(C) 2013 Jean-François Landry. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Landry J-F, Hebert PDN (2013) Plutella australiana (Lepidoptera, Plutellidae), an overlooked diamondback moth revealed by DNA barcodes. ZooKeys 327: 43–63. doi: 10.3897/zookeys.327.5831
The genus Plutella was thought to be represented in Australia by a single introduced species, P. xylostella (Linnaeus), the diamondback moth. Its status as a major pest of cruciferous crops, and the difficulty in developing control strategies has motivated broad-ranging studies on its biology. Prior genetic work has generally supported the conclusion that populations of this migratory species are connected by substantial gene flow. However, the present study reveals the presence of two genetically divergent lineages of this taxonin Australia. One shows close genetic and morphological similarity with the nearly cosmopolitan Plutella xylostella. The second lineage possesses a similar external morphology, but marked sequence divergence in the barcode region of the cytochrome c oxidase I gene, coupled with clear differences in genitalia. As a consequence, members of this lineage are described as a new species, P. australiana Landry & Hebert, which is broadly distributed in the eastern half of Australia.
Australia, karsholtella, new species, xylostella
The diamondback moth, Plutella xylostella (Linnaeus), is one of the most damaging insect pests, attacking cruciferous crops, such as cabbage and cauliflower, across its nearly cosmopolitan range. Because biological agents have proven ineffectual (
The present study was motivated by a large-scale DNA barcode study of Australian Lepidoptera (
Most of the Australian specimens of Plutella examined in this study were collected at UV light from 2004–2012 as a result of a sampling program to obtain specimens of Lepidoptera for DNA barcode analysis. The results from Australian specimens were placed in a broader perspective through the inclusion of sequence records from two specimens (when available) for each of five other non-Australian Plutella species and one species in the closely allied genus Eidophasia possessing coverage on BOLD (
DNA extracts were prepared from a single leg removed from each of 402 specimens of Plutella xylostella. DNA extraction, PCR amplification of the barcode region of COI, and subsequent sequencing followed standard protocols at the Canadian Centre for DNA Barcoding (
Details on the date and site of collection for each specimen, as well as a photograph are available through the following dataset (dx.doi.org/10.5883/DS-PLUT1). The same DOI provides access to the sequence records, trace files, and primer sequences used for PCR amplification, together with GenBank accession numbers.
Genitalia dissections and slide mounts followed
The configuration of the vinculum-saccus in Plutella male genitalia makes it difficult to spread the genitalia open in the standard manner for slide mounting without causing significant distortion. As a result, the differences between Plutella australiana and Plutella xylostella are not readily apparent if standard mounts are attempted, even if the unrolling technique is employed. To display them properly, the different parts of the male genitalia were separated and temporarily mounted in lactic acid on slides under cover slips raised with vinyl props as wedges to prevent distortion or flattening. After photography, genitalia parts were permanently embedded in Euparal. Genitalia were photographed with a Nikon DS-Fi1 digital camera mounted on a Nikon Eclipse 800 microscope at magnifications of 100×. Nikon’s NIS 2.3 Elements was used to assemble multiple photos of different focal planes into single deep-focus images.
AMS Australian Museum, Sydney, New South Wales, Australia
ANIC Australian National Insect Collection, CSIRO, Canberra, Australia
ASCU Agricultural Scientific Collections Unit, Orange Agricultural Institute, Orange, New South Wales, Australia
BIOUG Biodiversity Institute of Ontario, University of Guelph, Guelph, Ontario, Canada
BMNH The Natural History Museum, London, UK
CNC Canadian National Collection of Insects, Arachnids, and Nematodes, Ottawa, Ontario, Canada
USNM National Museum of Natural History, Smithsonian Institution, Washington, D.C., USA
ZMUC Zoological Museum, University of Copenhagen, Copenhagen, Denmark
Analysis revealed that individuals of the five non-Australian species of Plutella and the species of Eidophasia each formed a distinct sequence cluster in the NJ tree (Figure 1). Interspecific divergences between pairs of these taxa averaged 9.5% and ranged from 2.2%–14.0%. The lowest mean divergence value was between Plutella notabilis Busck and Plutella hyperborella Strand, while the greatest was between Plutella xylostella and Plutella geniatella Zeller (Table 1). Because of their deep divergence, each species was assigned to a different Barcode Index Number (BIN) (
NJ tree based on K2P distances for the barcode region of the cytochrome c oxidase I gene among seven species of the genus Plutella and one member of the closely allied genus Eidophasia. Because of the large number of specimens for both Plutella xylostella and Plutella australiana, the tree nodes have been collapsed and specimen records are plotted by state for Australia and by country of origin in other cases. The bracketed numerals indicate the number of specimens from each site. The type specimen of Plutella karsholtella is reassigned to Plutella xylostella and is the only specimen of this species from Spain (Canary Islands).
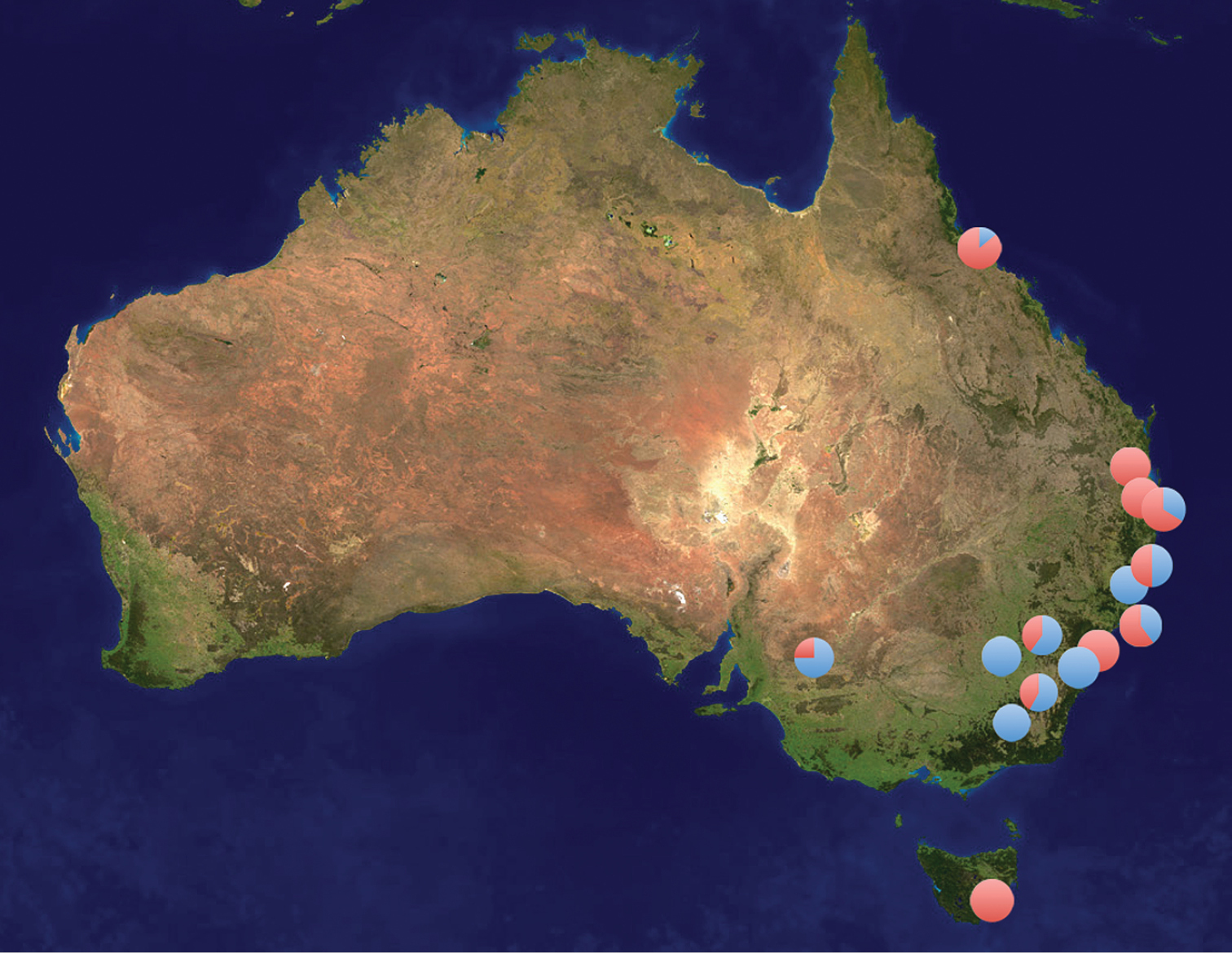
Sites in Australia where specimens of Plutella xylostella (red) and Plutella australiana (blue) have been collected. The pie diagrams show the proportion of the two species at each site. These records only include specimens identified through DNA barcode analysis.
Mean sequence divergences (K2P) for the barcode region of the COI gene for seven members of the genus Plutella and one member of the closely allied genus Eidophasia.
| Plutella armoraciae | Plutella geniatella | Plutella hyperboreella | Plutella xylostella | Plutella notabilis | Plutella porrectella | Eidophasia vanella | |
|---|---|---|---|---|---|---|---|
| Plutella geniatella | 0.071 | ||||||
| Plutella hyperboreella | 0.050 | 0.040 | |||||
| Plutella xylostella | 0.120 | 0.140 | 0.126 | ||||
| Plutella notabilis | 0.066 | 0.043 | 0.022 | 0.138 | |||
| Plutella porrectella | 0.079 | 0.071 | 0.058 | 0.138 | 0.066 | ||
| Eidophasia vanella | 0.098 | 0.121 | 0.107 | 0.115 | 0.110 | 0.124 | |
| Plutella australiana | 0.091 | 0.098 | 0.085 | 0.086 | 0.088 | 0.096 | 0.105 |
http://zoobank.org/20416523-A949-4784-BB18-00A8BA208B6D
http://species-id.net/wiki/Plutella_australiana
Barcode Index Number: BOLD:AAC6876
Figs 3–9, 17, 19, 21, 23, 25, 27, 29, 31Thirty males and 22 females were included in the type series. Five additional specimens were also barcoded but excluded from the type material due to their poor condition.
Holotype ♂, specimen # BIOUG00844-C06, labelled as follows: [label1] “Subset of: LOT# L#2010AUS-0039 | AUS: New South Wales [sic]: Canberra; Cook | 35.2612°S, 149.0591°E 632m asl 15-Oct-10 | coll. Christy Carr, Paul Hebert, Stephanie Kirk, | Jaclyn McCormick, Jayme Sones”; [label2, pale yellow] “Barcode of Life | DNA voucher specimen | Sample ID BIOUG00844-C06 | BOLD Proc. ID: PHLCA1136-11”; [label3, pale green] “genitalia slide | JFL1731 [male symbol]”; [label4, orange] “HOLOTYPE | Plutella | australiana | J.-F. Landry & Hebert”. Genitalia slide JFL1731. Condition of specimen: double-mounted, wings partly spread, left antenna missing, right hind and left mid- and hind legs removed for DNA barcoding. Deposited in ANIC.
PARATYPES: 29 males, 22 females. Australian Capital Territory: Canberra, Cook, 8 Moss Street, 35.261°S, 149.059°E, alt. 632 m, UV light, C. Carr, P.D.N. Hebert, S. Kirk, J. McCormick, J. Sones: 1♂, 1.X.2010, specimen # BIOUG00792-E09 (CNC); 2♂, 6.X.2010, specimen # BIOUG00831-A04 (ANIC), BIOUG00831-H06 (BIOUG); 5♂, 7.X.2010–8.X.2010, specimen # BIOUG00788-B01 (ANIC), BIOUG00788-F08 (ANIC), BIOUG00788-F11 (slide JFL1730) (CNC), BIOUG00788-F12 (ANIC); 1♀, 8.X.2010, specimen # BIOUG00829-H10 (CNC); 1♂, 9.X.2010, specimen # BIOUG00843-C02 (BIOUG); 1♂, 2♀, 15.X.2010, specimen # BIOUG00844-A09 (CNC), BIOUG00844-C03 (AMS), BIOUG00844-G03 (ANIC); 1♂, 2♀, 18.X.2010–20.X.2010, specimen # BIOUG00788-G04, slide JFL1740 (CNC), BIOUG00788-G06, slide JFL1736 (CNC), BIOUG00788-G05 (CNC); 1♂, 25.X.2010, specimen # BIOUG00788-H07 (CNC); 1♀, 27.X.2010, specimen # BIOUG00790-G12 (ANIC). Same locality, collected by P.D.N. Hebert: 2♂, 22.III.2011, specimen # BIOUG01025-G05 (ZMUC), BIOUG01025-G06 (CNC); 1♂, 1♀, 10.XI.2011, CCDB-12828-G04 (ZMUC), CCDB-12828-F10 (AMS); 1#m 12.XI.2011, specimen # BIOUG02125-G06 (CNC); 3♂, 1♀, 13.XI.2011, specimen # BIOUG02127-F12 (ANIC), BIOUG02127-G10 (BMNH), BIOUG02127-H01 (ANIC), BIOUG02127-H03 (ANIC); 1♂, 1♀, 16.XI.2011, CCDB-15380-G10 (BIOUG), CCDB-15380-E08 (AMS); 1♀, 18.XI.2011, BIOUG02123-E08 (USNM); 1♀, 23.X.2011, specimen # BIOUG02109-B09 (BMNH); 1♀, 24.X.2011, specimen # BIOUG02109-C07, slide JFL1741 (ANIC); 1♀, 5.XI.2011, specimen # BIOUG02112-F11 (AMS); 1♀, 6.XI.2011, specimen # BIOUG02108-C09, slide JFL1737 (CNC). Same locality, collected by P.D.N. Hebert, R. Labbee, V. Levesque-Beaudin, J. McCormick, J. Sones, J. Webb: 1♀, 29.III.2011–12.IV.2011, specimen # BIOUG01172-G03, slide JFL1738 (ANIC). Canberra, CSIRO property, 35.275°S, 149.111°E, alt. 588 m: 1♀, 14.XI.2011–21.XI.2011, Malaise trap, P.D.N. Hebert, specimen # BIOUG02239-A02 (BIOUG). New South Wales: Byron Bay, 28.658°S, 153.622°E, alt. 13 m: 1♀, 30.XII.2007, P.D.N. Hebert, specimen # 07-NSWBB-0046, slide JFL1684 (CNC). 2800 Pinnacle Rd., Lot 58, 33.297°S, 149.075°E, alt. 920 m., 1♀, 3.III.2005, H. Loecker, specimen # 05-NSW-00731 (ASCU). Orange, 353 Pinnacle Rd., UV light trap, 33.297°S, 149.075°E, 2♂, 1♀, 26.X.2010, H. Loecker, specimen # ww04709–ww04711 (ASCU). Smiths Lake, 32.377°S, 152.504°E, 1♂, 1♀, 24.XII.2010–24.XII.2010, P.D.N. Hebert, specimen # BIOUG00987-B02 (ANIC), BIOUG00987-E12 (slide JFL1735) (CNC). Weddin Mt. National Park/Bimbi State Forest, Grenfell, nr. “Seatons Farm”, 33.913°S, 147.947°E, at light, 1♀, 9.XI.2007, H. Loecker, specimen # AM 2272, slide JFL1739 (ASCU). Hat Head, 31.063°S, 153.052°E, alt. 36.58 m., 2♂, 28.XII.2008, P.D.N. Hebert, specimen # 08-NSWHH-1277 (slide JFL1689) (ANIC), 08-NSWHH-1340 (slide JFL1690) (CNC). South Australia: 1 km N Border Cliffs, near the banks, Renmark, 34.024°S, 140.89°E, 4♂, 25.XI.2011, P.D.N. Hebert, UV light trap, specimen # BIOUG02248-G03 (slide JFL1732) (CNC), BIOUG02248-F12 (ANIC), BIOUG02248-G01 (ANIC), BIOUG02248-G04 (USNM). Lyrup Forest Reserve, 34.274°S, 140.64°E, 1♀, 8.XII.2011, P.D.N. Hebert, UV trap by lake, specimen # BIOUG02246-B09 (ANIC). Pike Creek Woolshed, 34.278°S, 140.711°E, 1♂, 6.XII.2011, P.D.N. Hebert, mercury vapor light, specimen # BIOUG02120-H01 (ANIC).
Australian Capital Territory: Canberra, Manuka, 35.278°S, 149.166°E, 1 ex. (abdomen missing), 16.XII.2005, P. Hebert, specimen # 05-ACTC-285 (BIOUG). New South Wales: 2800 Pinnacle Rd., Lot 58, 33.297°S, 149.075°E, 1♀, 1 ex. (abdomen missing), 24.II.2005, P.D.N. Hebert, specimen # 05-NSW-00732 (ASCU). Ellenborough, Tom’s Creek Retreat, 31.459°S, 152.476°E, 1 ex (abdomen missing), 17.XII.2005, P.D.N. Hebert, specimen # 06-NSWE-00800 (BIOUG). Kosciuszko National Park, Charlottes Pass, 36.26°S, 148.2°E, alt. 1844 m., 1♂, 1♀, 08–09.III.2009, E.D. Edwards, specimens # am10299, am10372 (ANIC). Queensland: Townsville, Hermit Park, 19.283°S, 146.801°E, 1 ex., 01.X.2010, G. Cocks, specimen # gvc15526-1L (AMS).
In external appearance Plutella australiana is indistinguishable from Plutella xylostella. Both species exhibit significant, overlapping variation in forewing pattern (Figs 3–16). Most specimens of both species have the pale, scalloped band along the hind/dorsal margin typically used to recognize Plutella xylostella. That band varies from strongly marked to nearly indistinct (the latter particularly so in females) in both species. Here we illustrate only a selection of the variants, but intermediates in amount of dark specking and spotting, fading of scalloped dorsal band, and intensity of brown colouration, exist among specimens of both examined. No reliable external difference was observed that permits the separation of the two species. Genitalia must be examined and they afford several good characters.
Dorsal aspect of forewings of Plutella australiana and Plutella xylostella. SpecimenID (sample ID) in parentheses. Scale bar = 2 mm. 3 Plutella australiana, male holotype, Australia: Canberra (BIOUG00844-C06) 4 Plutella australiana, female paratype, Australia: New South Wales, Bimbi State Forest (AM 2272) 5 Plutella australiana, male paratype, Australia: Canberra (BIOUG00788-F11) 6 Plutella australiana, female, Australia: Canberra (BIOUG02123-E08) 7 Plutella australiana, female, Australia: Canberra (BIOUG02112-F11) 8 Plutella australiana, female, Australia: Canberra (BIOUG02108-C09) 9 Plutella australiana, male, Australia: Canberra (CCDB-15830-E08) 10 Plutella xylostella, male, Australia: Canberra (CCDB-12828-E05) 11 Plutella xylostella, male, Australia: Canberra (BIOUG02113-F05) 12 Plutella xylostella, female, Australia: Canberra (BIOUG01172-A09) 13 Plutella xylostella, female, Australia: Canberra (CCDB-12828-G07) 14 Plutella xylostella, female, Australia: Canberra (CCDB-12828-H01) 15 Plutella xylostella, female, Canada: Québec (CNCLEP00098486) 16 Plutella xylostella, female holotype of Plutella karsholtella, Canary Islands: Tenerife (ZMUC00401145).
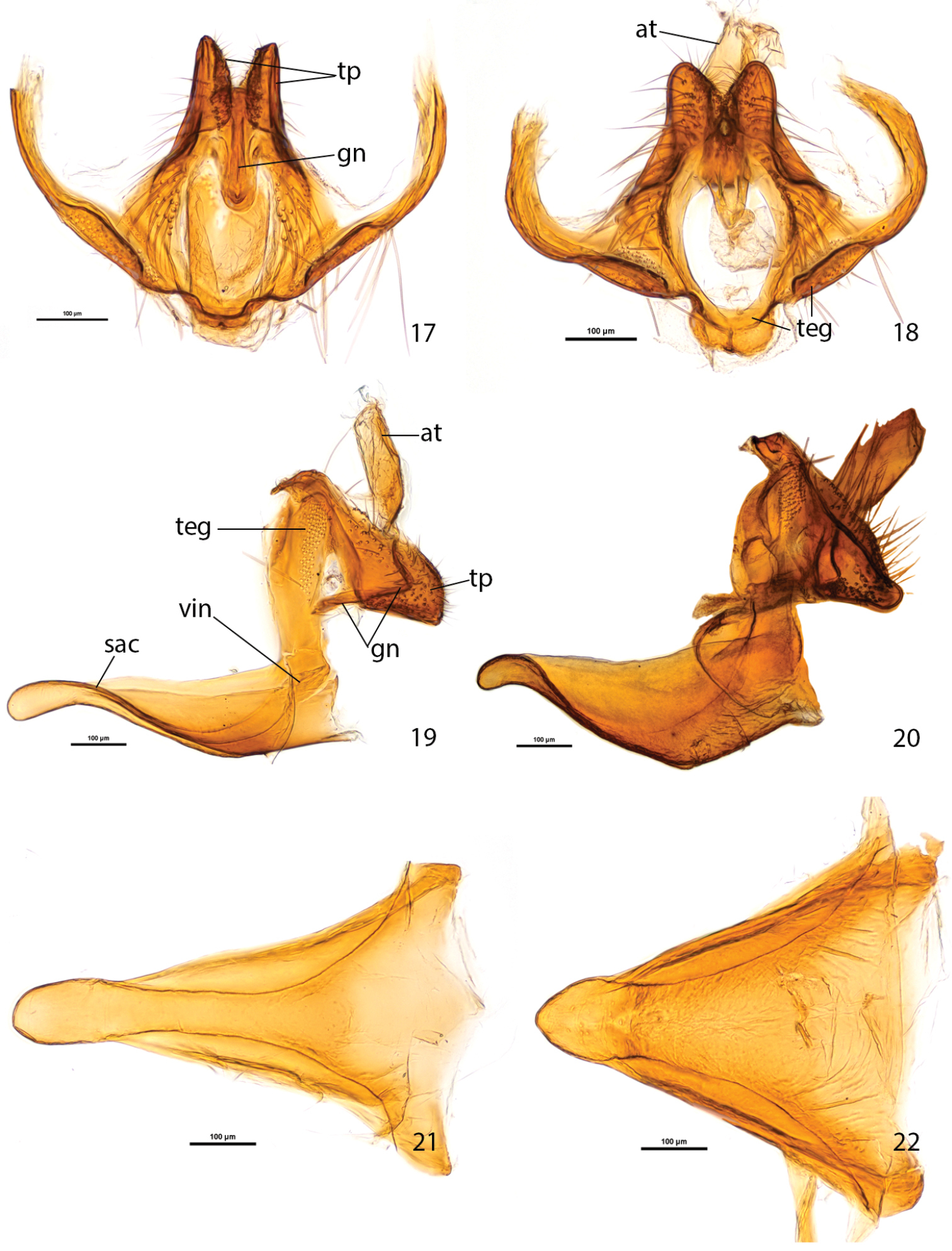
In Plutella australiana, the male genitalia appear overall more slender than in Plutella xylostella, particularly if viewed ventrally (Figs 27–28). The most easily observed difference involves the shape of the vinculum-saccus (Figs 19–22): in Plutella australiana it is slender with a slight medial constriction, a more protruded and inflated anterior apex, and is about 1.5× as long as wide; in Plutella xylostella it has a broader, more chunky aspect and in profile, is deeply concave, and is about as long as wide. The teguminal processes (Figs 17–18) are more slender and slightly separated medially in Plutella australiana, whereas they are broader and medially contiguous in Plutella xylostella. The valva (Figs 23–24) is evenly rounded with a slight sinuation in the ventral margin, and a zone of spiniform setae that is restricted to the medial area in Plutella australiana; whereas its ventro-distal margin is more or less distinctly angled and the zone of spiniform setae extends all the way to the angled apex in Plutella xylostella.
Male genitalia of Plutella australiana and Plutella xylostella. 17–18 tegumen-gnathos, ventral aspect 17 Plutella australiana (slide JFL1730, specimen BIOUG00788-F11) 18 Plutella xylostella (slide JFL1729, specimen BIOUG02113-A06). 19–20, tegumen-uncus-gnathos-vinculum complex, lateral aspect, valvae and phallus removed 19 Plutella australiana (slide JFL1732, specimen BIOUG02248-G03) 20 Plutella xylostella (slide JFL1733, specimen BIOUG02248-G02). 21–22 vinculum–saccus, ventral aspect 21 Plutella australiana (slide JFL1730) 22 Plutella xylostella (slide JFL1729). Scale bar = 100µ; all to same scale. at = anal tube; gn = gnathos; sac = saccus; teg = tegumen; tp = teguminal process; vin = vinculum.
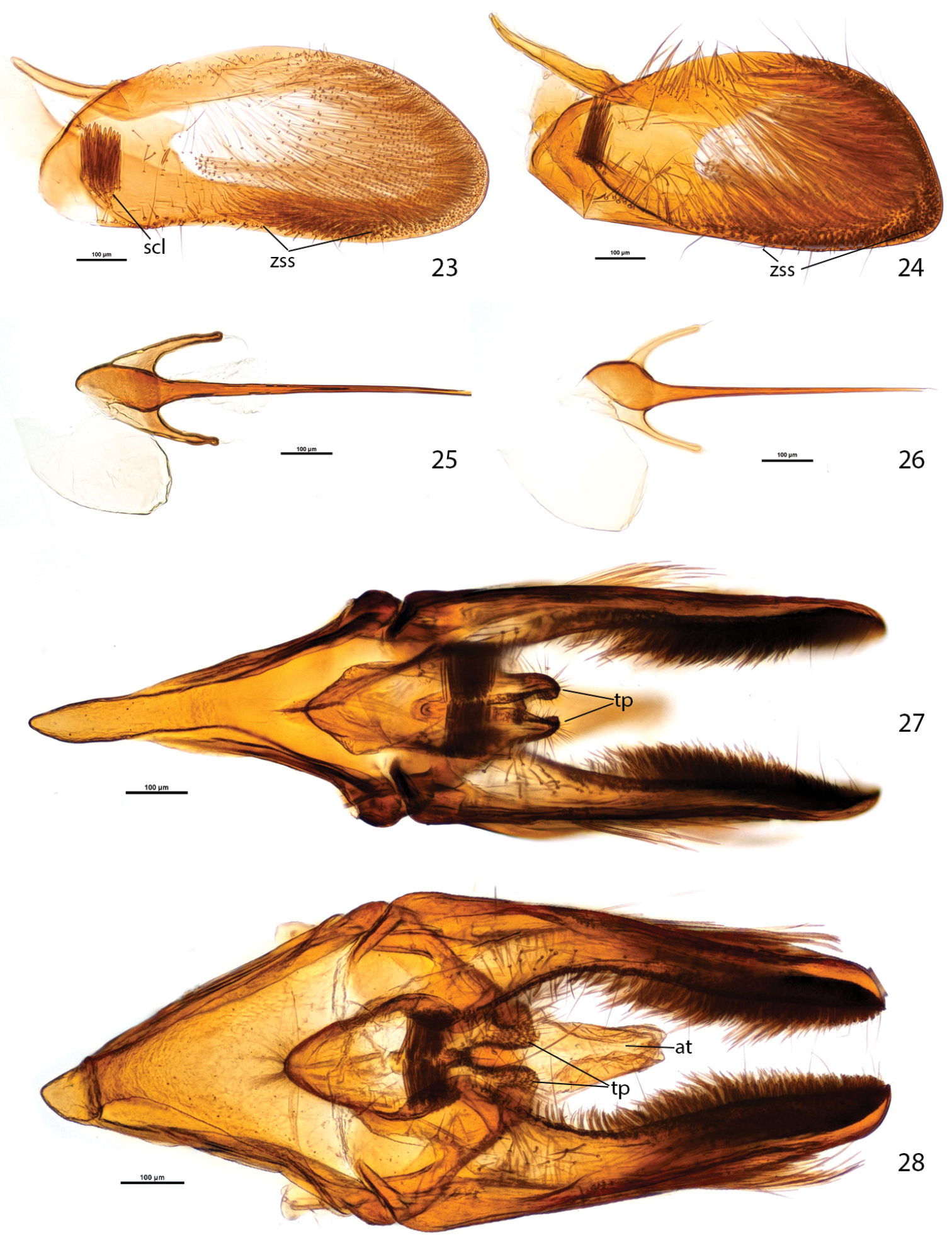
Male genitalia of Plutella australiana and Plutella xylostella. 23–24, valva, inner aspect 23 Plutella australiana (slide JFL1689, specimen 08-NSWHH-1277) 24 Plutella xylostella (slide JFL1733, specimen BIOUG02248-G02). 25–26, phallus, dorsal aspect 25 Plutella australiana (slide JFL1690, specimen 08-NSWHH-1340) 26 Plutella xylostella (slide JFL1688, specimen 06-TASB-01769). 27–28, genitalia with phallus removed, ventral aspect 27 Plutella australiana (slide JFL1735, specimen BIOUG00987-E12) 28 Plutella xylostella (slide JFL1734, specimen 09-NSWHH-1674). Scale bar = 100µ; all to same scale. at = anal tube; scl = sacculus; tp = teguminal process; zss = zone of spiniform setae.
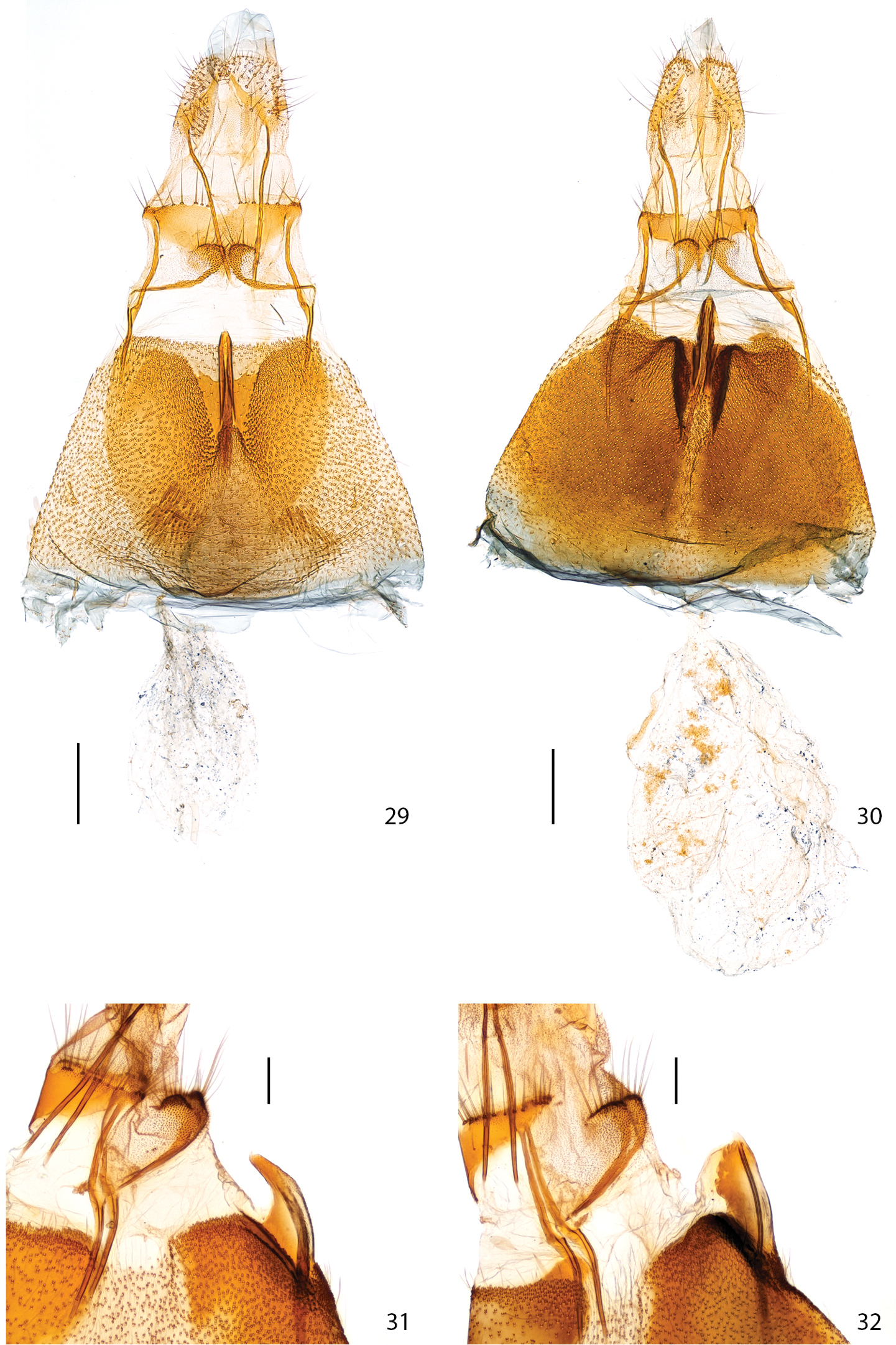
Female genitalia: In Plutella australiana, abdominal sternum 7 (S7) has a heart-shaped melanized area surrounding the antrum and it has a flat surface; the apex of the tubular projection of the antrum is barely extended beyond the posterior margin of S7 (0.15× length of S7) when viewed ventrally (Fig. 29), and has a constricted, curved apical half when viewed laterally (Fig. 31). In Plutella xylostella, the area of S7 surrounding the antrum is bordered by markedly raised pair of folds of the S7 wall which form two conical projections bracing the tubular projection of the antrum; the apex of the tubular projection of antrum is extended further out beyond posterior margin of S7 (0.5× length of S7) when viewed ventrally (Fig. 30), and is evenly broad and straight when viewed laterally (Fig. 32). The corpus bursae is proportionally smaller and about equal in length to S7 in Plutella australiana, whereas in Plutella xylostella it is proportionally larger and about 1.5× the length of S7.
Female genitalia. 29 Plutella australiana, ventral aspect (slide JFL1684, specimen 07-NSWBB-0046) 30 Plutella xylostella, ditto (slide JFL1685, specimen 07-NSWBB-0144) 31 Plutella australiana, lateral aspect of antrum projection (slide JFL1736, specimen BIOUG00788-G06) 32 Plutella xylostella, ditto (slide MIC6811, specimen BIOUG02113-D10). Scale: Figs 29–30 = 200µ; Figs 31–32 = 100µ.
Male (Figs 3, 5, 9). Head off-white, vertex pale greyish brown, area behind eye and beneath ocellus greyish brown. Labial palpus porrect, segment 2 with forward-directed triangular, pale greyish brown tuft, leading edge white; segment 3 upturned, as long as 2, greyish white to greyish brown. Antenna about two-thirds length of forewing; scape with dense off-white to pale greyish brown pecten; flagellum dorsally off-white with a few scattered brown rings in distal half. Mesoscutum off-white. Tegulae greyish brown to brown. Forewing upper surface with costal region brownish grey with sparse, darker speckles; medial and fold region brown to buff-brown; dorsal region off-white to pale buff, sinuous margin with two or three scallops and lined with thin dark brown line, forming a dorsal band of two or three diamonds when wings are folded, in several specimens with dorsal margin lined with a few scattered dark brown dots; apical area paler and mottled with a mix of pale grey, brown, and black suffusion; apical fringe with alternating white and dark brown thin bands.
Female (Figs 4, 6, 7, 8). As male except colouration more variable, with several individuals paler overall and with subdued contrast from brownish buff to pale whitish grey, tegula off-white, scalloped dorsal region of forewing indistinct, costal and disc area with dark speckles.
Forewing length: males, 5.4mm–6.9mm (mean 6.1mm, n=30); females, 5.6mm–6.9mm (mean 6.1mm, n=22).
Male genitalia (Figs 17, 19, 21, 23, 25, 27) (6 preparations examined). Abdominal tergite 8 (T8) subquadrate, anteriorly with round emargination and protruded antero-lateral corners. Pleural lobes large, about as long as T7+T8, their inner margin edged with thin, ribbon-like sclerotization. Coremata present, 3.5× length of T8. Tegumen a narrow, transverse arch with very narrow dorsal rim, anterior margin laterally notched, pedunculi fused with dorsal arms of vinculum. Uncus absent. Teguminal processes developed as pair of heavily sclerotized, setose lobes projected to one-third of valva length; in profile with dorsal edge broadly rounded and ventral edge straight; in ventral view outline conical with slight outward curvature. Anal tube with lightly sclerotized distal portion about length of teguminal processes. Gnathos a narrow band extended and bent downward at right angle between teguminal processes with apex pointed anteriorly. Vinculum arms dorsally extended and fused with tegumen, ventrally extended into slender, triangular, transversely arched and ventrally concave saccus; in ventral view saccus about 1.5× as long as wide, middle portion slightly constricted, in profile about 5× as long as high. Valva subelliptical in outline, costa and apex evenly rounded, ventral margin slightly concave, length/width about 2.3; spiniform setae of ventral margin arranged in stretched cluster restricted to medial area; sacculus situated in antero-ventral area, with tight cluster of spiniform setae, which medially abut each other below socii when valvae are closed; outer wall of valva with transparent suboval “window” bearing two tufts of long, fine scales, one in antero-ventral third, the other in dorso-distal third. Phallus thin, sharply pointed, needle-like, with bulbous base braced by pair of posteriorly directed, hook-like processes; bulbus ejaculatorius crescentic in outline, about the size of bulbous base.
Female genitalia (Figs 29, 31) (7 preparations examined). Sternite 7 (S7) with medio-posterior, heart-shaped, sclerotized area that is distinctly delineated from less melanized antero-lateral areas, posterior emargination forming channel surrounding tubular medial projection of antrum bearing apical ostium bursae, sides of channel flat, only medial anteriormost portion of channel slightly raised around base of antrum; tubular projection 0.4× length of S7, in ventral aspect with apex only slightly extended beyond posterior margin of S7 (0.15× length of S7), in lateral view with distal portion constricted and upcurved. Ductus bursae thin, delicate, anterior two-thirds membranous, posterior third (section inside antrum) sclerotized. Corpus bursae slightly shorter (0.9) than S7, thinly membranous, delicate, without signa. Posterior apophyses subequal in length to anterior ones. Anterior apophyses with a thin ventral branch extended transversally across S8 to lamella postvaginalis in middle; lamella postvaginalis composed of paired crescentic pads covered with sensilla trichodea and distally setose. Tergite 8 with transversely sclerotized distal third on which base of posterior apophyses are inserted, posterior margin lined with setae. Sternite 8+ovipositor subequal in length to S7.
Note about male genitalia. The gnathos of Plutella xylostella has not been described in recent publications (
The term ‘teguminal processes’ has been used by
This species name reflects the current restriction of this taxon to Australia. It is a noun in apposition.
Plutella australiana is so far known only from eastern Australia, in contrast to Plutella xylostella which is cosmopolitan in distribution. Plutella australiana and Plutella xylostella appear to have largely overlapping distributions within Australia as both species were collected in the ACT, NSW, QLD, and SA (Figure 2). They were collected together and on the same dates around Canberra on several occasions (Mar 2011, Apr 2011, Oct 2010 and 2011, Nov 2011), indicating that their adult flight periods and habitat requirements overlap. Plutella australiana may occur in other parts of Australia with the lack of records reflecting gaps in collecting. Although current records suggest that Plutella australiana is absent from Tasmania, further sampling is also required to confirm this fact. The two species appear to be roughly similar in abundance based on current records with 62 Plutella xylostella and 57 Plutella australiana among the haphazardly collected Australian specimens that have been sequenced.
Australia, Australian Capital Territory, Cook, 35.2612°S, 149.0591°E.
Plutella xylostella is thought to feed on a wide variety of cruciferous plants in Australia, including native and introduced species. However, Australian records are in question because of the past oversight of Plutella australiana. As a result, the host plants of both species are uncertain.
http://species-id.net/wiki/Plutella_xylostella
Barcode Index Number: BOLD:AAA1513
Figs 10–16, 18, 20, 22, 24, 26, 28, 30, 32The colouration of Plutella xylostella has been characterized as variable, with paler individuals in xeric regions (
In a taxonomic review of Hawaiian Plutella,
At least one of these Hawaiian races was included in a previous study of mtDNA variation in Plutella xylostella (as undescribed Plutella ‘UPA’ by
Plutella xylostella has long been regarded as a very common and widely distributed species within Australia (
The past oversight of the presence of two Plutella species in Australia likely explains the regional allozyme variation previously detected in Australian populations of Plutella xylostella (
There is a need to discover the host plant(s) of Plutella australiana to ascertain if it is also a crop pest. If so, its presence represents a new risk to international trade which should be evaluated. Examining known hosts of Plutella xylostella and other related Plutella may provide useful clues. Although Plutella xylostella has been recorded from a wide range of hosts across the world, records from plant families other than Brassicales are uncorroborated, with most being implausible (
The high genetic distance among taxa analyzed, and the placement of Eidophasia vanella between the Plutella australiana – Plutella xylostella cluster and other members of Plutella suggests that current generic limits need further assessment.
This work was supported by grants to PDNH from NSERC and from the Government of Canada through Genome Canada and the Ontario Genomics Institute in support of the International Barcode of Life Project. We are very grateful to colleagues at the Canadian Centre for DNA Barcoding for overseeing the sequence analysis. Special thanks are due to Andrew Mitchell, Australian Museum in Sidney and his colleagues at the Orange Agricultural Institute, New South Wales Department of Primary Industries for collecting specimens and their survey efforts to find the new species in NSW Brassica crops. We thank Ole Karsholt, University Museum, Copenhagen, for supplying the photograph and a leg for barcoding of the type of Plutella karsholtella. We are also grateful to Peter Huemer, Tiroler Landesmuseen, for allowing use of data on Plutella geniatella. Vazrick Nazari photographed the genitalia and assisted JFL with the morphological analysis, while Megan Milton aided with the sequence analysis. Two anonymous reviewers and the editor provided useful comments and suggestions.