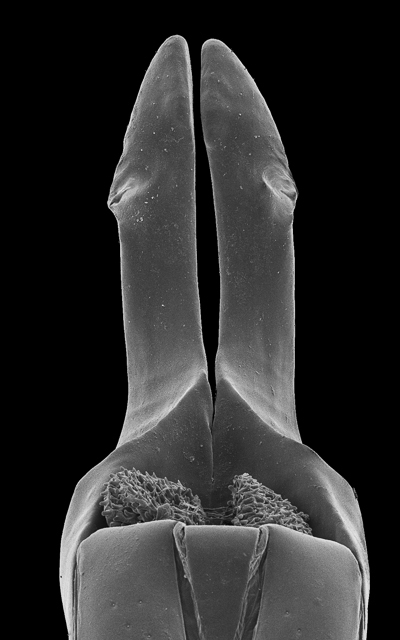
rSEM illustrating the distiphallus of Oxysarcodexia (Xylocamptopsis) fringidea (Curran & Walley) (Sarcophagidae, Diptera). The offline version of this rSEM can be explored by moving the mouse over the image in the x-axis and can be viewed using Adobe® Acrobat® Reader version 8 or higher, with Adobe® Flash® Player installed. Compatible with Windows and Mac OS X.6 (Snow Leopard, 2009) or newer. Image components were archived in MorphBank as objects #830953-830970.

rSEM illustrating the posterior gonopod of Ommatoiulus khroumiriensis Akkari & Enghoff (Julidae, Diplopoda). The offline version of this rSEM can be explored by moving the mouse over the image in the x-axis and can be viewed using Adobe® Acrobat® Reader version 8 or higher, with Adobe® Flash® Player installed. Compatible with Windows and Mac OS X.6 (Snow Leopard, 2009) or newer. Image components were archived in MorphBank as objects #831016-831034.
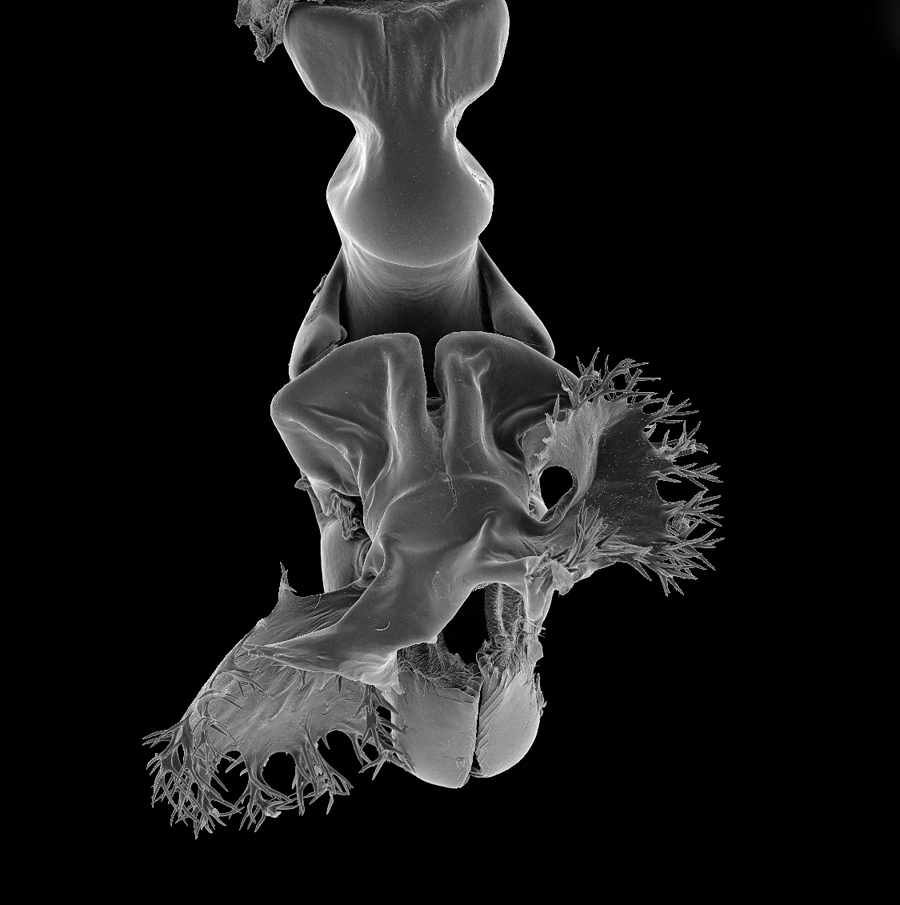
rSEM illustrating the median lobe (paramere removed) of Bolitogyrus sp. (Staphylinidae, Coleoptera). The offline version of this rSEM can be explored by moving the mouse over the image in the x-axis and can be viewed using Adobe® Acrobat® Reader version 8 or higher, with Adobe® Flash® Player installed. Compatible with Windows and Mac OS X.6 (Snow Leopard, 2009) or newer. Image components were archived in MorphBank as objects #830913, and 830920-830935.
The workflow described herein will generate interactive,
information-rich animations that facilitate both the study
and communication of complex morphology in systematics (Figs 3–5). rSEMs
combine sharply detailed SEM micrographs into a single,
holistic representation of a structure that may even
reveal diagnostic characters visible but yet unnoticed
when using a light microscope. The use of SEM is not
always optimal or appropriate as users may not have access
to facilities or cannot image large, fragile or unique
specimens. In these cases, morphological characters will
still need to be illustrated by line drawings or stacked
images, despite their potential complexity. Differences in
color and degree of sclerotization are also lost in SEM.
However, rSEMs can be created using technology and
software in widespread use and provide an accessible
alternative to other imaging methods such as µCT if the
user is focused on structural aspects of external
morphology. As HTML5 is becoming more prominent than
Flash® as a media development standard, an update is
planned to incorporate instructions for developing rSEM in
HTML5. rSEMs are integrated into a taxonomic revision for
the first time in a companion article (
With the development of Web 2.0 and Open Access
publishing, the demand for image quality and methods for
visualizing taxonomic traits has increased. Recently,
publishing 3D models and embedding multimedia files in
biomedical journals has become common practice (e.g.
Here, additional rSEMs of arthropod genitalia were provided as examples of complex morphology. However, this approach could easily be extended to structures than other genitalia, and could be applied to the illustration of a phylogenetically important character system (e.g., a rotatable insect head capsule). So far, we have only animated rSEM in the x-axis because our test structures varied mainly along the y-axis. However, illustration of structures along both x and y-axes of rotation, while requiring many more images, may be a powerful form of data delivery in certain situations. For example, if a finely controlled, electronic stage is available, full-color, rotatable habitus images could be produced using a dissecting microscope and stacking software. There are clearly far more applications of rSEM or rSEM-like animations than we have presented here, and we encourage readers to experiment with this technique in their own scientific exploration of Earth’s biodiversity.
We are grateful to Nikolaj Scharff (Copenhagen) for sharing some insights on SEM mounting techniques, Petr Vostrel for making his Reel plug-in freely available online. Daniel Mietchen, Pavel Stoev and one anonymous reviewer provided comments that greatly improved this manuscript.
New user guide to the creation and publication of rSEM (doi: 10.3897/zookeys.328.5768.app1) File format: Microsoft Word document (doc).
Explanation note: A step-by-step style of instructions for new users, involving the software used by the authors. Note: in the future, these instructions may not correspond exactly with the interfaces of the corresponding software.
rSEM illustrating the distiphallus of Oxysarcodexia (Xylocamptopsis) fringidea (Curran & Walley) (Sarcophagidae, Diptera); web-published using Magic 360TM script files. Click and drag to rotate the rSEM and point click to open and close the magnification tool. (doi: 10.3897/zookeys.328.5768.app2) File format: Hypertext Markup Document, archived (zip).
Explanation note: Example of rSEM published online using Magic 360TM script.