(C) 2013 Trip Lamb. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Lamb T, Pollard R, Bond JE (2013) Genetic variation corroborates subspecific delimitation in the Namib fog-basking beetle, Onymacris unguicularis (Haag) (Tenebrionidae, Coleoptera). ZooKeys 353: 47–60. doi: 10.3897/zookeys.353.6228
The fog-basking beetle, Onymacris unguicularis (Haag, 1875), is currently listed as a polytypic form comprising two subspecies. A flightless substrate specialist, the beetleis endemic to vegetationless dunes in the Namib, where southern populations constitute the nominate subspecies, O. u. unguicularis, and populations some 300 km to the north compose O. u. schulzeae Penrith, 1984. Their taxonomic descriptions are based on minor differences in pronotal and prosternal shape, and the phylogenetic validity of these subspecies has yet to be ascertained. Here we reassess the polytypic status of O. unguicularis by (1) examining diagnostic phenotypic characters in conjunction with a geometric morphometric analysis, and (2) conducting phylogenetic analysis of mitochondrial DNA sequences. Our results confirm pronotal and prosternal differences, which are complemented by geometric morphometric resolution of the subspecies. Phylogenetic analysis recovered two reciprocally monophyletic lineages that exhibit perfect phylogeographic congruence with phenotypic variation. Our genetic data identify southern and northern populations as distinct lineages, corroborate morphometric data regarding subspecific delimitation, and therefore support the recognition of O. u. unguicularis and O. u. schulzeae as valid taxa under the general lineage concept.
Subspecies, integrative taxonomy, Namib Desert, Onymacris, Tenebrionidae
Darkling beetles (family Tenebrionidae) figure prominently in the arthropod fauna of Africa’s Namib Desert, where they compose ~80% of all coleopterans (
Onymacris unguicularis has also been the subject of taxonomic investigation;
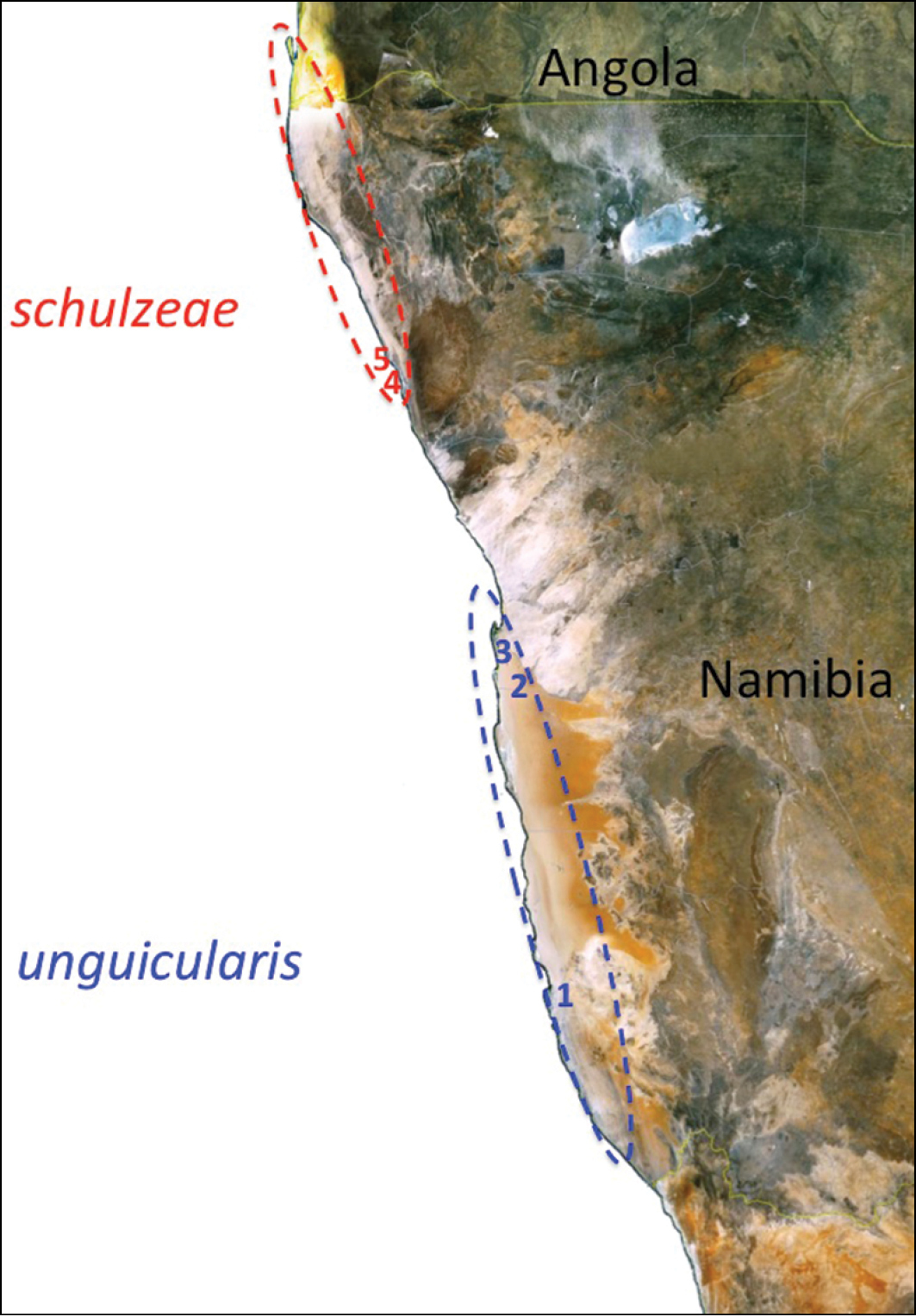
Map illustrating the range and disjunct distribution of Onymacris unguicularis in the Namib Desert. Subspecies distributions are approximated by oval overlays; localities for genetic sampling, listed from south to north, are: 1 Luderitz 2 Gobabeb 3 Walvis Bay, and 4, 5 Torra Bay.
Dorsal habitus of Onymacris unguicularis unguicularis (2) and Onymacris unguicularis schulzeae (3).
Efforts in subspecies delimitation mirror those of species delimitation conceptually if not methodologically and, similarly, engage controversy (
Subspecific taxa are now routinely reassessed using molecular phylogenetic analysis as a key component of integrative or coalescent approaches to recover evolutionarily independent lineages. These investigations generally yield one of two outcomes. Patterns of genetic variation may exhibit discordance with traditionally defined subspecies, either phenotypically or geographically, if not both (
In this study we examined morphological and genetic variation in Onymacris unguicularis, adopting
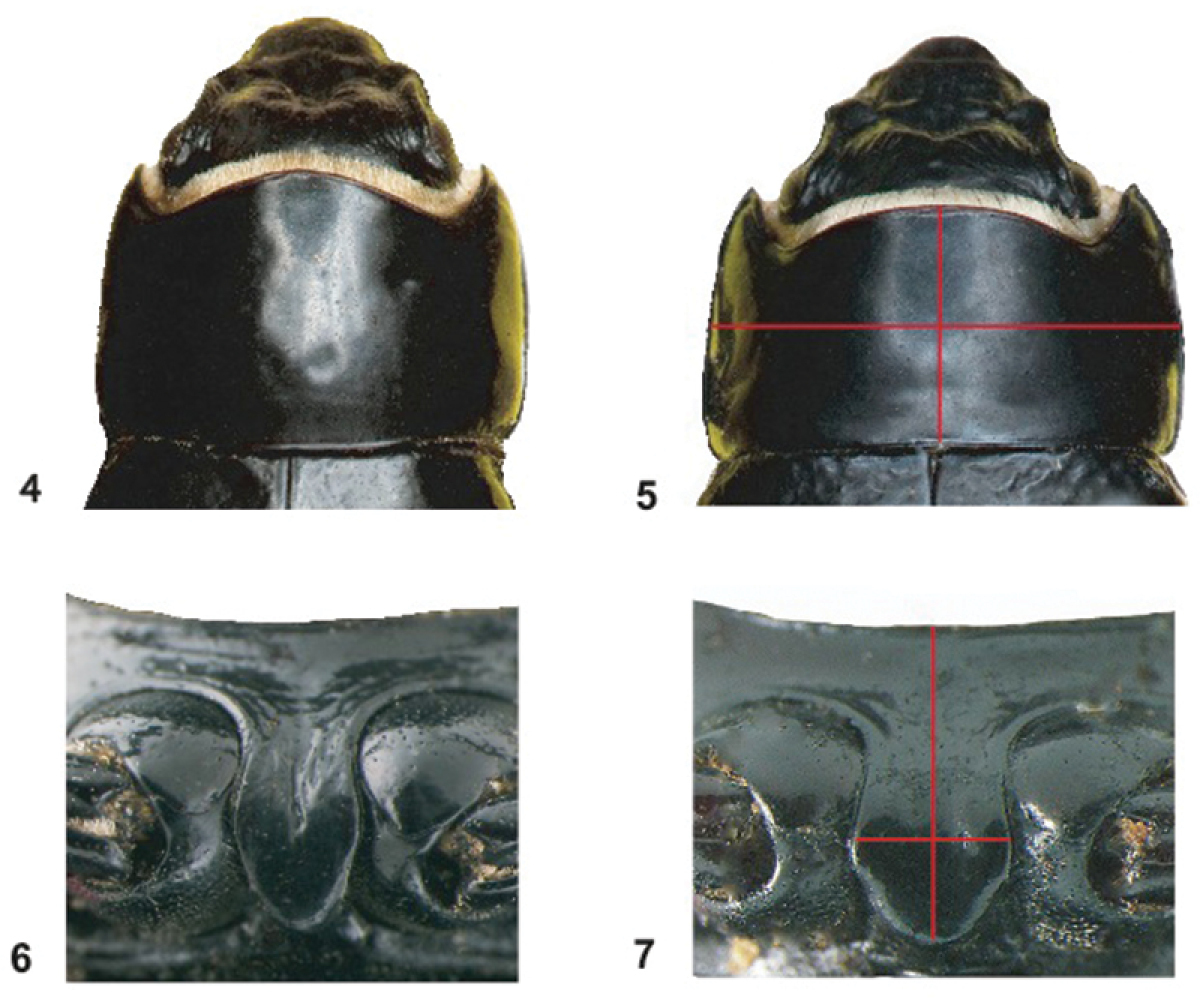
Pronotum (4–5) and prosternum (6–7) of Onymacris unguicularis unguicularis (4, 6) and Onymacris unguicularis schulzeae (5, 7). Measurements for ratio calculations are marked on 5 and 7.
Lateral aspects of the prosternal process depicting a blunt apex (8) in Onymacris unguicularis schulzeae and toothed apex (9) in Onymacris unguicularis unguicularis.
To reassess
We identified eleven dorsal landmarks—eight type 1 and three type 3 (
Landmarks for the geometric morphometric analysis of dorsal (elytral) shape.
Sixteen beetles were captured, preserved (100% ethanol), and processed for DNA analysis; the specimens included 12 Onymacris unguicularis unguicularis, representing three geographic localities, and four Onymacris unguicularis schulzeae, representing two relatively close localities (Fig. 1; Appendix). Onymacris laeviceps Gebien, 1938 and Onymacris plana, identified as sister taxa to Onymacris unguicularis in a generic-level phylogeny (
PCR primers and amplification conditions.
| Gene | Primer | Annealing | Cycles | Reference |
|---|---|---|---|---|
| cox1 | TY-J-1460 | 50oC | 35 | |
| TL2-N-3014 | ||||
| C1-J-2183 | sequencing only | |||
| cox2 | TL2-J-3037 | 50oC | 35 | |
| TK-N-3785 |
Amplification products were cleaned using exoSAP-IT (USB Corp.) and sequenced on an Applied Biosystems 3130 capillary sequencer. Sequences were edited and assembled in Sequencher 4.9 (GeneCodes, Ann Arbor, MI) and aligned using ClustalX ver. 2.0 (
We used Bayesian inference (BI) and maximum likelihood (ML) methods to analyze the concatenated gene (cox1-cox2) dataset. We used Kakusan 4 (
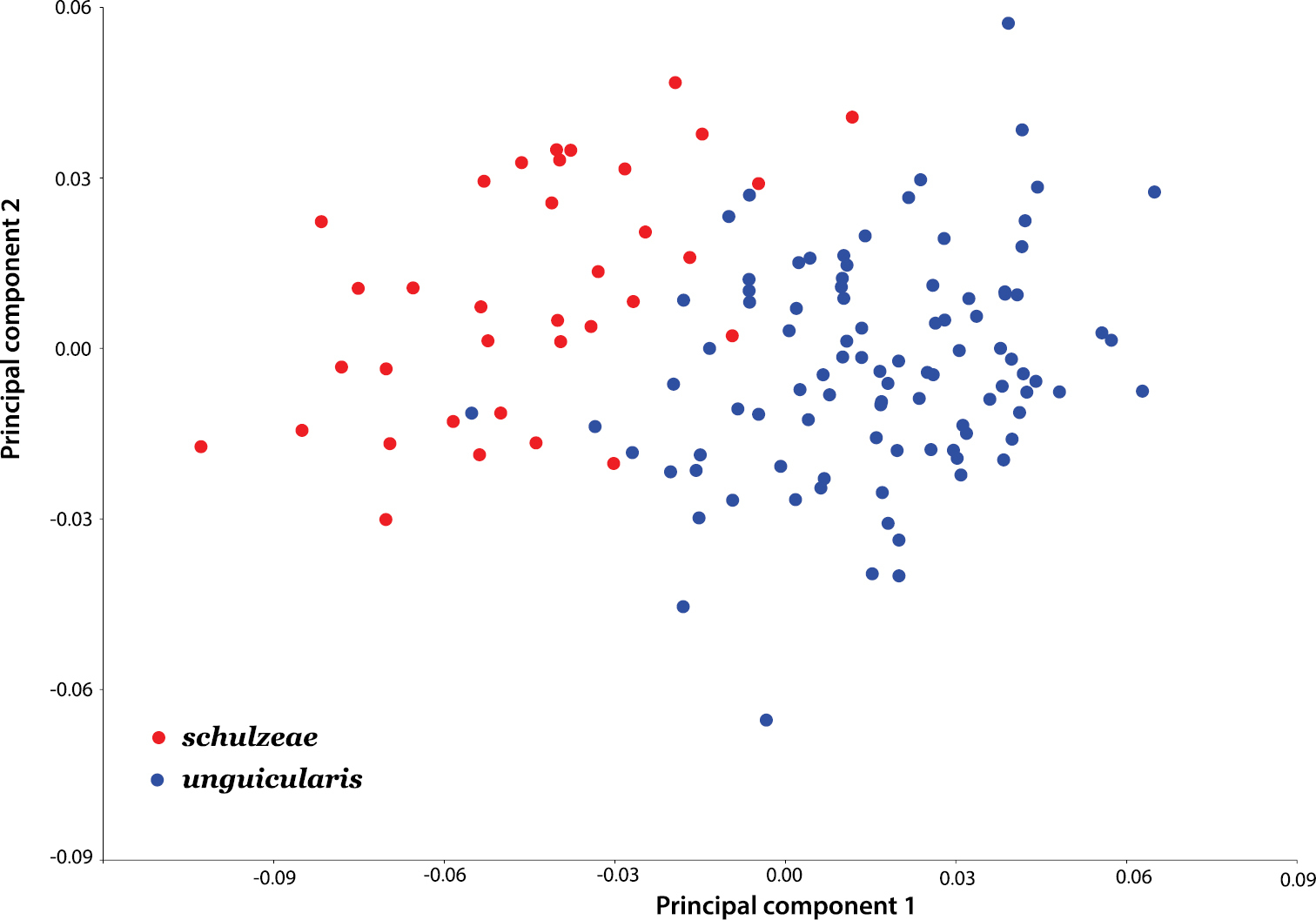
Ratios generated for both the protonal and prosternal datasets differed significantly between subspecies (p < 0.0001), with minimal overlap for each character (Table 2). In the geometric morphometric analysis, the first two principal components based on the non-uniform components of dorsal (elytral) shape account for 78.54% of the variation between subspecies. An ordination plot of PC1 and PC2 revealed that the two subspecies are relatively well separated along the PC1 axis (Fig. 11). Dorsal shape separation probably reflects proportionally longer elytra in Onymacris unguicularis unguicularis, which become apparent in side-by-side comparisons with Onymacris unguicularis schulzeae (Figs 2–3). In light of these findings, we measured elytral length and width (at the midpoint of elytral length) for all specimens to determine whether a simple ratio (EL/EW) might reflect the subspecific separation observed in our geometric morphometric analysis. We also noted the position of greatest elytral width for each specimen, scored as midpoint, anterior to midpoint, or posterior to midpoint. Despite broad overlap, elytral ratios differed significantly (p < 0.0001) between subspecies (Table 2); elytral width is widest anterior to midpoint in both subspecies but is positioned closer to the pronotal suture in Onymacris unguicularis schulzeae. Of the three ratios, we consider that for the pronotum to be the strongest diagnostic metric.
Pronotal, prosternal, and elytral ratio means and ranges.
| Character | Subspecies | N | Mean | Range |
|---|---|---|---|---|
| pronotum | Onymacris unguicularis unguicularis | 95 | 1.66 ± 0.08 | 1.47–1.83 |
| Onymacris unguicularis schulzeae | 35 | 1.97 ± 0.13 | 1.73–2.35 | |
| prosternum | Onymacris unguicularis unguicularis | 94 | 2.22 ± 0.17 | 1.86–2.71 |
| Onymacris unguicularis schulzeae | 33 | 2.01 ± 0.14 | 1.65–2.34 | |
| elytra | Onymacris unguicularis unguicularis | 95 | 1.44 ± 0.08 | 1.25–1.61 |
| Onymacris unguicularis schulzeae | 34 | 1.35 ± 0.07 | 1.24–1.47 |
Scatterplot of the principal component scores derived from geometric morphometric analysis of dorsal shape.
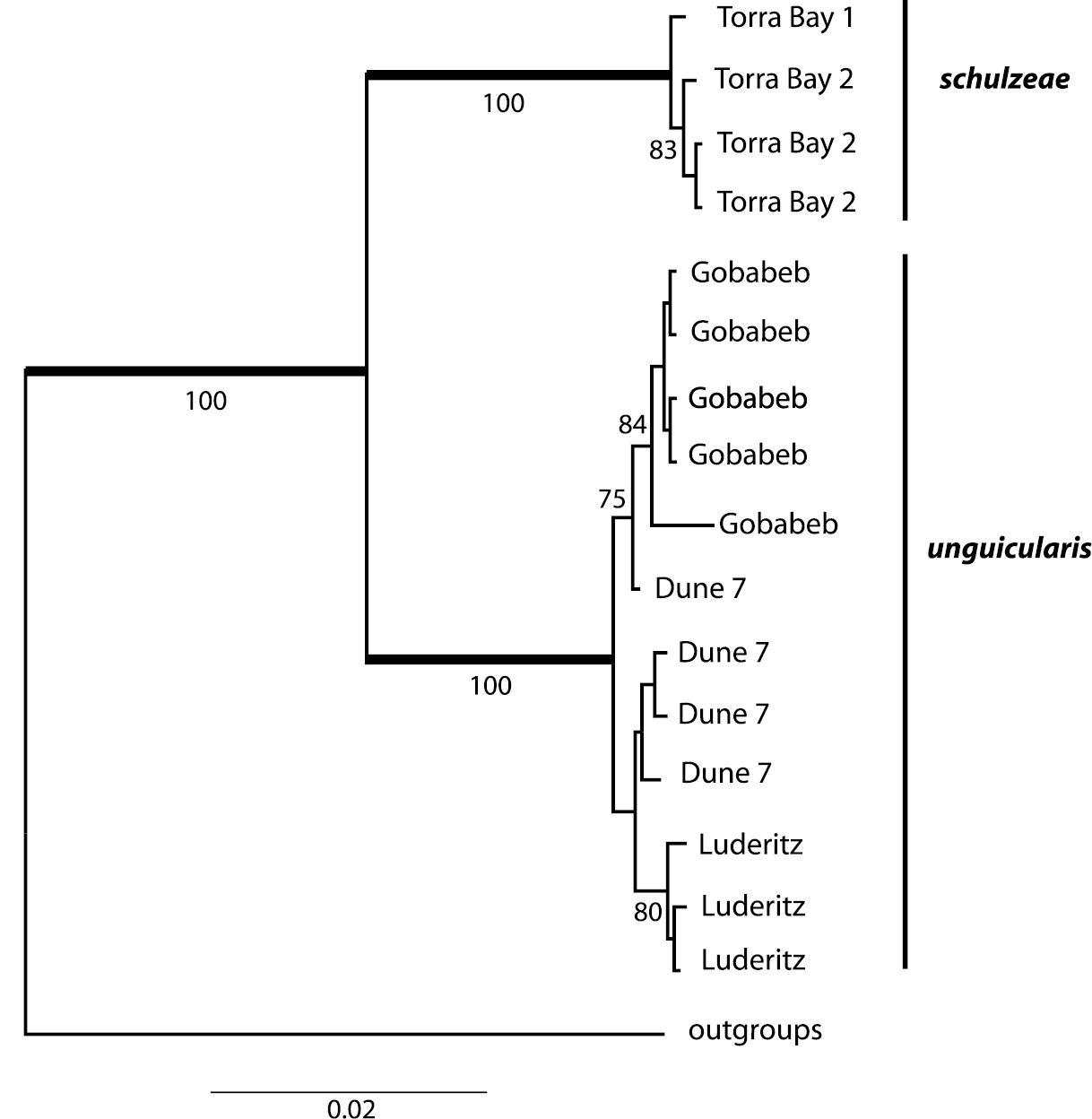
Concatenated sequence data for the cox1 (1574 bp) and cox2 (680 bp) genes yielded eleven haplotypes among the 16 beetles surveyed: eight haplotypes for Onymacris unguicularis unguicularis and three for Onymacris unguicularis schulzeae (Genbank accession numbers KF835703-KF835721). No haplotypes were shared between subspecies. Mean haplotype divergence (calculated from uncorrected pair-wise distance values) was limited across geographic localities for both Onymacris unguicularis unguicularis (cox1 = 0.046%; cox2 = 0.027%) and Onymacris unguicularis schulzeae (cox1= 0.008%; cox2 = 0.014%) but differed substantially between the two subspecies (cox1 = 3.87%; cox2 = 3.01%). The BI (harmonic mean –ln = 4690.15) and ML (–ln = 4172.41) analyses generated topologically identical trees in which subspecies were shown to be reciprocally monophyletic (Fig. 12). Moreover, subspecific monophyly was strongly supported (Bayesian posterior probabilities = 1.0; ML bootstraps = 100%), in contrast to the marginal to moderate support observed for haplotype relationships of geographic localities within subspecies (Fig. 12).
Bayesian consensus tree showing relationships within Onymacris unguicularis. Bold branches subtend nodes with Bayesian posterior probabilities of 1.0; numbers below the branches are ML bootstrap values.
We employed
We thank Ruth Müller of the Ditsong National Museum of Natural History, Pretoria, South Africa, for arranging specimen loan and shipment in a timely manner. Tom Fink graciously provided time and expertise with digital imagine capture and measurements. This project was supported in part by an East Carolina University Undergraduate Research Award and George T. Barthalmus Undergraduate Research Grant to Rachel Pollard. Specimens used for DNA sequencing were collected under a permit provided by Namibia’s Ministry of Environment and Tourism.
Localities for genetic and morphometric samples.
| Subspecies | Dataset | N | Locality | Voucher numbers |
|---|---|---|---|---|
| Onymacris unguicularis unguicularis | morphometric | 10 | Anigab | |
| 9 | Blauberg | |||
| 10 | Bogenfels | |||
| 1 | Chaneis | |||
| 12 | Gobabeb | |||
| 8 | Grillental | |||
| 9 | Hottentot Bay | |||
| 5 | Luderitz | |||
| 10 | Spencer Bay | |||
| 10 | Swakopmund | |||
| 11 | Walvis Bay | |||
| genetic | 4 | Dune 7, near Walvis Bay; -22.9691, 14.5946 | TL022 TL023 TL024 TL025 | |
| 5 | Gobabeb; -23.5691, 15.0424 | TL026 TL027 TL028 TL029 TL030 | ||
| 3 | 20 km E Luderitz; -26.7110, 15.2828 | TL031 TL032 TL033 | ||
| Onymacris unguicularis schulzeae | morphometric | 3 | near Foz du Cunene, Angola | |
| 4 | Lacrau, 13 km N Fos du Cunene, Angola | |||
| 11 | Kaokoveld coast, between Koichab and Unjab rivers | |||
| 12 | Unjab River, 8 km from mouth | |||
| genetic | 5 | near Torra Bay; -20.3345, 13.2929 | ||
| 3 | near Torra Bay; -20.3345, 13.2929 | TL034 TL035 TL036 | ||
| 1 | near Torra Bay; -20.2738; 13.2655 | TL037 |
*as coded in Genbank