(C) 2013 Prosanta Chakrabarty. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Chakrabarty P, Warren M, Page LM, Baldwin CC (2013) GenSeq: An updated nomenclature and ranking for genetic sequences from type and non-type sources. ZooKeys 346: 29–41. doi: 10.3897/zookeys.346.5753
An improved and expanded nomenclature for genetic sequences is introduced that corresponds with a ranking of the reliability of the taxonomic identification of the source specimens. This nomenclature is an advancement of the “Genetypes” naming system, which some have been reluctant to adopt because of the use of the “type” suffix in the terminology. In the new nomenclature, genetic sequences are labeled “genseq, ” followed by a reliability ranking (e.g., 1 if the sequence is from a primary type), followed by the name of the genes from which the sequences were derived (e.g., genseq-1 16S, COI). The numbered suffix provides an indication of the likely reliability of taxonomic identification of the voucher. Included in this ranking system, in descending order of taxonomic reliability, are the following: sequences from primary types – “genseq-1, ” secondary types – “genseq-2, ” collection-vouchered topotypes – “genseq-3, ” collection-vouchered non-types – “genseq-4, ” and non-types that lack specimen vouchers but have photo vouchers – “genseq-5.” To demonstrate use of the new nomenclature, we review recently published new-species descriptions in the ichthyological literature that include DNA data and apply the GenSeq nomenclature to sequences referenced in those publications. We encourage authors to adopt the GenSeq nomenclature (note capital “G” and “S” when referring to the nomenclatural program) to provide a searchable tag (e.g., “genseq”; note lowercase “g” and “s” when referring to sequences) for genetic sequences from types and other vouchered specimens. Use of the new nomenclature and ranking system will improve integration of molecular phylogenetics and biological taxonomy and enhance the ability of researchers to assess the reliability of sequence data. We further encourage authors to update sequence information on databases such as GenBank whenever nomenclatural changes are made.
GenBank, genetics, molecular phylogenetics, systematics, taxonomy
The use of genetic sequences has been increasing with each passing year (
Although an institutional catalog number for the specimen (the ‘voucher’) from which a sequence is obtained is often requested when a sequence is submitted, it is not obligatory. Most sequences available on GenBank lack this information. Sequences available from databases such as GenBank have little reference to the source of the genetic materials other than the title and authors of the original publication. Unfortunately, original publications also often lack information about the original specimens necessary to validate their identification. To remedy this deficiency and to remind researchers about the importance of providing and accurately identifying DNA voucher specimens, we propose a new genetic nomenclature based on a ranking of various source specimens. We also suggest various ways in which the link between specimens and genetic sequences can be made more transparent.
The GenSeq nomenclature combines the term “genseq” with a hyphen and a number from 1 to 5 reflecting the reliability ranking we provide in Table 1. Sequences from primary type specimens are referred to as genseq-1, with the 1 reflecting the highest reliability rank. In addition to these terms, the gene region(s) should be reported with the GenSeq reference; for example, “genseq-1 mitogenome, genseq-2 16S, ND2”, or “genseq-5 UCE chr11_2436”. The hyphen between “genseq” and the number must be included to allow search engines such as Google to search the entire text string because these searches treat hyphens as spaces. Note that in reference to the nomenclatural program we use capital letter “G” and “S” (e.g., GenSeq), and all lowercase letters (e.g., genseq-1) when referring to specific sequences.
Ranking Sequence Reliability. Ranking of source materials of genetic sequences based on reliability of taxonomic identification. Examples of the source material are listed in the third column with the last column providing the corresponding GenSeq nomenclature.
| Reliability Ranking | Source Materials | Examples | Corresponding GenSeq Nomenclature |
|---|---|---|---|
| Highest 1ST | Primary Types | Holotype, Lectotype, Syntype, Isosyntype, Neotype, Isotype | genseq-1 |
| 2nd | Secondary Types | Paratype, Paralectotypes, etc. | genseq-2 |
| 3rd | Topotypes (vouchered), or non-type specimens listed in original description or redescription | Topotype, Non-type specimen listed in original description or redescription | genseq-3 |
| 4th | Collections-vouchered non-types (not from original description or redescription) | Vouchered specimen | genseq-4 |
| 5th | Photo voucher only | No specimen voucher but photo voucher available | genseq-5 |
| Lowest | No voucher | Non-vouchered | No classification |
Many GenSeqs can be created from a single specimen and can be from a single gene fragment, multiple fragments, or an entire genome; for instance, “genseq-2 COI” and “genseq-2 COII, ND2, ” could be added from the same paratype voucher at a later date, as could GenSeqs from other specimens of this species (e.g., other paratypes, the holotype) from which DNA was extracted. This nomenclature is simply a flag to alert molecular biologists and taxonomists that sequences are available from type specimens and some confidently identified non-types (see below). We suggest that researchers preferentially use these sequences in molecular evolutionary analyses, as doing so should bolster confidence in conclusions based on the sequence data (Fig. 1). Tabulating the GenSeq nomenclature with GenBank numbers and catalog numbers for vouchers will provide subsequent workers with easy access to this information (Table 2). We suggest that authors report the GenSeq in either the Systematic Accounts section or Materials and Methods section of a manuscript. For these sections, an additional example of how the GenSeq nomenclature could be reported for type specimens is: “One of the paratypes (USNM 139024) was sequenced (GenBank accession number JZ254935) and therefore constitutes a genseq-2 cytochrome b.”
Example Reporting Table. Examples of how links between genetic sequences and vouchers in institutional collections could be displayed as a table in publications reporting new sequences.
| Species | Specimen Catalog # | GenBank # | GenSeq Nomenclature | |
|---|---|---|---|---|
| COI | ND1 | |||
| Typhleotris mararybe | LSUMZ 13636 (a holotype) | HM590594 | HM590606 | genseq-1 COI, ND1 |
| Paretroplus tsimoly | AMNH 229558 (a paratype) | JZ590596 | NA | genseq-2 COI |
| Nandopsis haitiensis | UMMZ 236321 (a topotype) | BK590595 | BK590607 | genseq-3 COI, ND1 |
| Halieutichthys intermedius | FMNH 96353 (a non-type specimen voucher) | AY722169 | AY722306 | genseq-4 COI, ND1 |
| Equulites absconditus | NMNH 12345PV2 (a photo voucher) | NA | BG34621 | genseq-5 ND1 |
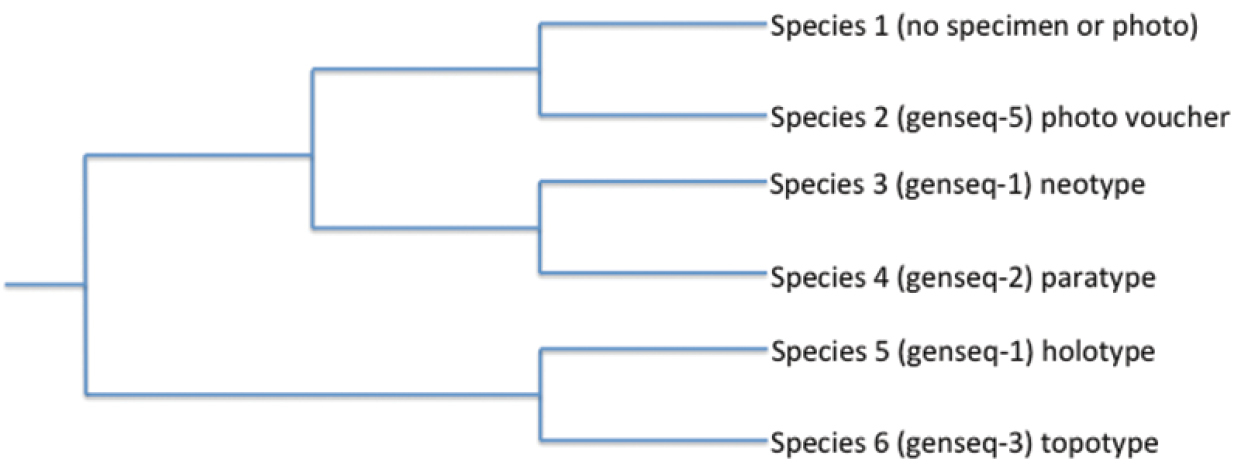
Example of how the GenSeq ranking system of sequences from various sources (Table 1) can be used to assess the trustworthiness of data used to reconstruct phylogenetic relationships. The rankings (the # in the “genseq-#”) make it clear that the relationship recovered between Species 3 and Species 4, from primary and secondary types, should be trustworthy because the taxonomic identifications of the voucher specimens are considered to be highly reliable. In contrast, the recovered sister relationship between Species 1 and Species 2 may be less trustworthy because of the weak reliability rankings of these sequences from non-types. Species 1 lacks both a specimen or photo voucher and therefore does not have a GenSeq ranking.
The “genseq” suffix will flag genetic sequences in any manuscript so that authors can better assess the reliability of the taxonomic identification of specimens used to obtain those sequences. A sequence from a holotype provides the highest reliability of taxonomic identification and is therefore awarded the highest ranking in the system (genseq-1; Table 1). Although secondary types are not name-bearing specimens, in most cases identifications of secondary types are at least as trustworthy, and generally more so, than those from non-types, which may have been identified by non-experts or that may lack vouchered specimens. For this reason, sequences from secondary types are given the second highest ranking (genseq-2). Sequences from vouchered topotypes—individuals collected from the type locality of a species—are given the third highest ranking as are individuals not designated as types in the original description but are identified as belonging to the new taxon in that same manuscript (both genseq-3).
The genseq-3, -4, and -5 categories (Table 1) represent a departure and expansion from the previous Genetypes nomenclature. Because most species included in molecular analyses will not have type specimens available for sequencing, it is important to expand the genetic nomenclature and ranking to forecast the reliability of other sources of sequences. A sequence from a vouchered specimen that was not included in the type series but that is identified in the original description of the species as a member of the new taxon should be flagged as “genseq-3.” The “3” is used as a suffix in this case again to reflect that it belongs to the third highest category of reliability (Table 1). Sequences from vouchered specimens from a redescription of a resurrected species should also be flagged “genseq-3.” Sequences from non-type specimens that are not mentioned in the original description, or redescription, but that are confidently identified by an expert should be flagged “genseq-4” with the “4” suffix again reflecting the 4th highest ranking of reliability. Finally, “genseq-5” is a flag for sequences that lack any specimen voucher but that have a well-documented and publically available photo voucher. A photo voucher is not ideal but is necessary when the organism is still alive, highly endangered, extremely large, or extremely small (e.g., a larva where the entire sample must be used to obtain sufficient DNA).
In cases where the term “genseq-[3, 4, or 5]” category is used to identify sequences that are from a non-type specimen, the specimen should be identified by an authority and deposited in a reputable natural history collection. Both specimen and photo vouchers would be included in the “hologenophore” category of biological vouchers described by
To better understand how sequences from type specimens are currently being reported in the scientific literature, we conducted a survey of recent publications describing new species of fishes that included DNA data. Fishes were chosen in part because we are ichthyologists, but also because fishes are described at a rate that is the highest among vertebrates (
We used a Google Scholar search (www.scholar.google.com) to find papers published between 2010 and 2011 using the search term “new fish species DNA”. The retrieved papers were reviewed for any mention of sequences obtained from a holotype or paratypes. Because many papers did not link the GenBank #’s with the voucher’s catalog number, we conducted a corresponding search on GenBank (http://www.ncbi.nlm.nih.gov/) to determine whether those catalog numbers were reported there.
Each species recovered from the Google Scholar search was entered into the GenBank “nucleotide” search-engine field. If a catalog number of a holotype or paratype(s) was recovered in either the original paper or with GenBank, it was reported in Table 3. In cases where the link between voucher and sequence was unclear, authors of the descriptions were contacted to clarify the link. We only report examples where the genetic sequences could be positively linked to the catalog number of the holotype or paratypes. (Other forms of GenSeqs were not searched for in this initial case study.)
Results of Search for Sequences from Types. GenSeq nomenclature applied to DNA sequences of fishes described from 2010–2011. The data were mined from GenBank and Google Scholar. Institutional abbreviations follow Sabaj-Perez (2012) except GSDNA which is the Natural History Gallery of Casalina. ◆ indicates that the catalog number of the voucher was reported with the genetic sequences in the published original description. ○ indicates that the catalog number of the voucher was recorded in GenBank with the sequences. Lack of either symbol indicates that the authors were e-mailed to find the link between a voucher and a sequence.
| Species (Group) | Citation | Type of type | Voucher catalog | GenBank # | GenSeq |
|---|---|---|---|---|---|
| Bathygobius antilliensis (Teleostei: Gobiidae) | Holotype | AMNH 251650 | HM748393 | genseq-1 COI | |
| Paratypes | 16 examples | 16 examples | genseq-2 COI | ||
| Bathygobius geminatus (Teleostei: Gobiidae) | Holotype | USNM 398105 | HM748368 | genseq-1 COI | |
| Paratypes | AMNH 251648 | HM748389 | genseq-2 COI | ||
| USNM 398102 | HM748357 | ||||
| USNM 398103 | HM748365 | ||||
| USNM 398106 | HM748369 | ||||
| USNM 398107 | HM748373 | ||||
| USNM 398109 | HM748375 | ||||
| USNM 398112 | HM748379 | ||||
| Chimaera opalescens (Chondrichthyes: Holocephali) | Paratypes | MNHN 2007-1557 | GU244532 | genseq-2 COI | |
| MNHN:2007-1555 | GU244533 | ||||
| MNHN:2007-1567 | GU244534 | ||||
| MNHN:2007-1579 | GU244531 | ||||
| Callopanchax sidibei (Nothobranchiidae: Epiplateinae) | Holotype | ZFMK 41613 | GU553012 | genseq-1 16S | |
| Hypsolebias guanambi (Cyprinodontiformes: Rivulidae) | Paratypes | UFRJ 6782.1 | HQ833483 | genseq-2 CytB | |
| UFRJ 6782.2 | HQ833484 | ||||
| UFRJ 6782.3 | HQ833485 | ||||
| UFRJ 6782.4 | HQ833486 | ||||
| Petrocephalus similis (Osteoglossomorpha: Mormyridae) | Holotype | CU 95318 | JF438961 | genseq-1 CytB | |
| Paratypes | CU 93218.1 | JF438960 | genseq-2 CytB | ||
| CU 93218.2 | JF438962 | ||||
| CU 93218.3 | JF438964 | ||||
| CU 93218.4 | JF438963 | ||||
| CU 93219 | JF438965 | ||||
| Starksia sangreyae (Teleostei: Labrisomidae) | Holotype | USNM 398932 | HQ600872 | genseq-1 COI | |
| Paratypes | USNM 398939 | HQ600865 | genseq-2 COI | ||
| USNM 398933 | HQ600873 | ||||
| USNM 398936 | HQ600868 | ||||
| USNM 398934 | HQ600875 | ||||
| USNM 398935 | HQ600874 | ||||
| USNM 398938 | HQ600866 | ||||
| USNM 398940 | HQ600864 | ||||
| Starksia springeri (Teleostei: Labrisomidae) | Paratypes | USNM 399658 | HQ600878 | genseq-2 COI | |
| USNM 399659 | HQ600876 | ||||
| Starksia weigti (Teleostei: Labrisomidae) | Paratypes | USNM 399649 | HQ600886 | genseq-2 COI | |
| USNM 399653 | HQ600934 | ||||
| USNM 399652 | HQ600935 | ||||
| USNM 399651 | HQ600936 | ||||
| USNM 399656 | HQ600927 | ||||
| USNM 399655 | HQ600932 | ||||
| Starksia williamsi (Teleostei: Labrisomidae) | Paratypes | USNM 397396 | HQ543039 | genseq-2 COI | |
| Starksia robertsoni (Teleostei: Labrisomidae) | Paratypes | USNM 399909 | HQ600961 | genseq-2 COI | |
| USNM 399911 | HQ600960 | ||||
| Starksia greenfieldi (Teleostei: Labrisomidae) | Paratypes, Topotypes | USNM 398922 | HQ600924 | genseq-2 COI, genseq-3 COI | |
| USNM 398921 | HQ600925 | ||||
| USNM 398920 | HQ600947 | ||||
| Esox flaviae (Esociformes, Esocidae) | Holotype | GSDNA1 | HM563688.1 (COI) | genseq-1 COI, CytB | |
| JN190460 (cytB) | |||||
| Milyeringa brooksi (Teleostei: Gobiiformes) | Paratype | LSUMZ 13637 | HM590607 (ND2), HM590601 (cytB), HM590595 (COI) | genseq-2 ND2, CytB, COI | |
| Leptoderma macrophthalmum (Otocephala: Alepocephalidae) | Holotype | ZMUB 19686 | AP011500 | genseq-1 mitogenome | |
| Sparisoma rocha (Actinopterygii: Labridae) | Paratype | ZUEC 6349 | GU985520 (16S) | genseq-2 16S, 12S | |
| GU985521 (12S) | |||||
| Halichoeres rubrovirens (Perciformes: Labridae) | Paratype | CIUFES 0317 | GU938858 | genseq-2 CytB | |
| CIUFES 1279 | GU938859 | ||||
| CIUFES 1474 | GU938860 | ||||
| CIUFES 1475 | GU938861 | ||||
| USNM 397005 | GU938862 | ||||
| Betadevario ramachandrani (Cyprinidae: Danioninae) | Paratype | NRM 57780 | GU327623 (cytB), GU327622 (Rho) | genseq-2 CytB, rhodopsin | |
| Crenicichla hu (Teleostei: Cichlidae) | Paratype | MACN-ict 9430.1 | GQ328038 (trnQ, trnM, ND2, trnW, trnA) | genseq-2 trnQ, trnM, ND2, trnW, trnA | |
| MACN-ict 9430.2 | GQ328039 (trnQ, trnM, ND2, trnW, trnA) |
The Google Scholar search produced 47 publications from 2010 and 2011 that included descriptions of new species of fishes and used sequence data. Only 13 of those papers indicated that sequences were derived from a type or non-type specimen (Table 3). Of the remaining 34 publications there was either no clear link between catalog numbers of vouchers and sequences (even after a query e-mail was sent to a corresponding author), or, rarely, it was made clear in the paper that no types were among those sequenced.
Of the 13 publications in Table 3, only three reported the catalog number of the type specimens along with the GenBank #’s, both in the manuscript and on GenBank. Six others reported both numbers only in the paper, and two reported the catalog number solely on GenBank. The two remaining papers (of the 13) were verified to have sequences from a primary or secondary type only after a query e-mail to the corresponding author. These authors did not supply the catalog numbers of the voucher specimens from which GenBank sequences were obtained either in their manuscript or on GenBank.
This is not a complete list of descriptions of new species of fishes with genetic sequences from type specimens. There are likely some publications that were not found via GoogleScholar or that would have been found in other search engines because of the nature of the scripts used in those searches. This search on descriptions of new species of fishes using sequence data is only a rough proxy for other groups of organisms.
GenSeq is a nomenclatural label for sequence data from confidently identified vouchered specimens. By explicitly flagging gene sequences from type materials, the new nomenclature will enable researchers to utilize sequences from the best-identified specimens when available. In particular, “genseq-1” and “genseq-2” flags will highlight sequences (see Table 1) from GenBank that, due to their direct link to primary and secondary type specimens, will be more credible than sequences from specimens with less certain identifications. Type materials remain essential for taxonomic comparisons, but sequence data from type materials have not been fully incorporated into these comparisons (see references in
The burden of linking specimens (even type specimens) to sequences from the publications of others is one reason for the creation of this expanded nomenclature. Authors often do not provide a clear link between voucher specimens and the sequences obtained from them. Presumably authors publishing on taxonomy and molecular phylogenetics would be much better at providing a clear link between the two, but as made evident from the results of this study, these authors often fail to make this link. Unfortunately, many authors simply make a statement similar to the following: “the sequences obtained from this study were given the GenBank #’s XX12428-XX12531, ” which tells no one which sequence belongs to which specimen (or even which species). These data are even more poorly reported on GenBank, where few researchers provide catalog numbers of the vouchers from which the sequences were obtained. A clear link between the specimens’ catalog number and the sequences’ GenBank #’s should either be made in a table (as in Table 2), or the voucher’s catalog numbers should be listed in GenBank with the genetic sequences. Ideally, both tasks should be done to maximize transparency.
The GenSeq nomenclature also incorporates sequences from non-type materials because many species will never have their type specimens sequenced. This could be because some collections will not want the morphological integrity of the type specimen to be diminished by the removal of a subsample for DNA extraction, or because the specimen has been fixed in formalin (as is the case for most fish, reptile, and amphibian specimens), or by some other preservation method and will no longer yield sufficient amounts of DNA. An example of how one of the non-type flags (viz., genseq-3, genseq-4, genseq-5) can be used is in cases of a taxonomic treatment in which a formerly synonymized species is resurrected and there is not a type specimen that will yield DNA. Sequences from a fresh specimen for the resurrected species should be flagged as “genseq-3.” Because the identification of the voucher is tied to the work of a taxonomist resurrecting a species (i.e., the first reviser), other researchers should have high confidence in its correct identification. Although not type material, such specimens and sequences from them should be regarded as highly likely to be correctly identified.
A Google Scholar search of the usage of the former Genetypes nomenclature, revealed 24 citations from 2010–2013 for sponges, fungi, fishes, amphibians, birds, mammals, and insects. Without a search term (in this case Genetypes), finding sequences that are derived from type specimens requires reading original publications or looking up sequences in a database. As our results indicate, authors are often inconsistent in how this information is reported, if they choose to report it at all. The benefits of having a search term like “genseq-#” embedded in a manuscript can be demonstrated by doing a simple Google Scholar search on a similar label, such as “holotype;” such a search can be rendered even more specific by adding a scientific name, e.g., “holotype Typhleotris mararybe.” Our new genetic terminology will enable researchers to conduct searches such as “genseq-# + Genus species, ” which will help them locate genetic sequences from well-documented, and likely properly identified, vouchered specimens.
Ultimately, the GenSeq approach will benefit all forms of taxonomic research as molecular phylogenetics becomes integrated with taxonomy and as technology improves in molecular biology. We remind researchers about the importance of vouchers and reporting taxonomic changes to databases such as GenBank. Taxonomic changes, misidentifications, and other changes to sequences need to be reported before they are perpetuated erroneously in the literature. If a species has sequences on GenBank and that species is later split into two species, the taxonomy should be updated by the authors on GenBank. Without this update, the original GenBank sequences that represent the new species in the split, rather than the existing one, may be used erroneously by unsuspecting researchers.
To expand usage of GenSeq flags for genetic sequences, a summary of this nomenclature should be incorporated into the “Instruction to Authors” for taxonomic journals.
Sequence data: Manuscripts containing novel amino acid sequences (e.g. primer sequences) will only be accepted if they carry an International Nucleotide Sequence Databases (INSD) accession number from the European Biology Laboratory (EMBL), GenBank Data Libraries (GenBank) or DNA Data Bank of Japan (DDBJ). [Name of Journal] strongly recommends that authors include institutional catalog numbers for specimens preserved in collections, and information identifying sequences that are derived from type specimens (see below) when they deposit data in genetic databanks. Database [GenBank] accession [catalog] numbers should be included in the Materials and Methods section. If specimens were not vouchered (tissued specimens should be vouchered when possible!), photographs and collection locality data for tissued specimens must be provided.
A nomenclature for genetic sequences for types and confidently identified non-type specimens has been proposed by Chakrabarty et al. (2013); a sequence from a holotype is identified as genseq-1, one from a paratype is identified as genseq-2, one from a topotype is genseq-3, etc. The genetic marker(s) used should also be incorporated into the nomenclature (e.g. genseq-2 COI).
Authors who wish to report GenSeqs in a web interface (in addition to in a published manuscript and on GenBank) may choose the widely used Animal Diversity Web (ADW; http://animaldiversity.ummz.umich.edu/). The editors of this website suggest that the “Other Comments” field can be used to report GenBank links of GenSeqs (Tanya Dewey pers. comm.). For help creating an ADW page for a new taxon using the GenSeq nomenclature, please contact the first author of this paper.
In the future, we hope the GenSeq nomenclature will be widely used and eventually incorporated into GenBank and other large genetic databases. Incorporation is not currently possible with GenBank’s user-driven interface which would allow too much human error in labeling sequences (pers. com., Scott Federhen, GenBank;
We thank all the authors who were contacted and provided additional information about the sequences in Table 3. Two anonymous reviewers provided excellent suggestions that have been incorporated into this final publication. We would like to acknowledge Dr. Tanya Dewey (Animal Diversity Web) and Andy Bentley (University of Kansas) for their comments. We would like to also acknowledge the Louisiana State University Office of Strategic Initiatives 2012 Summer Research Experience for Undergraduates Program and the LSU Initiative for Maximizing Diversity for allowing MW to participate in this project. Financial support for this project was obtained from a National Science Foundation Grant to PC (DEB 0916695).