Citation: Rakauskas R, Havelka J, Zaremba A, Bernotienė R (2014) Mitochondrial COI and morphological evidence for host specificity of the black cherry aphids Myzus cerasi (Fabricius, 1775) collected from different cherry tree species in Europe (Hemiptera, Aphididae). ZooKeys 388: 1–16. doi: 10.3897/zookeys.388.7034
Partial sequences of the mitochondrial COI gene of forty eight European and two Turkish population samples of Myzus cerasi from different winter hosts (Prunus spp.) were subjected to phylogenetic analyses. The analysed M. cerasi samples emerged as paraphyletic relative to a Myzus borealis sample used as an out-group, and formed two major clades in neighbor joining, maximum parsimony, maximum likelihood and Bayesian inference trees, corresponding to subspecies living specifically on Prunus avium and P. cerasus. Multivariate discriminant analysis (method of canonical variates) was applied to find out if morphological variation of samples correlated with mitochondrial COI and host plant information. Mean scores on the first two canonical variables clustered samples fully in accordance with their COI haplotypes and host plants confirming the existence of two morphologically similar winter host - specific subspecies of M. cerasi in Europe. No single morphological character enabled satisfactory discrimination between apterous viviparous females of the two subspecies. A three-character linear discriminant function enabled 92.37% correct identification of apterous viviparous females of M. cerasi cerasi (n = 118) and 93.64% of M. cerasi pruniavium (n = 110). A key for the morphological identification of the two subspecies is presented and their taxonomic status is discussed.
Molecular systematics, cherry aphids, morphological key to subspecies
Black cherry aphid Myzus cerasi (Fabricius, 1775) is reported to be a serious pest of cherries all over the world (
A similar species complex is found in the mealy plum aphid, Hyalopterus spp. (
The Myzus cerasi complex has not yet been the subject of detailed molecular study, although certain DNA sequences have been isolated (
Fifty population samples of apterous viviparae of black cherry aphids from nine European countries and Turkey were collected in 2004–2013, mostly from Prunus cerasus, Prunus avium, but also from three other Prunus species (Table 1). Twenty samples were used for morphology - based stepwise discriminant analysis. The remaining 30 samples were used for subsequent evaluation of the derived discrimination functions. Samples of Myzus borealis Ossiannilsson, 1959 and Myzus persicae (Sulzer, 1776) (GenBank Accession No AB506741) were used as out-groups for the phylogenetic analyses.
Aphid material used in the present study. Samples used for morphology-based discriminant analysis are given in bold.
| Place, date, collection number; (number of individual apterae per sample) | GenBank Accession No |
|---|---|
| Myzus cerasi on Prunus cerasus L. | |
| Jieznas, Prienai distr., Lithuania, 2012.05.30, 12-25; (8) | KF754311 |
| Alytus, Lithuania, 2012.05.30, 12-30; (8) | KF754310 |
| Daugai, Alytus distr., Lithuania, 2012.05.30, 12-32; (8) | KF754303 |
| Skirgiškės, Vilnius distr., Lithuania, 2012.06.05, 12-37; (8) | KF754304 |
| Eišiškės, Šalčininkai distr., Lithuania, 2012.06.13, 12-43; (7) | KF754329 |
| Labanoras, Švenčionys distr., Lithuania, 2012.06.19, 12-70; (8) | KF754306 |
| Molėtai, Lithuania, 2012.06.19, 12-72; (8) | KF754314 |
| Kraujaliai, Molėtai distr., Lithuania, 2012.07.10, 12-120; (8) | KF754305 |
| Žičkai, Molėtai distr., Lithuania, 2012.07.13, 12-132; (8) | KF754321 |
| Nida, Neringa, Lithuania, 2012.08.09, 12-176; (8) | KF754322 |
| Smiltynė, Klaipėda, Lithuania, 2012.08.12, 12-191; (8) | KF754309 |
| Preila, Neringa, Lithuania, 2012.08.13, 12-203; (8) | KF754330 |
| Bagnolo Mella, Brescia prov., Italy, 2013.05.01, 13-27; (8) | KF754338 |
| Poncarale, Brescia prov., Italy, 2013.05.02, 13-33; (8) | KF754337 |
| Suginčiai, Molėtai distr., Lithuania, 2013.06.15, 13-83; (8) | KF754345 |
| Akmeniai, Lazdijai distr., Lithuania, 2013.05.30, 13-57; (8) | KF754341 |
| Karsava, Ludza distr., Latvia, 2013.07.17, 13-133; (8) | KF754344 |
| Gorodok, Vitebsk distr., Belarus, 2008.06.17, 08-6; (6) | KF754302 |
| Zadrachje, Vitebsk distr., Belarus, 2008.06.18, 08-18; (8) | KF754325 |
| Riga, Latvia, 2008.07.03, 08-73; (8) | KF754328 |
| Skirgiškės, Vilnius distr., Lithuania, 2011.06.15, 11-46; (8) | KF754316 |
| Cluj Gilau, Romania, 2012.06.19, Z12-112; (8) | KF754327 |
| Cluj Gilau, Romania, 2012.06.19, Z12-116; (8) | KF754319 |
| Poncarale, Brescia prov., Italy, 2013.05.02, 13-30; (8) | KF754346 |
| Mezöpeterd, Hajdu-Bihar distr., Hungary, 2012.06.20, Z12-122; (8) | KF754333 |
| Myzus cerasi on Prunus avium L. | |
| Skirgiškės, Vilnius distr., Lithuania, 2012.06.05, 12-39; (7) | KF754332 |
| Šalčininkai, Lithuania, 2012.06.13, 12-48; (6) | KF754317 |
| Bratoniškės, Vilnius distr., Lithuania, 2012.06.14, 12-56; (7) | KF754313 |
| Blagoevgrad, Bulgaria, 2012.06.26, 12-83; (7) | KF754318 |
| Frankfurt/Main, Germany, 2012.06.30, 12-104; (8) | KF754307 |
| Kraujaliai, Molėtai distr., Lithuania, 2012.07.10, 12-111; (8) | KF754308 |
| Stirniai, Molėtai distr., Lithuania, 2012.07.12, 12-128; (8) | KF754320 |
| Juodkrantė, Neringa, Lithuania, 2012.08.10, 12-182; (8) | KF754323 |
| Pervalka, Neringa, Lithuania, 2012.08.11, 12-188; (7) | KF754336 |
| Preila, Neringa, Lithuania, 2012.08.13, 12-199; (7) | KF754335 |
| Rondo, Katowice, Poland, 2011.05.13, 11-10; (8) | KF754349 |
| Tekir, Karamanmarash distr., Turkey, 2011.05.21, 11-25; (5) | KF754312 |
| Göksun, Karamanmarash distr., Turkey, 2011.05.21, 11-27; (8) | KF754315 |
| Kairėnai, Vilnius, Lithuania, 2010.07.01, 10-3; (7) | KF754324 |
| Zafferana, Catania, Italy, 2004.06.28, 04-49; (5) | KF754339 |
| Costinesti, Romania, 2012.06.15, Z12-90; (8) | KF754326 |
| Galata, Varna, Bulgaria, 2012.06.18, Z12-102; (7) | KF754351 |
| Burgas, Bulgaria, 2012.06.19, Z12-110; (8) | KF754331 |
| Cluj Gilau, Romania, 2012.06.19, Z12-117; (7) | KF747679 |
| Carpendolo, Brescia prov., Italy, 2013.04.27, 13-12; (7) | KF754340 |
| Akmeniai, Lazdijai distr., Lithuania, 2013.05.30, 13-60; (8) | KF754347 |
| Wojslawice, Lower Silesia, Poland, 2013.06.20, 13-98; (8) | KF754342 |
| Myzus cerasi on Prunus serrulata Lindl. | |
| Wojslawice, Lower Silesia, Poland, 2013.06.20, 13-97; (8) | KF754348 |
| Myzus cerasi on Prunus maackii Rupr. | |
| Dobele, Latvia, 2013.07.03, 13-119; (8) | KF754343 |
| Myzus cerasi on Prunus mahaleb L. | |
| Medias, Sibiu distr., Romania, 2012.06.19, Z12-113; (8) | KF754334 |
| Myzus borealis on Galium rubioides L. | |
| Zmejinyje ostrova, Kanev distr., Cherkasy reg., Ukraine, 2006.06.16, 06-74 | KF754350 |
For molecular analysis, a single aphid individual from one sampled plant was considered as a unique sample. Total genomic DNA was extracted from a single aphid using the DNeasy Blood & Tissue kit (Qiagen), which involved at least a 2 h digestion of tissue with proteinase K. Partial sequences of mitochondrial COI gene were PCR-amplified using earlier published primers (
In addition to the sequences from 50 samples of Myzus cerasi, COI sequences of Myzus borealis from subgenus Myzus sensu stricto (the same subgenus as Myzus cerasi) and Myzus persicae from subgenus Nectarosiphon Schouteden, 1901 were selected as out-groups for the phylogenetic analyses, which included neighbor joining (NJ), maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference in phylogeny (BI). NJ, MP and ML analyses were performed using MEGA 5 (
Samples representing different clades in the molecular tree and haplotype network were used for stepwise discriminant analysis followed by canonical analysis: 10 samples from sour cherry, Prunus cerasus, and 10 samples from sweet cherry, Prunus avium (shown in bold in Table 1).
Based on earlier taxonomic work (
Measurements of the slide-mounted apterous viviparous females were performed by means of interactive measurement system Micro-Image (Olympus Optical Co. GmbH). STATISTICA 8 version software (
Morphological characters that contributed most to canonical discrimination functions were evaluated as having potential for separation of taxa. An identification key was constructed based on these discrimination functions. The key was then tested on the 30 aphid samples that had not been used in its construction (listed in normal font in Table 1).
Fifty partial COI sequences of Myzus cerasi and one of Myzus borealis from 11 countries were included in analysis. The alignment contained 616 bases in the final set with three variable sites, all of which appeared parsimony informative. The average base composition was A = 34.0%, C = 12.7%, G = 12.3% and T = 41.0%. The overall transition/transversion ratio R = 1.221 for all sites.
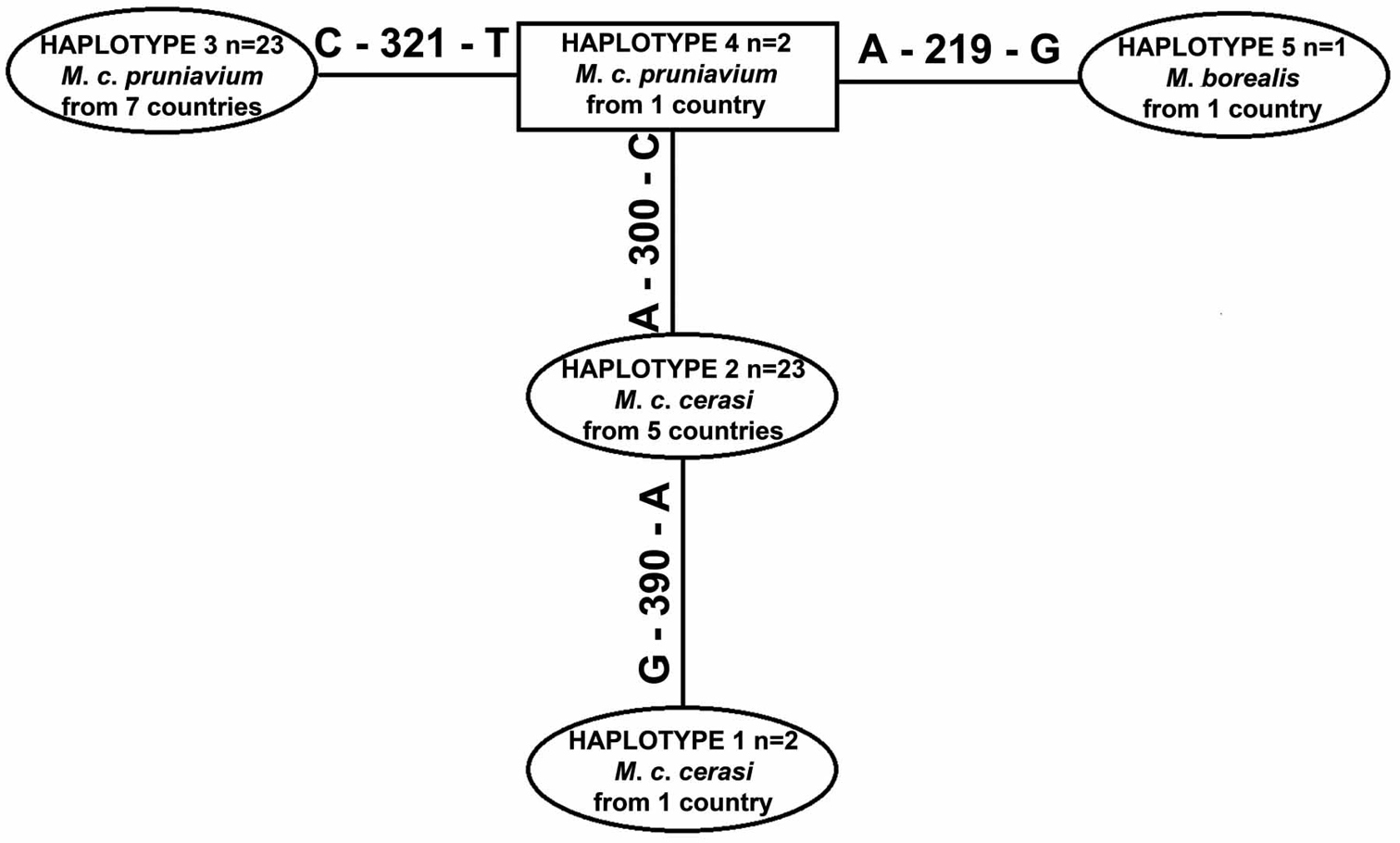
Five COI haplotypes were detected (Fig. 1): one for Myzus borealis, two for samples from Prunus cerasus and two for samples from Prunus avium (Table 2). Aphids collected from Prunus mahaleb, Prunus maackii and Prunus serrulata had the same haplotype (No. 3) as the majority of samples from Prunus avium. COI haplotypes detected among samples from Prunus cerasus (No. 1 and 2) and the remaining Prunus species (No. 3 and 4) differed in the following nucleotide positions of 616 bp alignment: 300 (between No. 1–2 and No. 3–4), 321 (between No. 3 and No. 4) and 390 (between No. 1 and No. 2). The range of the intraspecific pairwise sample divergences (K2P model) was 0.0 – 0.5% (average 0.2%), whilst interspecific pairwise sample divergences between three species of Myzus ranged from 0.2 to 6.8% (Table 3).
Haplotype network (TCS 1.21 software:
COI haplotypes of three Myzus taxa revealed by construction of haplotype network. Sample numbers are the same as given in Table 1.
| Haplotype number | Number of sequences | Sequence length (bp) | Sample numbers |
|---|---|---|---|
| Myzus cerasi cerasi (collected from Prunus cerasus) | |||
| 1 | 2 | 616 | 13-33; 13-27. |
| 2 | 23 | 616 | 08-6;12-32; 12-176; 12-30; 08-18; z12-122; 12-120; z12-112; 12-132; 12-25; 12-43; 12-191; 12-203; 12-70; 12-72; 08-73; 13-83; 13-133; 11-46; z12-116; 12-37; 13-30; 13-57 |
| Myzus cerasi pruniavium (collected from Prunus avium, except where otherwise noted) | |||
| 3 | 23 | 616 | 11-10; 12-39; z12-110; 12-182; 12-104; 12-56; 12-83; 12-111; 12-199; z12-90; z12-102; 12-48; 12-188; z12-113 (Prunus maackii); 04-49; 12-128; 10-03; 13-60; 13-98; 13-97 (Prunus serrulata); 13-12; 13-119 (Prunus mahaleb); z12-117 |
| 4 | 2 | 616 | 11-27; 11-25 |
| Myzus borealis (collected from Galium rubioides) | |||
| 5 | 1 | 616 | 06-74 |
Range of pairwise interspecific sample divergences of mitochondrial COI gene fragment (K2P model) for three species of Myzus (number of samples used is in parentheses).
| Species 1 | Species 2 | Range of divergence, % |
|---|---|---|
| Myzus cerasi (50) | Myzus borealis (1) | 0.2 – 0.5 |
| Myzus cerasi (50) | Myzus persicae (1) | 6.6 – 6.8 |
| Myzus borealis (1) | Myzus persicae (1) | 6.8 |
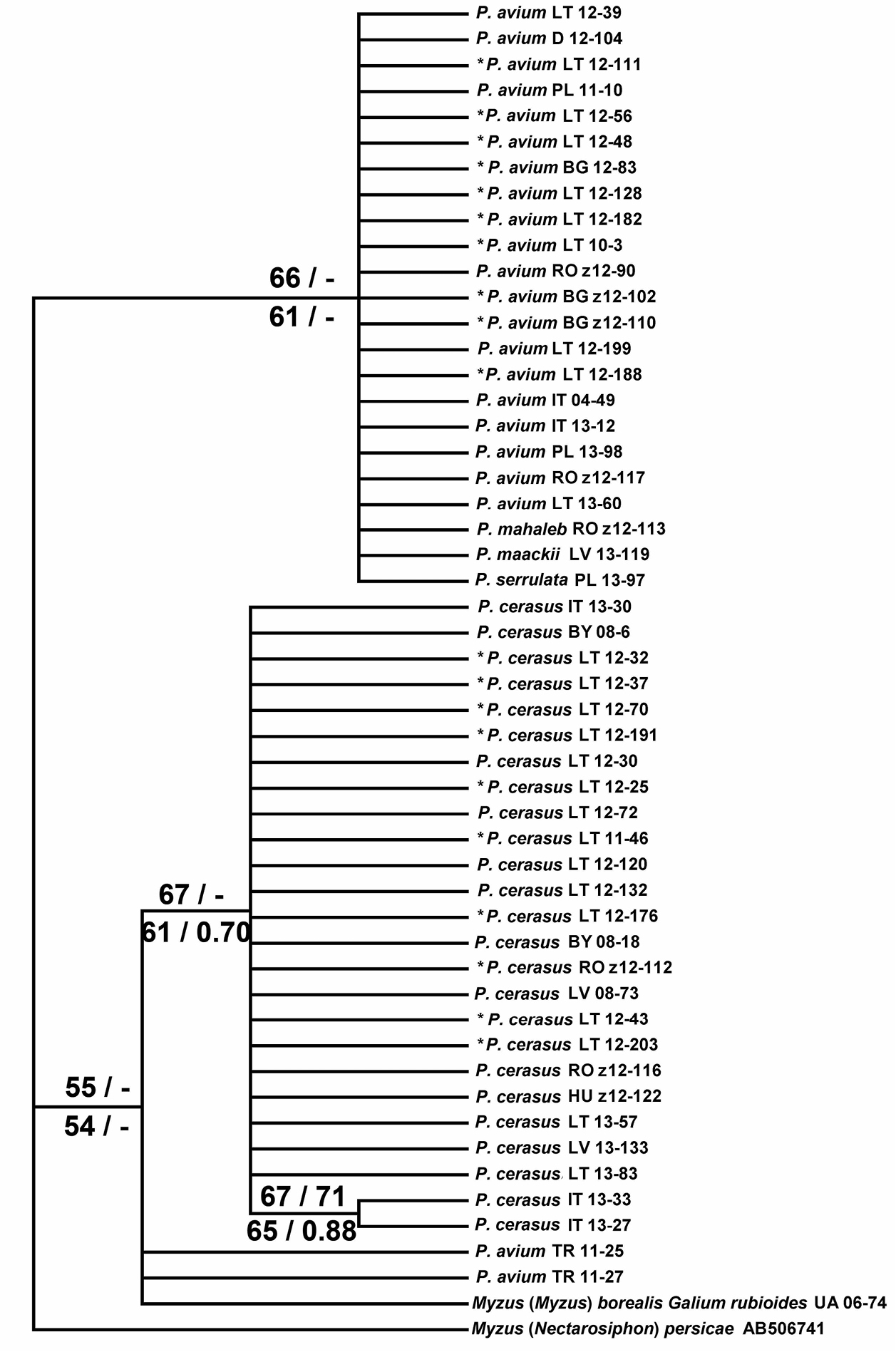
The maximum parsimony (MP) analysis of partial COI sequences resulted in 930 equally parsimonious trees (length = 43, CI = 1.00, RI = 1.00). The ML tree (T92 model) showed similar topology, as did NJ (K2P distances) and BI (HKY+I+G model) analyses. NJ, MP and ML bootstrap values over 50% together with BI posterior probabilities over 0.50 are given at respective nodes of the same tree in Fig. 2. Thus the Myzus cerasi samples form two major clades corresponding to two host-specific black cherry aphid taxa. One clade consists of all but two of the samples from Prunus avium, plus aphids collected from Prunus mahaleb, Prunus maackii and Prunus serrulata. The other clade contains all samples from Prunus cerasus and also includes the sample of Myzus borealis collected from Galium rubioides.
Maximum likelihood (ML) tree showing phylogenetic relationships among Myzus cerasi based on partial sequences of mitochondrial COI (616 positions in final set). Numbers above branches indicate support of NJ (left, > 50%) and MP (right, > 50%) bootstrap test with 1000 replicates, and numbers below branches indicate support of ML (left, > 50%) bootstrap test with 1000 replicates and posterior probabilities of BI analysis (right, > 0.50). Samples used for the discriminant analysis with a priori specified group membership followed by the construction of identification key are asterisked (*). The remaining samples were used for the post hoc classification. Sample numbers are the same as given in Table 1, together with the abbreviated symbol of respective country BG – Bulgaria, BY – Belarus, D – Germany, HU – Hungary, IT – Italy, LV – Latvia, LT – Lithuania, PL – Poland, RO – Romania, TR – Turkey, UA – Ukraine.
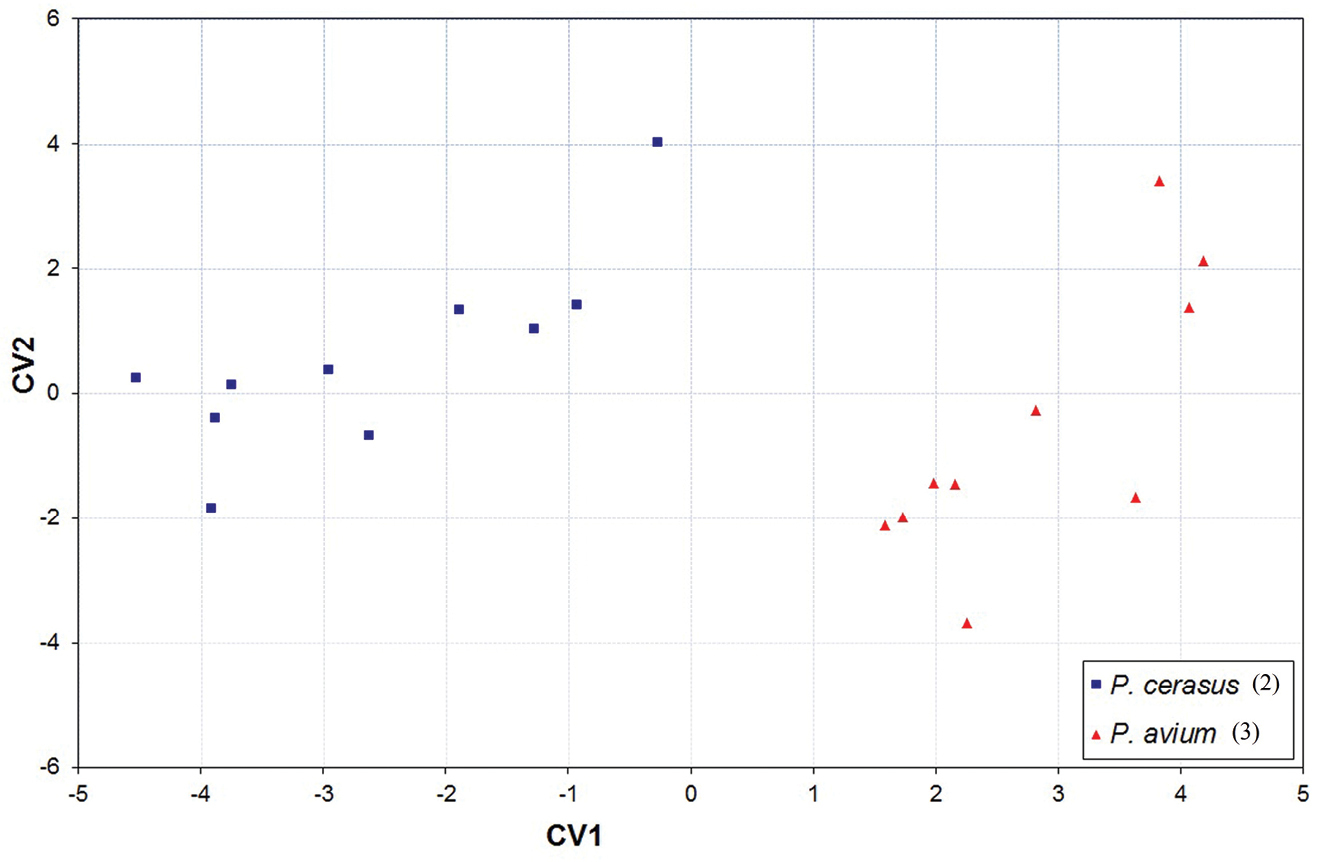
When morphometric data of apterous viviparous females from 20 different geographical localities were subjected to discriminant analysis with sample collection number as the grouping variable, the first two canonical variates (Fig. 3) clearly separated sour cherry samples (COI haplotype No. 2) from those collected from sweet cherry (COI haplotype No. 3). Length of terminal process of antennal segment 6 (A6TPL), length of siphunculus (SL) and maximal length of the ventral body hairs (VBSLmax) appeared to be important predictors for separation of the two taxa (Table 4).
Plot of the mean scores of the first two canonical variates for 20 samples of Myzus cerasi (for specimen numbers per sample see Table 1). Samples cluster in accordance with winter host plant and COI haplotype (haplotype number is given in parentheses, see Table 2 for other haplotypes).
Contributions of 11 morphological characters to the canonical function discriminating 20 samples of Myzus cerasi. Character abbreviations are the same as in the text (Material and methods).
| Wilks’ Lambda | Partial Wilks’ Lambda | F-remove | p-level | Toler. | 1-Toler. (R-Sqr.) | |
|---|---|---|---|---|---|---|
| A6TPL | 0.34 | 0.65 | 72.00 | 0.00 | 0.46 | 0.54 |
| SL | 0.30 | 0.73 | 49.26 | 0.00 | 0.31 | 0.69 |
| VBSLmax | 0.27 | 0.83 | 28.31 | 0.00 | 0.79 | 0.21 |
| Bwant3 | 0.24 | 0.91 | 12.99 | 0.00 | 0.71 | 0.29 |
| URL | 0.23 | 0.93 | 9.51 | 0.00 | 0.65 | 0.35 |
| A6BL | 0.23 | 0.94 | 8.69 | 0.00 | 0.47 | 0.53 |
| DT3L | 0.23 | 0.97 | 4.62 | 0.01 | 0.49 | 0.51 |
| FF | 0.23 | 0.97 | 4.23 | 0.02 | 0.86 | 0.14 |
| VBSLmin | 0.23 | 0.97 | 3.70 | 0.03 | 0.85 | 0.15 |
| CL | 0.22 | 0.98 | 2.38 | 0.10 | 0.57 | 0.43 |
| CW | 0.22 | 0.99 | 1.75 | 0.18 | 0.56 | 0.44 |
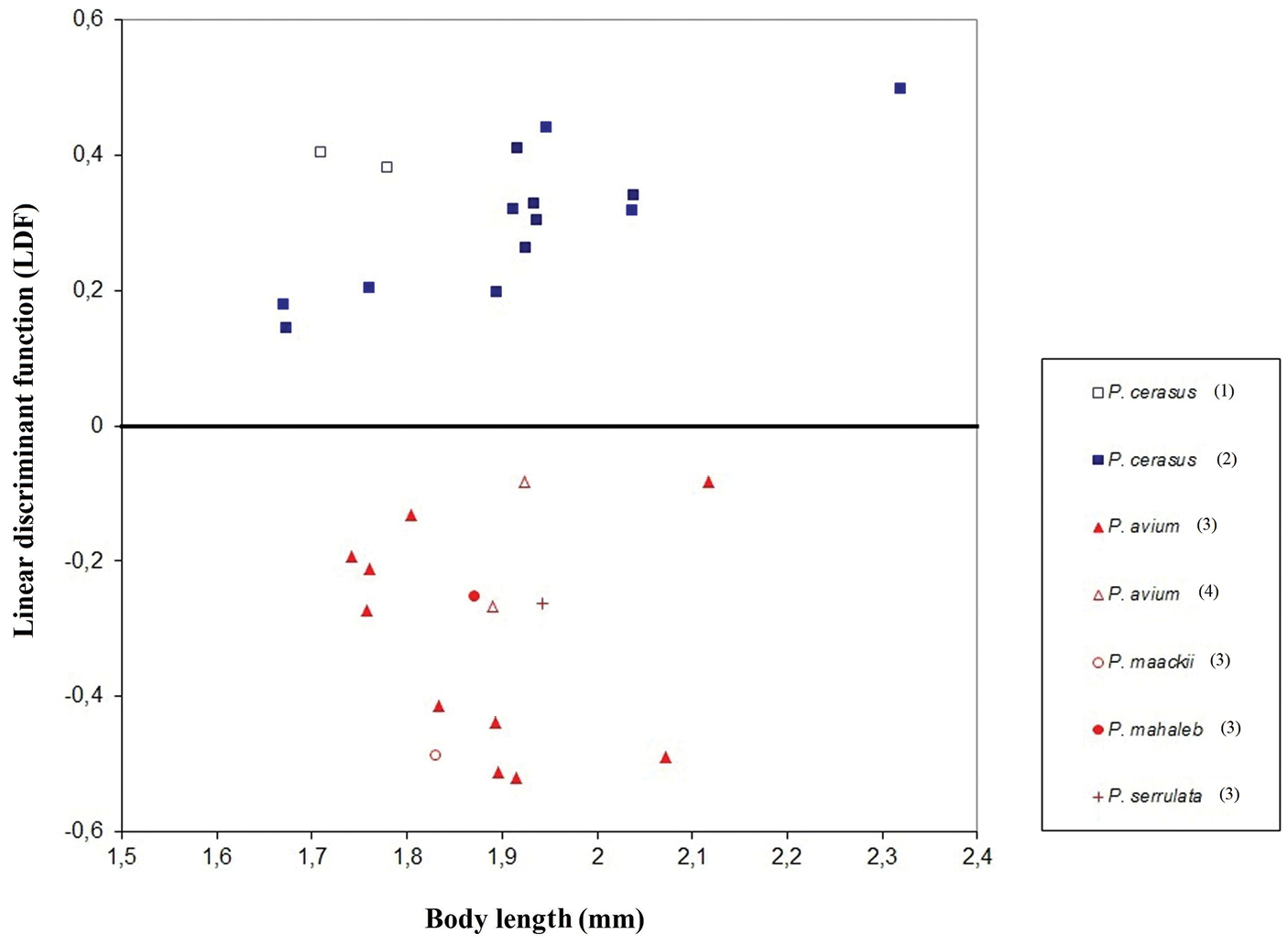
To discriminate between apterous viviparous females of host-specific black cherry aphid samples representing different clades in the haplotype network and the phylogenetic tree (Figs 1–2), the following linear discriminant function (LDF) was obtained: 3.924682 × SL - 5.6667 × A6TPL - 32.5504 × VBSLmax + 1. Using this LDF, 97.37% individuals from the whole dataset were reclassified correctly into their a priori specified groups with host plant species as grouping variable, including 96.2% of apterous viviparous females from Prunus avium (n = 79) and 98.6% from Prunus cerasus (n = 73). The post hoc classification of the remaining thirty samples gave 92.37% correct specimen identification of Myzus cerasi cerasi (n = 118) and 93.64% of Myzus cerasi pruniavium (n = 110). The scatterplot of the mean LDF and body length values calculated for each of 30 samples representing different host specific subspecies of Myzus cerasi is shown in Fig. 4. The following key is therefore suggested for the identification of apterous viviparous females of the two subspecies of Myzus cerasi when sampled from winter hosts.
Plot of the mean scores of the individual LDF values (number of specimens per sample is given in Table 1) plotted against the mean body length for 30 samples of Myzus cerasi (normal font in Table 1) used to evaluate effectiveness of the eventual identification key. The icons are color-coded to match the COI haplotypes. Samples cluster in accordance with winter host plant and COI haplotype (haplotype number is given in parentheses, see Table 2 for haplotype information).
| 1 | Value of LDF [3.92× (length of siphunculus) - 5. 67×(length of terminal process of antennal segment 6) - 32.55×(maximal length of the ventral body hairs) + 1] greater than zero. On Prunus cerasus (and sometimes on Prunus avium) | Myzus cerasi cerasi |
| – | Value of LDF less than zero. On Prunus avium, Prunus maackii, Prunus mahaleb, Prunus serrulata | Myzus cerasi pruniavium |
The combination of genetic distance evaluation with phylogenetic tree-building methods and multivariate analyses of morphometric data has been successfully applied to solve taxonomic problems in aphids, particularly in the genera Hyalopterus (
Pairwise sample divergences of 1145 bp mitochondrial COI gene fragment (K2P model) between three subspecies of Aphis fabae (number of sequences used is in parentheses).
| Subspecies 1 | Subspecies 2 | Mean and range of divergence, % |
|---|---|---|
| Aphis fabae fabae (3) | Aphis fabae cirsiiacanthoidis (12) | 1.2 (0.97–1.42) |
| Aphis fabae fabae (3) | Aphis fabae mordvilkoi (3) | 0.15 (0.00–0.26) |
| Aphis fabae cirsiiacanthoidis (12) | Aphis fabae mordvilkoi (3) | 1.05 (0.97–1.15) |
Myzus borealis is clearly closely related to Myzus cerasi and differs by only 0.2–0.5% of the COI sequences involved in the analysis. This suggests that it may also belong to the same species level taxon. More samples of Myzus borealis are needed to confirm this hypothesis. However, it should be noted that partial COI sequences of two biologically distinct Macrosiphum species, Macrosiphum rosae (Linnaeus, 1758) and Macrosiphum knautiae Holman, 1972 are very similar (
This research was funded by a grant (No LEK-04/2012) from the Research Council of Lithuania. Kind assistance of S. Barbagallo and G. Cocuzza (Catania, Sicily), V. Zhuravlev (Kyiv, Ukraine), S. Buga (Minsk, Belarus), V. Spuņģis (Riga, Latvia), A. Stalažs (Dobele, Latvia), M. Badini (Bagnolo Mella, Italy), A. A. Işıkber and M. M. Aslan, (Kahramanmaraş, Turkey) during field sampling in respective countries cannot be overestimated.