(C) 2013 José L. Fernández-Triana. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Fernández-Triana JL, Cardinal S, Whitfield JB, Hallwachs W, Smith MA, Janzen DH (2013) A review of the New World species of the parasitoid wasp Iconella (Hymenoptera, Braconidae, Microgastrinae). ZooKeys 321: 65–87. doi: 10.3897/zookeys.321.5160
The New World species of Iconella (Hymenoptera: Braconidae, Microgastrinae) are revised. Iconella andydeansi Fernández-Triana, sp. n., Iconella canadensis Fernández-Triana, sp. n., and Iconella jayjayrodriguezae Fernández-Triana, sp. n., are described as new. Iconella isolata (Muesebeck, 1955), stat. r., previously considered as a subspecies of Iconella etiellae (Viereck, 1911), is here elevated to species rank. All species have different, well defined geographic distributions and hosts. Taxonomic keys are presented in two formats: traditional dichotomous hardcopy versions and links to electronic interactive versions (software Lucid 3.5). Numerous illustrations, computer-generated descriptions, distributional information, host records (mostly Lepidoptera: Crambidae and Pyralidae), and DNA barcodes (where available) are presented for every species. Phylogenetic analyses of the barcoding region of COI indicate the possibility that Iconella is not monophyletic and that the New World species may not form a monophyletic group; more data is needed to resolve this issue.
Iconella, Microgastrinae, New World, taxonomic review, host caterpillars, DNA barcoding, Area de Conservacion Guanacaste, parasitoid wasps
The genus Iconella was erected by
Until now, only one species was known from the New World, Iconella etiella (Viereck, 1911). However, ongoing research on the Microgastrinae fauna of Area de Conservacion de Guanacaste (ACG) in northwestern Costa Rica (
It should be noted that
Iconella is rare in collections, and even when present, specimens tend to be categorized as “unidentified Microgastrinae”. In the New World, with the exception of one species, the genus seems to be rare in nature as well. Even long-term and comprehensive rearing programs –such as those in ACG, Costa Rica- have recovered relatively few specimens. This study is mostly based on the examination of unidentified Iconella specimens housed in the CNC, representing close to 40 specimens; 11 specimens reared by the ACG inventory -and housed in the CNC and the Illinois Natural History Survey, Champaign, Illinois, United States (INHS); one specimen from the Laurentian Forestry Center, Ste.-Foy, Quebec, Canada (LFS); and the holotype of Iconella etiellae from the Smithsonian Institution, Washington DC, United States (NMNH).
Morphological terms and measurements of structures are mostly as used by
The species descriptions are based mostly on the holotype female, with other specimens studied (when available) for intraspecific variation. When the holotype could not be examined (Iconella isolata) or it was a male (Iconella etiellae), female specimens were used for these redescriptions.
Lucid 3.5.4 (http://www.lucidcentral.com/) software was used to generate automatic descriptions of the species and to prepare Lucid identification keys. A dataset of 20 characters and 126 character-states was used to provide uniform descriptions for all species treated. Description format includes one sentence per character, with the character (in italics) mentioned first and the character-state following after a colon, e.g., “Pterostigma color: mostly brown, with yellowish-white spot at anterior 0.2 ×”.
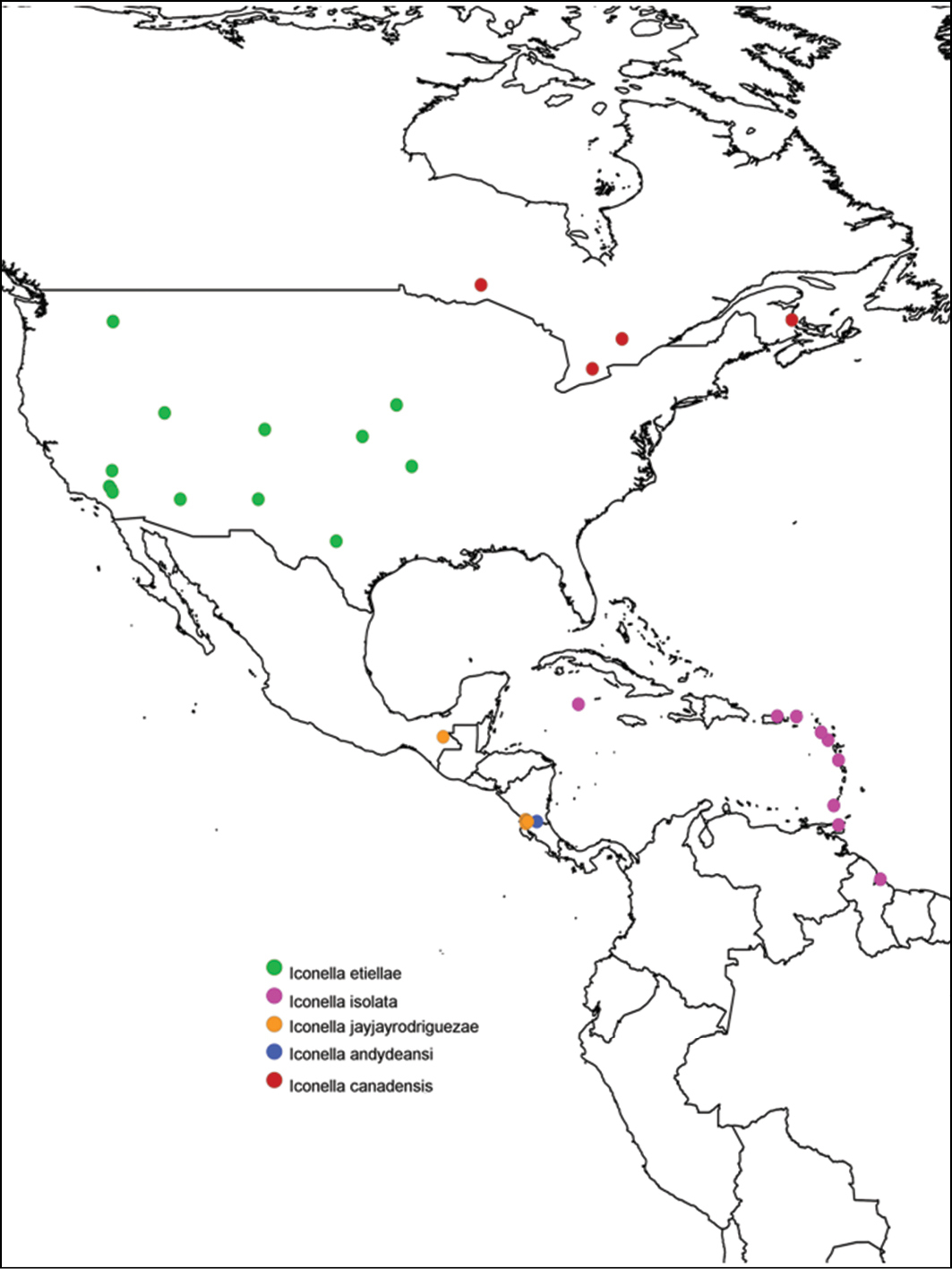
A map with the distribution of all New World species of Iconella was generated using SimpleMappr (
In the taxonomic treatment of species, “Specimens Examined” presents the specimen’s information in the following format: “Number of females/males, acronym of the storing institution between parentheses, COUNTRY: State/Province, city, other locality details, coordinates (in Decimal Degrees, abbreviated as “Lat:” and “Long:”), date, collector name, biological information on host (starting with “ex”), ACG database codes (in the format “yy-SRNP-xxxxxx” or “DHJPARxxxxxxx”, with the former being the voucher code of the host and the latter being the voucher code of the parasite). For states of the United States and for Canadian provinces/territories, acronyms consisting of two capital letters are used, following Canada Post (http://www.canadapost.ca/tools/pg/manual/PGaddress-e.asp).
DNA barcodes for all specimens that were barcoded were obtained using DNA extracts prepared from single legs using a glass fibre protocol (
DNA barcode sequences were aligned in Geneious Pro 6.0.5 (
Many of the Iconella specimens in BOLD were collected before 1979. Characteristic of such moderately-aged specimens, the COI fragments generated were less (~160bp) than the full length characteristic of the standard DNA barcode (>600bp) (
To investigate the phylogenetic relationships of the species, the aligned dataset was analyzed in MrBayes v. 3.2.1 (
As with many other taxa, the generic status of Iconella will only be solved when a comprehensive phylogeny of Microgastrinae is carried out. In the meantime we think is best to keep it as a valid genus, based on the available morphological evidence.
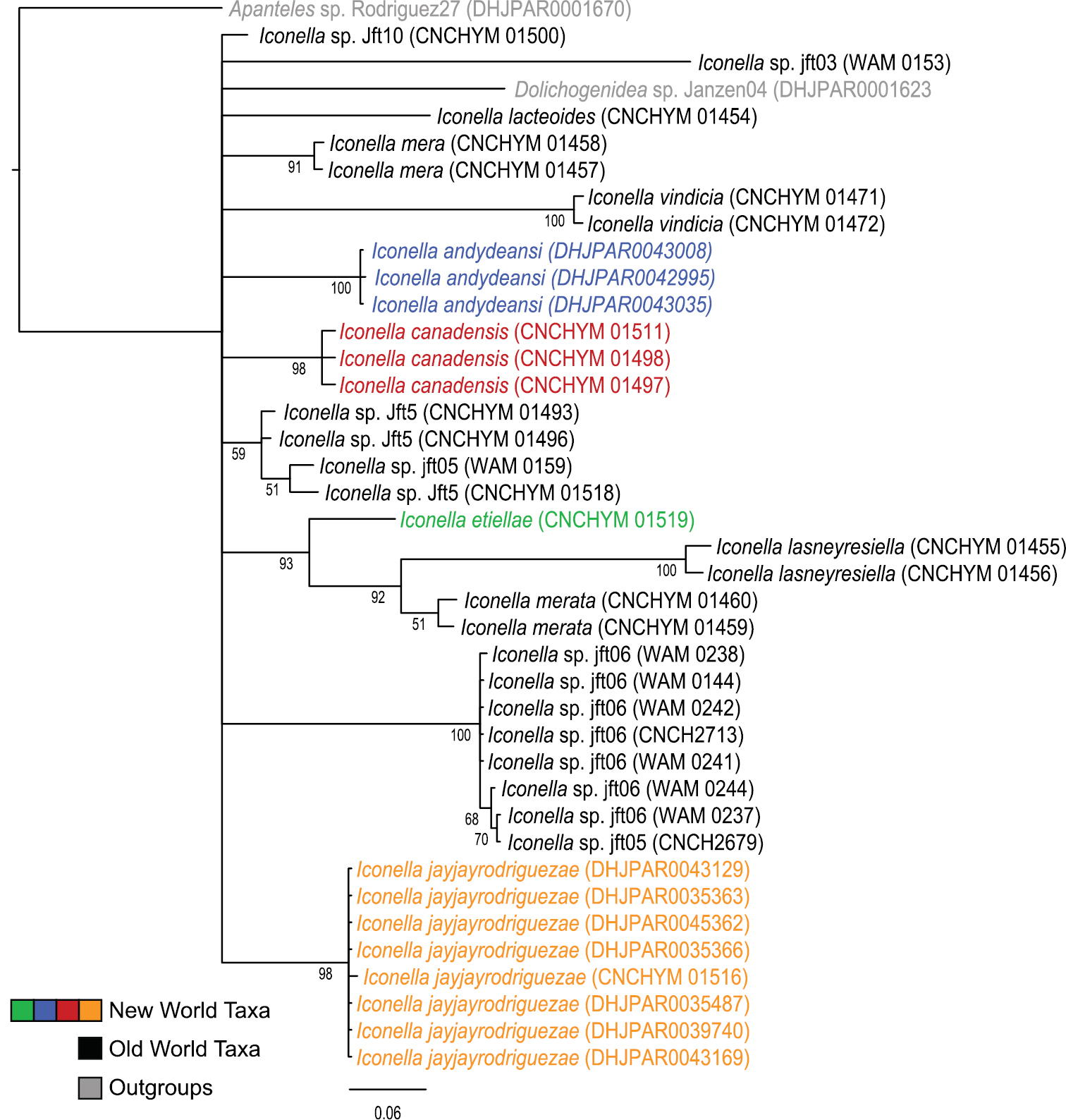
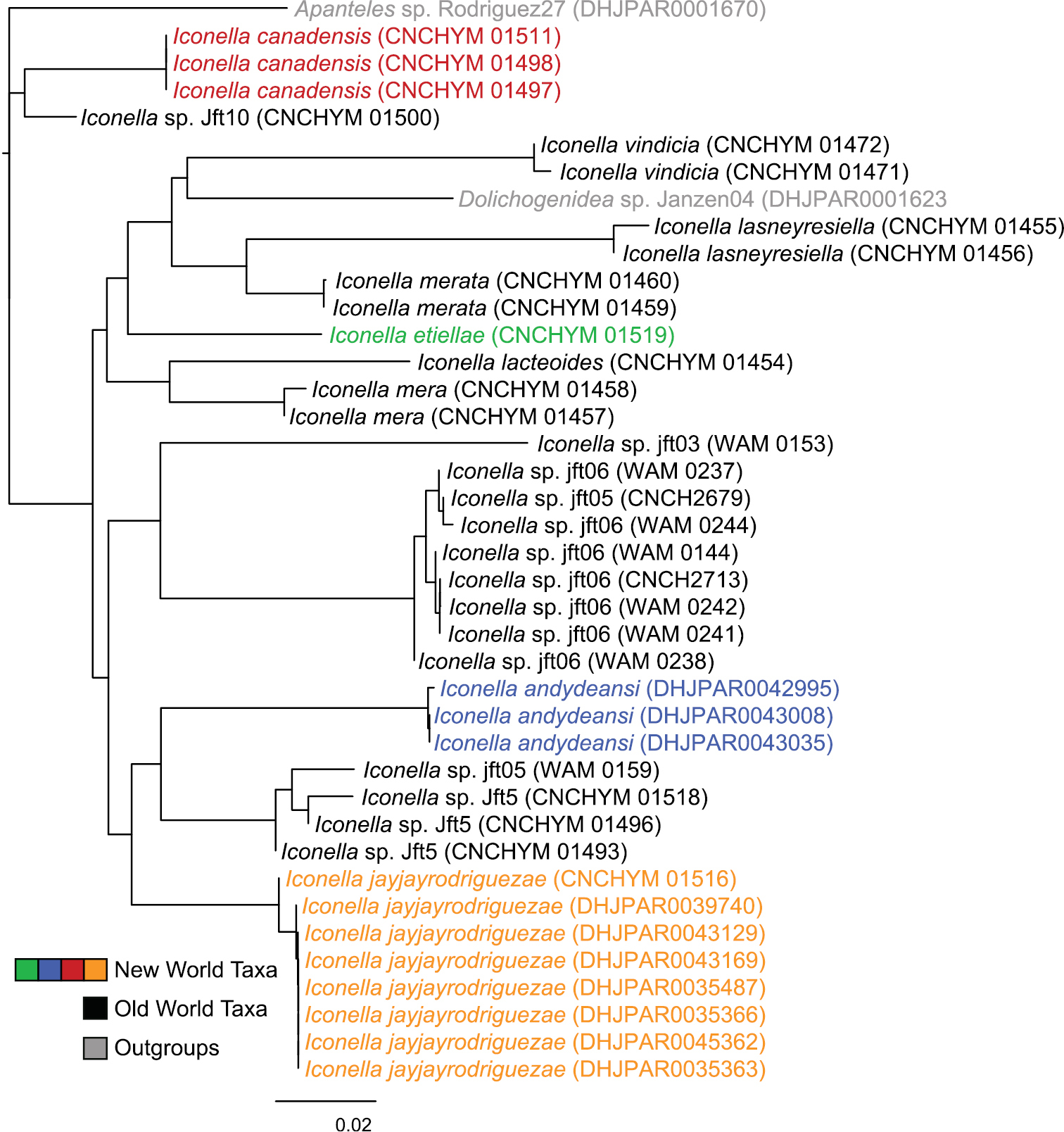
Phylogenetic analyses of the COI DNA, however, are inconclusive as to whether the genus Iconella is monophyletic (Figs 1 and 2). All Bayesian (only the tree resulting from analysis of the full dataset is shown in Fig. 1), ML (tree not shown), and parsimony (tree not shown) analyses failed to recover monophyly of the genus. However, the model-based methods do not offer strong support against monophyly either. The highest posterior probability (PP) supporting non-monophyly was 0.55 when 3rd codon positions were removed from the Bayesian analysis, and BS support against monophyly in the ML analysis was only 0.21.
Majority rule consensus tree calculated from the posterior tree samples of the Bayesian analysis of the full Iconella COI fragment dataset. Values below the branches are posterior probabilities. The New World species are individually color coded.
Neighbor-joining tree of the full Iconella COI fragment dataset using the TN93 model. The New World species are individually color coded.
We recognize five species of Iconella in the New World, including the three new species described in the present paper, and one that was previously considered as a subspecies (
All of the described species have a different, well delimited geographic range (Fig. 3) and also differ in their known hosts. Most of the host species in the Western Hemisphere belong to the Lepidoptera families Pyralidae and Crambidae (with appropriate caution that what was called Pyralidae in the past is in part called Crambidae at present). The NJ tree (Fig. 2) as well as Bayesian (Fig. 1), ML, and parsimony based phylogenetic analyses of the barcodes support the validity of all New World species recognized herein (there is no molecular data available for Iconella isolata). However, the barcoding dataset did not contain a strong enough phylogenetic signal to resolve the phylogenetic relationships among Iconella species.
Distribution of New World species of Iconella. Species are individually color coded.
While these DNA barcodes and short COI fragments (mini-barcodes (
| 1 | Propodeum mostly smooth and polished (as in Fig. 21), except for sparse punctures on the anterior 0.2 × of propodeum, and rather small carinae radiating from median longitudinal carina; metatibia mostly yellow, at most with very small and faint brown spot on posterior 0.1 or less, and metatarsus mostly yellow, except for brown area on posterior half of first tarsomerus (Figs 17, 19, 23, 25); fore wing with most veins transparent or white, vein margins of same color than interior of vein (Figs 18, 24) | 2 |
| – | Propodeum with anterior 0.2 × covered with punctures, posterior 0.8 covered with mix of punctures, striated sculpture and carinae (as in Fig. 15); metatibia with brown to black coloration on posterior 0.2–0.4, and metatarsus mostly dark brown, except for yellowish area on anterior half of first tarsomerus (as in Fig. 33); fore wing with at least some veins with thin brown margins and interior of veins yellow to light brown (Figs 5, 11, 31) | 3 |
| 2 | Pterostigma almost completely brown, with only small whitish spot anteriorly (Fig. 18); humeral complex half yellow, half brown; profemur almost completely dark brown (yellow area absent or limited to posterior 0.2); interocellar distance 2.4 × or more posterior ocellus diameter (Fig. 20); mediotergite 2 width at posterior margin 4.6 × or less its length (Fig. 22); larger species, body length (head to apex of metasoma) 3.0 mm or more, and fore wing length 3.3 mm or more. [Western and central United States: AR, AZ, CA, CO, IA, NM, OK, and WA. Hosts: Etiella zinckenella, Olycella junctolineella, Psorosina hammondi, and Ufa rubedinella (Pyralidae)] | Iconella etiellae (Viereck, 1911) |
| – | Pterostigma mostly transparent or whitish, with only thin brown margins (Fig. 24); humeral complex yellow to white; profemur mostly yellow, dark brown area limited to anterior 0.2 or less; interocellar distance 2.1 × or less posterior ocellus diameter; mediotergite 2 width at posterior margin 5.0 × or more its length (Fig. 26); smaller size, body length (head to apex of metasoma) 3.0 mm or less, and fore wing length 3.2 mm or less. [Caribbean islands and northern part of South America: British Virgin Islands, Cayman Islands, Dominica, Grenada, Guyana, Montserrat, Puerto Rico, Saint Kitts & Nevis, Trinidad & Tobago. Host: Ancylostomia stercorea (Pyralidae)] | Iconella isolata (Muesebeck, 1955), stat. r. |
| 3- | Ocular-ocellar line 1.6 × posterior ocellus diameter; humeral complex half yellow, half brown; mediotergite 1 width at anterior margin 2.2 × or less its width at posterior margin (Fig. 13); ovipositor sheaths length 0.8 × or less metatibial length (Fig. 12); larger species, body length (head to apex of metasoma) 3.5 mm or more (rarely 3.2 mm) and fore wing length 3.5 mm or more; an extra-tropical species distributed in North America north of 40° N (Canada). [Eastern Canada: NB, ON, and QC. Host: Epinotia solandriana (Tortricidae) and, likely, Acrobasis betulella (Pyralidae)] | Iconella canadensis Fernández-Triana, sp. n. |
| - | Ocular-ocellar line 2.0 × or more posterior ocellus diameter; humeral complex fully yellow to white; mediotergite 1 width at anterior margin 3.1 × or more its width at posterior margin (Figs 9, 36); ovipositor sheaths length 1.1 × metatibial length (Figs 6, 32); smaller size, body length (head to apex of metasoma) 3.0 mm or less, and fore wing length 3.3 mm or less; tropical species from Central America south of 17° N (Mexico and Costa Rica) | 4 |
| 4 | Profemur mostly yellow, dark brown area limited to anterior 0.2 or less; meso- and meta- femora mostly dark brown, with proximal 0.1–0.2 yellow to orange; mesoscutellar disc sculpture centrally smooth with few, scattered punctures near margins (Fig. 36); mediotergite 2 width at posterior margin 4.1 × or less its maximum length medially (Fig. 36); body length (head to apex of metasoma) 2.9–3.0 mm; fore wing length 3.2–3.3 mm. [Costa Rica (ACG) and Mexico (Chiapas). Host: undescribed species of Phycitinae (Pyralidae)] | Iconella jayjayrodriguezae Fernández-Triana, sp. n. |
| – | Profemur dark brown on anterior half, yellow on posterior half; meso- and meta- femora usually fully dark brown to black; mesoscutellar disc sculpture mostly with punctures scattered all over disc surface (Fig. 9); mediotergite 2 width at posterior margin 4.4 × its maximum length medially (Fig. 9); body length (head to apex of metasoma) 2.7–2.8 mm; fore wing length 3.0 mm. [Costa Rica (ACG). Host: undescribed species of Spilomelinae (Crambidae)] | Iconella andydeansi Fernández-Triana, sp. n. |
http://zoobank.org/C9173C9E-1E3C-46A8-A459-97DDF2ABF1DF
http://species-id.net/wiki/Iconella_andydeansi
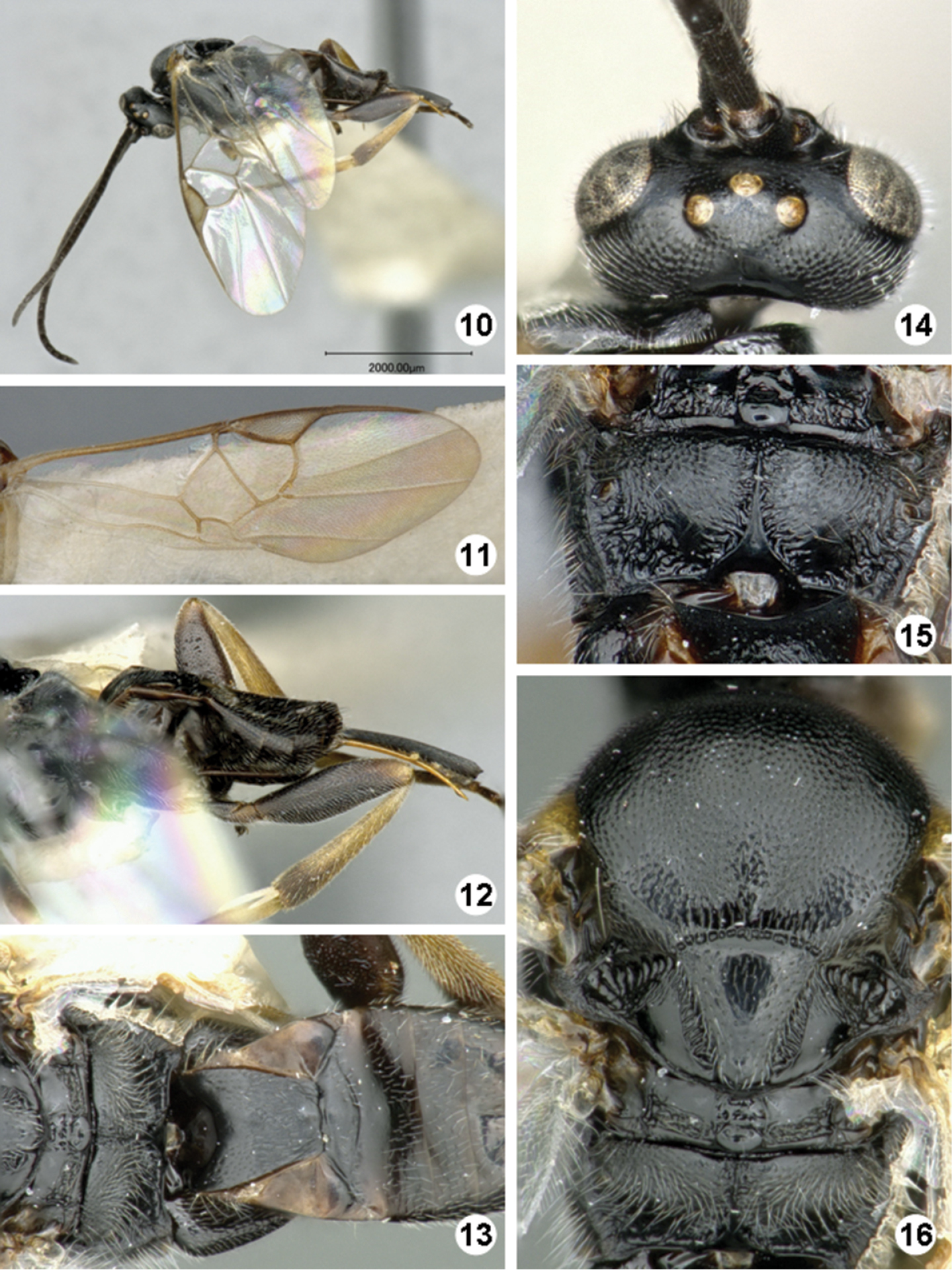
Figures 4–9COSTA RICA, Alajuela, Area de Conservacion Guanacaste, Sector Rincon Rain Forest, Camino Rio Francia, 410m. Lat: 10.90425, Long: -85.28651.
♀, CNC. First label: DHJPAR0043035. Second label: Voucher: D.H.Janzen & W.Hallwachs, DB: http://janzen.sas.upenn.edu, Area de Conservacion Guanacaste, COSTA RICA, 11-SRNP-41294. Collecting date of caterpillar host: 20.iii.2011, collection date (eclosion date) of wasp: 05.iv.2011.
Paratypes: 1 ♀, 1 ♂ (CNC) Costa Rica, same locality than holotype. Specimens voucher codes: DHJPAR0042995 and DHJPAR0043008.
Promefur color: dark brown on anterior half, yellow on posterior half. Meso- and meta- femur color: fully dark brown to black (Fig. 4). Metatibia and metatarsus color: Metatibia with brown to black coloration on posterior 0.2–0.4 ×; metatarsus mostly dark brown, except for yellowish area on anterior half of first tarsomerus (Fig. 6). Tegula and humeral complex color: tegula and humeral complex fully yellow to yellowish-white (Fig. 9). Pterostigma color: centrally yellow-white, with thin brown margins (Fig. 5). Fore wing veins color: at least some veins with thin brown margins and interior of veins yellow to light brown. Body length (head to apex of metasoma): 2.7 mm or 2.8 mm. Fore wing length: 3.0 mm. Ocular-ocellar line/posterior ocellus diameter: 2.2 ×. Interocellar distance/posterior ocellus diameter: 2.1 × (Fig. 7). Antennal flagellomere 2 length/width: 2.5 × or 2.7 ×. Antennal flagellomere 14 length/width: 1.3 ×. Length of flagellomere 2/length of flagellomere 14: 2.4 ×. Metafemur length/width: 3.1 ×. Mesoscutellar disc: mostly with punctures scattered all over disc surface (Fig. 9). Number of pits in scutoscutellar sulcus: usually 12 or less, ocasionally reaching up to 14 pits. Propodeum background sculpture: anterior 0.2-0.4 × with rather dull puntures; posterior 0.6–0.8 × mostly sculptured, with mix of small puntures and carinae (mostly radiating from strong, longitudinal median carina) (Fig. 8). Mediotergite 1 width at anterior margin/width at posterior margin: 3.4 ×. Mediotergite 2 width at posterior margin/length: 4.4 × (Fig. 9). Ovipositor sheaths length/metatibial length: 1.1 × (Fig. 6).
Male. As female, although sculpture is slightly smoother.
Iconella andydeansi. 4 Habitus, lateral view 5 Fore wing 6 Ovipositor sheats and metatibia 7 Head, dorsal view 8 Propodeum 9 Meso- and metasoma (partially), dorsal view.
We analyzed three full 658 bp barcodes for this species.
Host: An undescribed species of Phycitinae (Pyralidae) with provisional name “phyjanzen021 Janzen855” in the ACG database (http://janzen.sas.upenn.edu/caterpillars/database.lasso). Caterpillar collected while feeding on the foliage of Lepidoploa salzmannii (Asteraceae).
Only known from the holotype locality, in Sector Rincon Rain Forest of ACG at 410 m.
The species has been reared only in one place from three caterpillars collected at the same time on the same species of food plant, out of 12, 000+ rearings of ACG Pyralidae of more than 200 species. A single additional specimen, identified by DNA barcoding, has been reared in 2012 from the same species of host caterpillar in the same place and on the same food plant, but was not available for study. Iconella andydeansi is sympatric with Iconella jayjayrodriguezae in ACG, the latter being a species with a slightly larger ACG rain forest distribution but equally narrow food plant record, and also known from Chiapas, Mexico. Those are the only two species of New World Iconella that are known to be sympatric, and that was revealed through extensive collecting in ACG and the use of DNA barcodes. It is likely that further collecting in other areas of the Neotropics, as well as the barcoding of more fresh specimens, will reveal additional species.
This species is named in honor of Andy R. Deans (Pennsylvania State University, United States) in recognition of his major contribution to the taxonomy of the many species in the microgatrine genus Alphomelon that occur in Area de Conservación Guanacaste (e.g.,
http://zoobank.org/C3E8D164-ABBC-4823-9D56-50E302065F55
http://species-id.net/wiki/Iconella_canadensis
Figures 10–16CANADA. Ontario, Black Sturgeon Lake. Lat: 49.368333, Long: -88.881944.
♀, CNC. First label: Black Sturgeon Lake, Ontario, Em. 1-6-viii-1961, Insectary. Second label: Nest 97, Cell4ex provisions. Third label: W61319. Fourth label: Host either A. betullela or Rh. hasta. Fifth label: DNA Voucher CNCHYM 01498.
Paratypes: 3 ♀ (CNC) Canada: ON, Black Sturgeon Lake, 21–29.vii.1961, 26.vii.1962, and 2.viii.1962, ex: Provisions Nests 52 and 66, one specimen with DNA Voucher CNCHYM01497; 1 ♀ (CNC) Canada: ON, Galt, 11.vii.1952; 1 ♀ (CNC) Canada: ON, Whitney, 4.vii.1949, ex: Phalaenidae; 1 ♀, 1 ♂ (CNC) Canada: NB, Kouchibouguac National Park, 30.viii.1967, code-6060B, DNA Voucher CNCHYM01511 and CNCHYM01512; 1 ♀ (LFS) Canada: QC, Saint-Cléophas-de-Brandon, 4.vii.1968, ex: Epinotia solandriana on Betula papyrifera. Collecting dates of specimens examined: July and August (1949–1967).
Promefur color: dark brown on anterior half, yellow on posterior half. Meso- and meta- femur color: mostly dark brown but with proximal 0.1–0.2 × yellow to orange (Fig. 12). Metatibia and metatarsus color: Metatibia with brown to black coloration on posterior 0.2–0.4 ×; metatarsus mostly dark brown, except for yellowish area on anterior half of first tarsomerus (Fig. 10). Tegula and humeral complex color: tegula and anterior half of humeral complex yellow to yellowish-white, posterior half of humeral complex light brown to dark brown. Pterostigma color: centrally yellow-white, with thin brown margins, rarely mostly brown, with yellowish-white spot at anterior 0.2 × (Fig. 11). Fore wing veins color: at least some veins with thin brown margins and interior of veins yellow to light brown. Body length (head to apex of metasoma): 3.5 mm, 3.6 mm, 3.7 mm, rarely 3.2 mm. Fore wing length: 3.8 mm, 3.9 mm, 4.0 mm, 4.1 mm or 4.2 mm. Ocular-ocellar line/posterior ocellus diameter: 1.6 ×. Interocellar distance/posterior ocellus diameter: 1.9 × (Fig. 14). Antennal flagellomere 2 length/width: 3.0 ×. Antennal flagellomere 14 length/width: 1.6 ×. Length of flagellomere 2/length of flagellomere 14: 2.1 ×. Metafemur length/width: 3.2 ×, 3.3 ×, rarely 3.4 ×. Mesoscutellar disc: mostly smooth with few, scattered punctures near margins (Fig. 16). Number of pits in scutoscutellar sulcus: usually 16, ocasionally only 14 pits. Propodeum background sculpture: anterior 0.2-0.4 × with rather dull puntures; posterior 0.6-0.8 × mostly sculptured, with mix of small puntures and carinae (mostly radiating from strong, longitudinal median carina) (Fig. 15). Mediotergite 1 width at anterior margin/width at posterior margin: 2.1 × or 2.2 ×. Mediotergite 2 width at posterior margin/length: 3.6 ×, 3.8 × or 4.4 × (Fig. 13). Ovipositor sheaths length/metatibial length: 0.7 × or 0.8 × (Fig. 12).
Male. As female.
Iconella canadensis. 10 Habitus, lateral view 11 Fore wing 12 Ovipositor sheats, mesofermur, and metatibia 13 Propodeum, mediotergites 1–4, dorsal view 14 Head, dorsal view 15 Propodeum 16 Mesosoma, dorsal view.
We analyzed three short 164 bp COI sequences from the DNA barcode region.
Host: Epinotia solandriana (Tortricidae) and likely Acrobasis betulella (Pyralidae) (see Comments below).
Eastern Canada: NB, ON, QC.
The holotype has a label stating that it emerged from either Acrobasis betulella (Pyralidae) or Rheumaptera hasta (Geometridae). Based on the known biology of the genus Iconella in the world, the second alternative is unlikely, and thus we consider Acrobasis betulella as the potential host in that case. However, the pyralid host cannot be taken as definitive until more reared specimens confirm the decision.
The name refers to the known distribution of the species, in Eastern Canada.
http://species-id.net/wiki/Iconella_etiellae
Figures 17–22UNITED STATES, Washington, Pullman.
♂, NMNH (examined).
3 ♀, 3 ♂ (CNC) United States: CA (Apple Valley, Helendale, Kramer Hills and Panamint Valley) and UT (Dugway Proving Ground). Collecting dates of specimens examined: May and June (1953–1961).
Promefur color: almost completely dark brown (yellow area absent or limited to posterior 0.2 x) (Fig. 17). Meso- and meta- femur color: mostly dark brown but with proximal 0.1-0.2 × yellow to orange (Fig. 19). Metatibia and metatarsus color: Metatibia mostly yellow, at most with very small and faint brown spot on posterior 0.1 × or less; metatarsus mostly yellow, except for brown area on posterior half of first tarsomerus (Fig. 19). Tegula and humeral complex color: tegula and anterior half of humeral complex yellow to yellowish-white, posterior half of humeral complex light brown to dark brown. Pterostigma color: mostly brown, with yellowish-white spot at anterior 0.2 × (Fig. 18). Fore wing veins color: most of veins transparent or at most yellowish-white, margins of same color than interior of vein. Body length (head to apex of metasoma): 3.0 mm or 3.1 mm. Fore wing length: 3.3 mm or 3.4 mm. Ocular-ocellar line/posterior ocellus diameter: 1.6 × or 1.7 ×. Interocellar distance/posterior ocellus diameter: 2.4 × or 2.6 × (Fig. 20). Antennal flagellomere 2 length/width: 2.6 × or 2.7 ×. Antennal flagellomere 14 length/width: 1.4 × or 1.7 ×. Length of flagellomere 2/length of flagellomere 14: 2.2 ×. Metafemur length/width: 3.2 × or 3.3 ×. Mesoscutellar disc: mostly smooth with few, scattered punctures near margins (Fig. 22). Number of pits in scutoscutellar sulcus: usually 14 or more, ocasionally reaching up to 16 pits. Propodeum background sculpture: anterior 0.2–0.4 × with fine puntures; posterior 0.6–0.8 × mostly smooth, at most with some small carinae (mostly radiating from strong, longitudinal median carina) (Fig. 21). Mediotergite 1 width at anterior margin/width at posterior margin: 2.4 × or 2.5 ×. Mediotergite 2 width at posterior margin/length: 5.0 × or 5.1 × (Fig. 22). Ovipositor sheaths length/metatibial length: 1.1 × (Fig. 19).
Male. As female.
Iconella etiellae. 17 Habitus, lateral view 18 Fore wing 19 Ovipositor sheats, mesofermur, and metatibia 20 Head, dorsal view 21 Propodeum 22 Meso- and metasoma (partially), dorsal view.
We analyzed one 607 bp barcode for this species.
Hosts: Etiella zinckenella, Olycella junctolineella, Psorosina hammondi, and Ufa rubedinella (Pyralidae).
United States: AR, AZ, CA, CO, IA, NM, OK, WA.
There is a record from Virginia, United States (
http://species-id.net/wiki/Iconella_isolata
Figures 23–29TRINIDAD & TOBAGO, St. Augustine.
♀, NMNH (not examined). Paratypes. 2 ♀ and 1 ♂, CNC (examined).
11 ♀, 5 ♂ (CNC) British Guiana, Cayman islands (Grand Cayman, Georgetown), Grenada, Montserrat, Saint Kitts & Nevis (Nevis, Round Hill), Trinidad & Tobago (Paradise Mt., St. Augustine, and Tacarigua). Collecting dates of specimens examined: February, March and December (1950–1965).
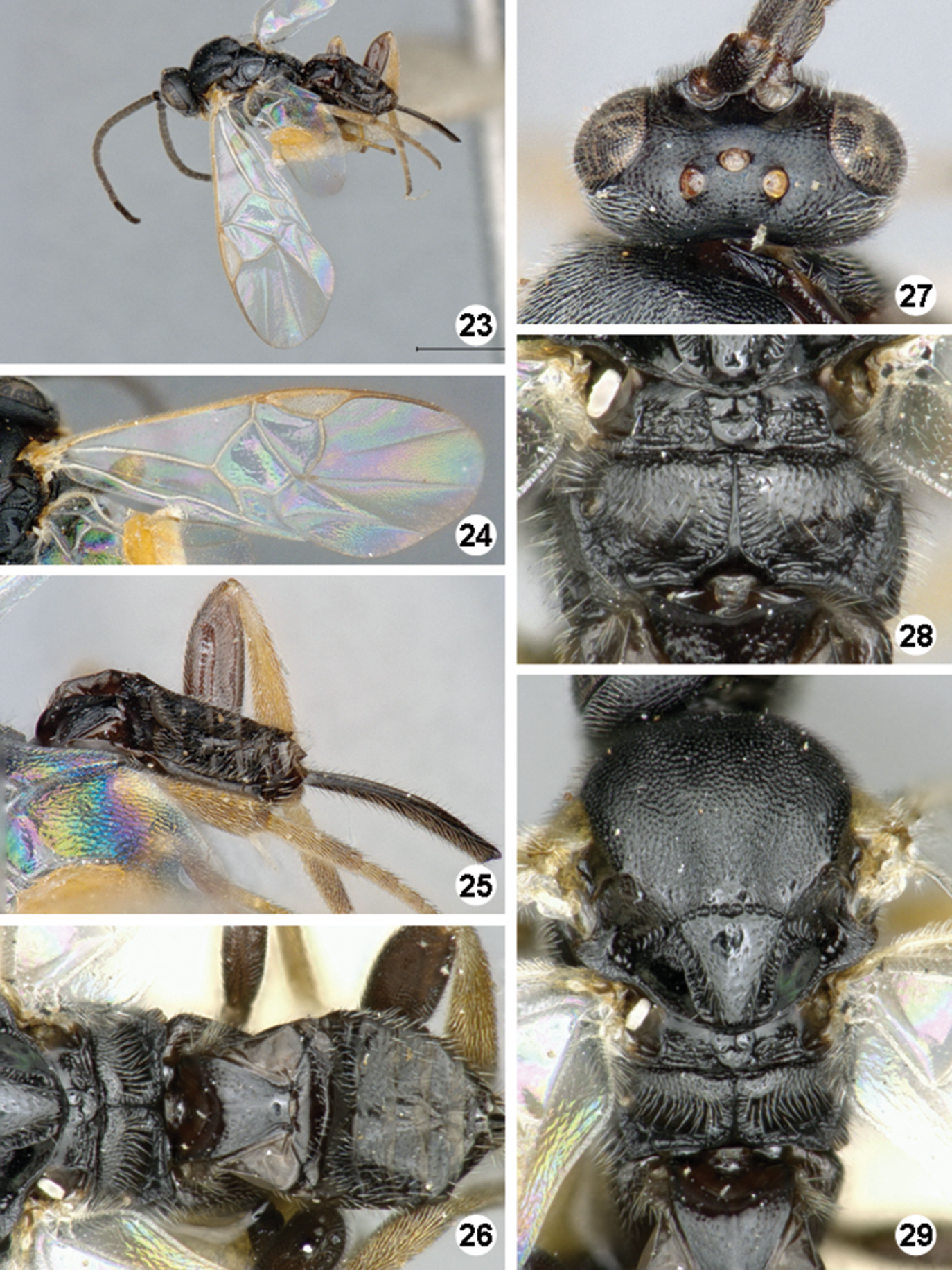
Promefur color: mostly yellow, dark brown area limited to anterior 0.2 or less, rarely dark brown on anterior half, yellow on posterior half. Meso- and meta- femur color: mostly dark brown but with proximal 0.1–0.2 × yellow to orange. Metatibia and metatarsus color: Metatibia mostly yellow, at most with very small and faint brown spot on posterior 0.1 × or less; metatarsus mostly yellow, except for brown area on posterior half of first tarsomerus. Tegula and humeral complex color: tegula and humeral complex fully yellow to yellowish-white (Fig. 29). Pterostigma color: centrally transparent, with yellow-white margins (Fig. 24). Fore wing veins color: most of veins transparent or at most yellowish-white, margins of same color than interior of vein. Body length (head to apex of metasoma): 2.4 mm, 2.5 mm, 2.6 mm, 2.7 mm, 2.8 mm or 2.9 mm. Fore wing length: 2.8 mm, 2.9 mm, 3.0 mm, 3.1 mm or 3.2 mm. Ocular-ocellar line/posterior ocellus diameter: 1.9 × or 2.0 ×. Interocellar distance/posterior ocellus diameter: 1.9 ×, 2.0 × or 2.1 × (Fig. 27). Antennal flagellomere 2 length/width: 2.2 ×, 2.3 × or 2.5 ×. Antennal flagellomere 14 length/width: 1.3 × or 1.4 ×. Length of flagellomere 2/length of flagellomere 14: 2.3 ×, 2.4 × or 2.5 ×. Metafemur length/width: 3.2 × or 3.3 ×. Mesoscutellar disc: mostly smooth with few, scattered punctures near margins (Fig. 29). Number of pits in scutoscutellar sulcus: usually 12 or less, ocasionally reaching up to 14 pits. Propodeum background sculpture: anterior 0.2–0.4 × with fine puntures; posterior 0.6–0.8 × mostly smooth, at most with some small carinae (mostly radiating from strong, longitudinal median carina) (Fig. 28). Mediotergite 1 width at anterior margin/width at posterior margin: 2.1 ×, 2.6 × or 2.8 ×. Mediotergite 2 width at posterior margin/length: 3.6 ×, 3.7 ×, 3.8 ×, 4.5 × or 4.6 × (Fig. 26). Ovipositor sheaths length/metatibial length: 1.0 × (Fig. 25).
Male. As female.
Iconella isolata. 23 Habitus, dorso-lateral view 24 Fore wing 25 Ovipositor sheats, mesofermur, and metatibia 26 Propodeum and metasoma, dorsal view 27 Head, dorsal view 28 Propodeum 29 Mesosoma and mediotergite 1 dorsal view.
No DNA barcode sequence was available for this species, the only one among the New World species without molecular data.
Host: Ancylostomia stercorea (Pyralidae).
British Virgin Islands, Cayman Islands, Dominica, Grenada, Guyana, Montserrat, Puerto Rico, Saint Kitts & Nevis, Trinidad & Tobago.
Until now, Iconella isolata had been considered to be a subspecies of Iconella etiellae. After study of numerous specimens, we have found consistent and significant differences in morphology, hosts and geographical distribution and thus consider Iconella isolata as a different species on its own. Cayman Islands and Saint Kitts & Nevis are new locality records, based on CNC specimens. The specimen from Cayman Islands represents the westernmost locality, and it expands considerably the previously known distribution of the species in the Caribbean islands.
http://zoobank.org/A1E2E3FC-6C2D-4E6F-83C0-AD388D55DCB5
http://species-id.net/wiki/Iconella_jayjayrodriguezae
Figure 30–36COSTA RICA, Alajuela, Area de Conservacion Guanacaste, Sector Rincon Rain Forest, Sendero Venado, 420m. Lat: 10.89678, Long: -85.27001.
♀, NMNH. First label: DHJPAR0039740. Second label: Voucher: D.H.Janzen & W.Hallwachs, DB: http://janzen.sas.upenn.edu, Area de Conservacion Guanacaste, COSTA RICA, 09-SRNP-41791. Collecting date of caterpillar host 21.vii.2009, collection date (eclosion date) of wasp 10.viii.2009.
Paratypes: 3 ♀, 1 ♂ (CNC) Costa Rica, Alajuela, ACG, Sector San Cristobal, Rio Blanco Abajo, 500m, Lat: 10.90037 Long: -85.37254; 1 ♀ (CNC) Mexico, Chiapas, 16°58'N, 91°47'W, 23–25.viii.1978. Collecting dates of specimens examined: January, March, April, June, July, and September (2009-2011) for Costa Rican specimens; August (1978) for Mexican specimen.
Promefur color: mostly yellow, dark brown area limited to anterior 0.2 or less. Meso- and meta- femur color: mostly dark brown but with proximal 0.1–0.2 × yellow to orange. Metatibia and metatarsus color: Metatibia with brown to black coloration on posterior 0.2–0.4 ×; metatarsus mostly dark brown, except for yellowish area on anterior half of first tarsomerus (Fig. 33). Tegula and humeral complex color: tegula and humeral complex fully yellow to yellowish-white. Pterostigma color: centrally transparent, with yellow-white margins, rarely centrally yellow-white, with thin brown margins (Fig. 31). Fore wing veins color: at least some veins with thin brown margins and interior of veins yellow to light brown. Body length (head to apex of metasoma): 2.9 mm or 3.0 mm. Fore wing length: 3.2 mm or 3.3 mm. Ocular-ocellar line/posterior ocellus diameter: 2.0 ×. Interocellar distance/posterior ocellus diameter: 1.9 × (Fig. 34). Antennal flagellomere 2 length/width: 2.3 ×, 2.4 × or 2.5 ×. Antennal flagellomere 14 length/width: 1.3 ×, rarely 1.1 × or 1.5 ×. Length of flagellomere 2/length of flagellomere 14: 2.3 ×, 2.4 ×, 2.5 × or 2.6 ×. Metafemur length/width: 3.2 ×, 3.3 × or 3.4 ×. Mesoscutellar disc: mostly smooth with few, scattered punctures near margins (Fig. 36). Number of pits in scutoscutellar sulcus: usually 12 or less, ocasionally reaching up to 14 pits. Propodeum background sculpture: anterior 0.2–0.4 × with rather dull puntures; posterior 0.6–0.8 × mostly sculptured, with mix of small puntures and carinae (mostly radiating from strong, longitudinal median carina) (Fig. 35). Mediotergite 1 width at anterior margin/width at posterior margin: 3.1 ×, 3.2 × or 3.3 ×. Mediotergite 2 width at posterior margin/length: 3.7 ×, 3.8 ×, 3.9 × or 4.1 × (Fig. 36). Ovipositor sheaths length/metatibial length: 1.1 × (Fig. 32).
Male. As female.
Iconella jayjayrodriguezae. 30 Habitus, lateral view 31 Fore wing 32 Ovipositor sheaths 33 Hind leg 34 Head, dorsal view 35 Propodeum 36 Meso- and metasoma (partially), dorsal view.
We analyzed eight 650–658 bp barcodes for this species, one from Mexico and seven from Costa Rica.
Host: An undescribed species of Spilomelinae (Crambidae) with provisional name “spiloBioLep01 BioLep414” in the ACG database (http://janzen.sas.upenn.edu/caterpillars/database.lasso). Caterpillar collected while feeding on the foliage of Lepidoploa salzmannii and Lepidoploa tortuosa (Asteraceae).
Costa Rica (ACG), and Mexico (Chiapas). In ACG it has been reared from four rainforest localities between 420–500m, 2-10 km apart, in six different months.
Although morphologically similar to Iconella andydeansi, Iconella jayjayrodriguezae is known from two very widely separated places in Central America, and the barcoding sequences of the two species differ by 8.7% (57 bp).
This species is named in honor of Josephine J. Rodriguez (National Center for Ecological Analysis and Synthesis, University of California, Santa Barbara, United States) in recognition of her outstanding enthusiasm for studying the taxonomy and biology of the microgastrine wasps of ACG (e.g.,
We gratefully acknowledge the support of the Guanacaste Dry Forest Conservation Fund, the Wege Foundation, the International Conservation Fund of Canada, the JRS Biodiversity Foundation, Jessie Hill, and the University of Pennsylvania for funding portions of the research. JFT gratefully acknowledge the support and sponsoring of the Lucid team (http://www.lucidcentral.com/). This study was also supported by NSF DEB 0515699 to DHJ and by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant to MAS. Laboratory analyses of these sequences were funded by the Government of Canada through Genome Canada and the Ontario Genomics Institute (2008-0GI-ICI-03). Kees van Achterberg (Naturalis, Leiden) and an anonymous reviewer helped to improve the quality of the manuscript.
Lucid key to the New World species of the parasitoid wasp Iconella (Hymenoptera, Braconidae, Microgastrinae). (doi: 10.3897/zookeys.321.5160.app) File format: Lucid Key Data (lk4).