(C) 2013 Shasha Yu. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Yu S, Wang Y, Rédei D, Xie Q, Bu W (2013) Secondary structure models of 18S and 28S rRNAs of the true bugs based on complete rDNA sequences of Eurydema maracandica Oshanin, 1871 (Heteroptera, Pentatomidae). In: Popov A, Grozeva S, Simov N, Tasheva E (Eds) Advances in Hemipterology. ZooKeys 319: 363–377. doi: 10.3897/zookeys.319.4178
The sequences of 18S and 28S rDNAs have been used as molecular markers to resolve phylogenetic relationships of Heteroptera for two decades. The complete sequences of 18S rDNAs have been used in many studies, while in most studies only partial sequences of 28S rDNAs have been used due to technical difficulties of amplifying the complete lengths. In this study, we amplified the complete 18S and 28S rDNA sequences of Eurydema maracandica Oshanin, 1871, and reconstructed the secondary structure models of the corresponding rRNAs. In addition, and more importantly, all of the length variable regions of 18S rRNA were compared among 37 families of Heteroptera based on 140 sequences, and the D3 region of 28S rRNA was compared among 51 families based on 84 sequences. It was found that 8 length variable regions could potentially serve as molecular synapomorphies for some monophyletic groups. Therefore discoveries of more molecular synapomorphies for specific clades can be anticipated from amplification of complete 18S and 28S rDNAs of more representatives of Heteroptera.
rRNA, secondary structure, Heteroptera, molecular synapomorphy, Eurydema maracandica
Each cluster of rDNA in turn contains external transcribed spacer (ETC), 18S rDNA, internal transcribed spacer 1 (ITS1), 5.8S rDNA, ITS2 and 28S rDNA (
As far as the rDNAs are concerned, complete sequences of 18S rDNA have been used in various phylogenetic studies on Heteroptera (
Reconstruction of the complete secondary structure model of 28S rRNA can only be achieved by full sequencing of 28S rDNA. According to the results of comparative studies of 18S rRNAs of Insecta (
According to the currently accepted classification of Heteroptera (
The specimen was collected from Yining, Xinjiang Uyghur Autonomous Region, China (43°56'N, 81°19'E, 570m) on July 26, 2011 by Qiang Xie. In the alcohol-kept insects’ collection of Nankai University, the ID number for the voucher specimen is NKU0150177. The species was identified based on the available literature and comparative material, its identity was confirmed by P. Kment. The specimen perfectly fits the original description (
Genomic DNA was extracted from thoracic tissue of ethanol-preserved specimen. Total genomic DNA was isolated using the CTAB-based method (
The secondary structure of 18S rRNA was slightly revised from the universal model of insects (
The data underpinning the analyses reported in this paper are deposited in the Dryad Data Repository at doi: 10.5061/dryad.j8kp5
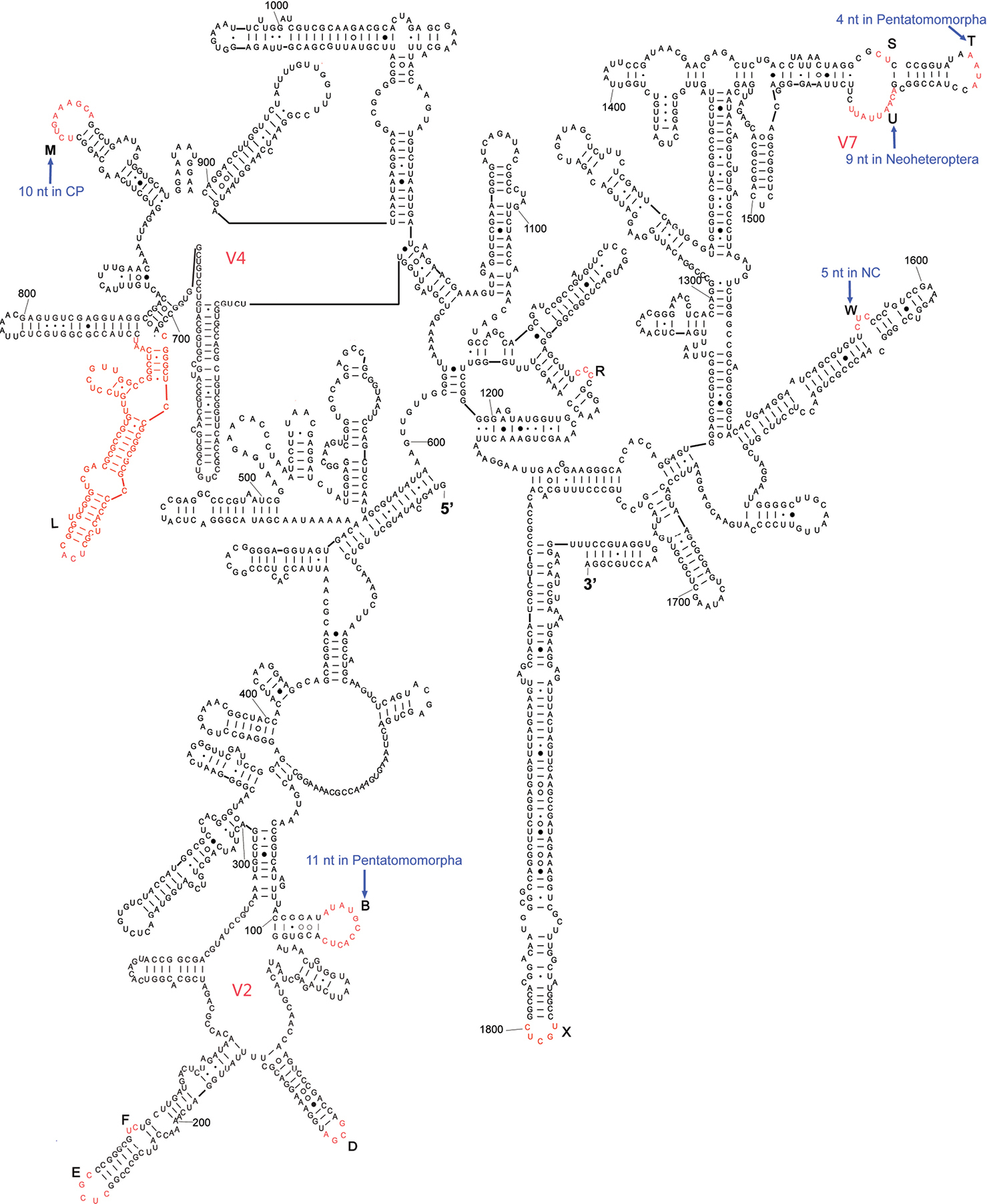
The complete sequence of 18S rDNA of Eurydema maracandica is 1, 894 bp. In the secondary structure of the corresponding rRNA (Fig. 1) the LVRs L and X regions were specifically optimized. The 18S rDNA sequence of Eurydema maracandica was aligned with other 140 orthologous sequences of 37 families in Heteroptera. The alignment result suggested that the local length variations of 18S rDNA are localized in 13 independent LVRs except for various indels. Most LVRs were restrained in three domains, which were previously named as V2, V4 and V7 (
Secondary structure model of 18S rRNA of Eurydema maracandica. The bases marked in black represent length-conservative regions, and the bases labeled as capital letters B to W in red represent 13 LVRs. CP and NC represent monophyletic groups Cimicomorpha+Pentatomomorpha and Naboidea+Cimicoidea, respectively. Base pairing is indicated as follows: standard canonical pairs by lines (G–C, A–U), wobble G:U pairs by dots (G·U), A:G or A:C pairs by open circles (A○G, A○C), and other non-canonical pairs by filled circles (e.g., A●A).
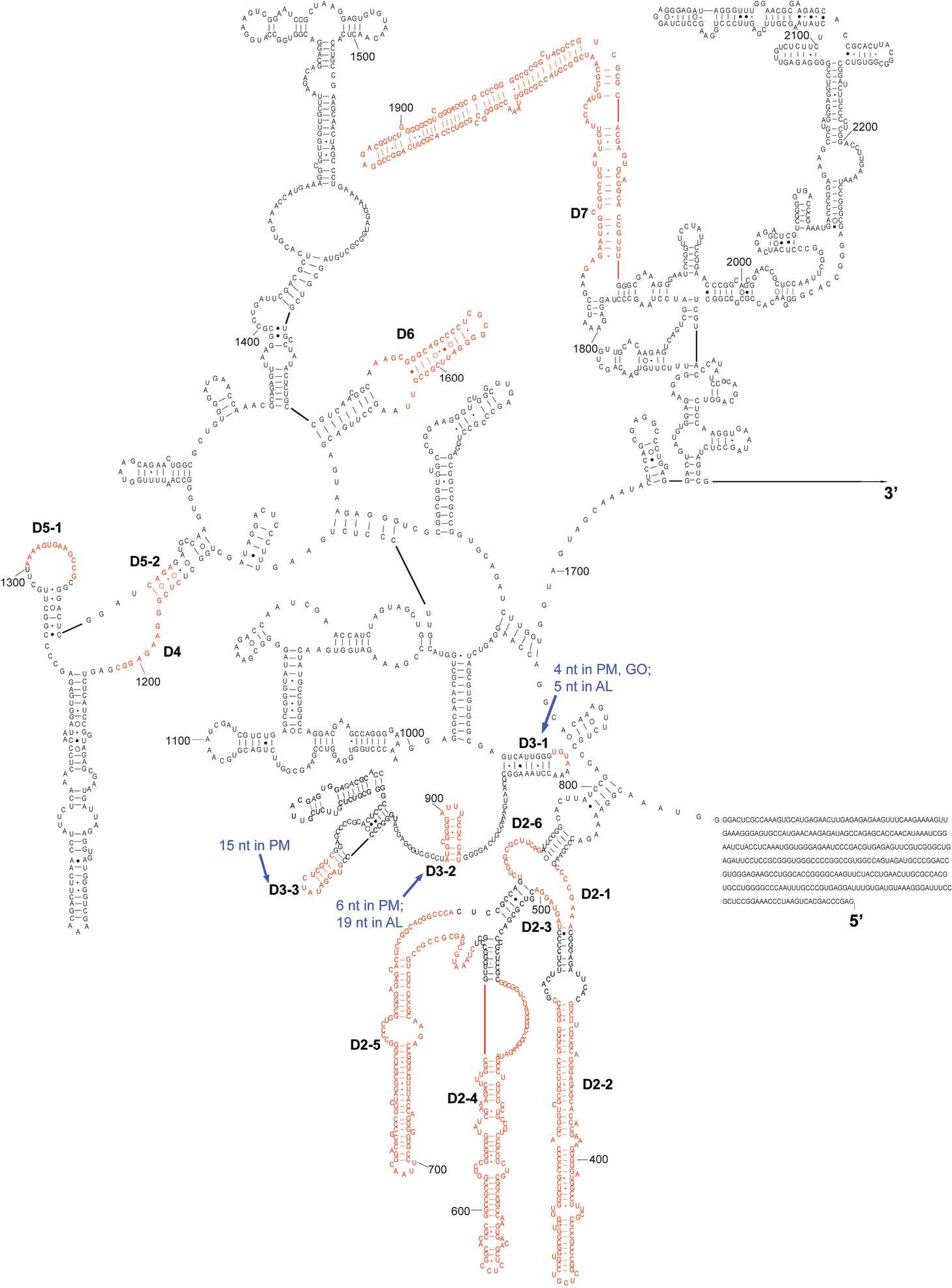
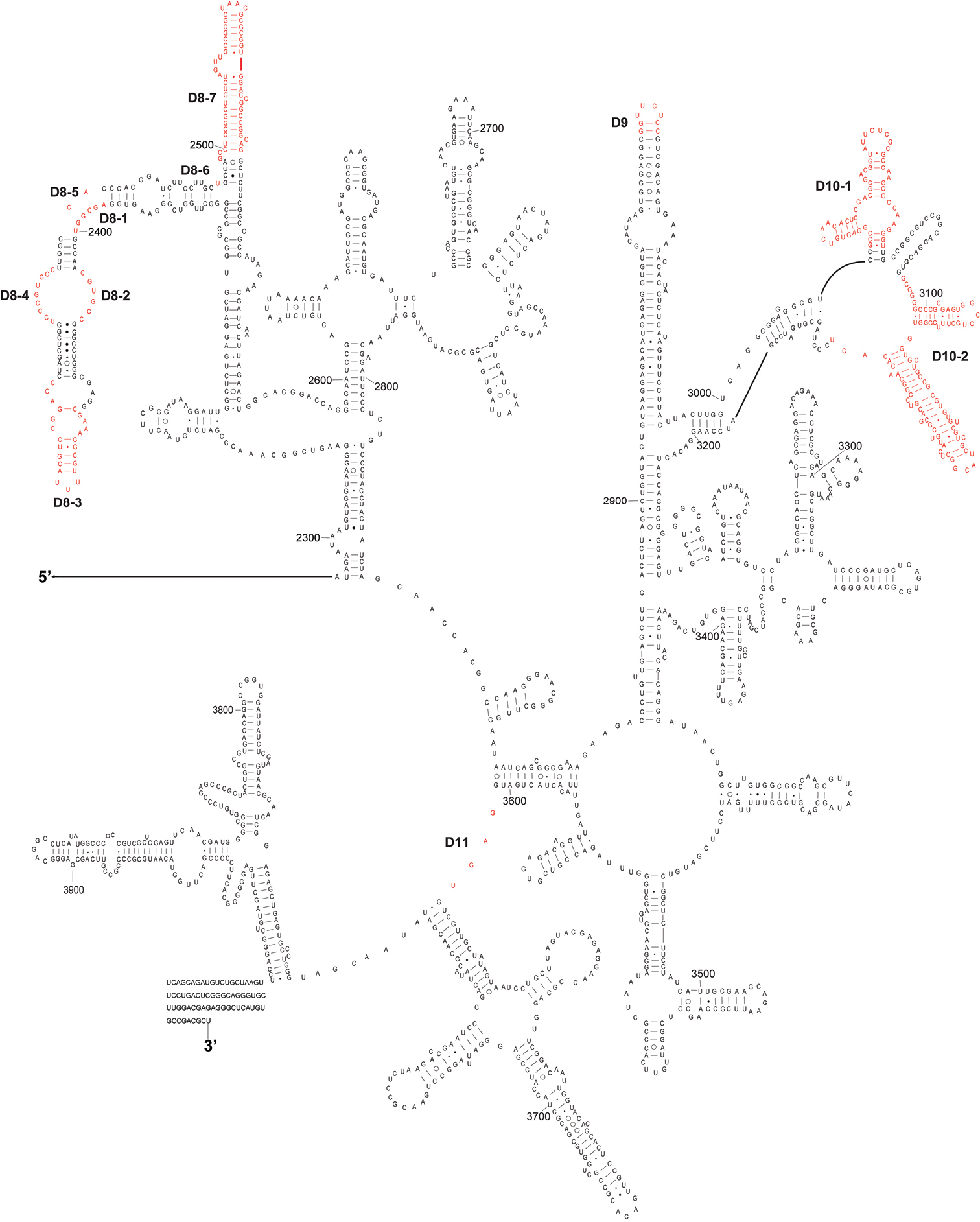
The complete sequence of 28S rDNA of Eurydema maracandica is 4, 030 bp. In the secondary structure of corresponding rRNA (Figs 2, 3), the LVRs D2, D3, D7, D8, D10, and D11 were specifically optimized. Compared with the secondary structure of the universal model of insect 28S rRNA, LVRs of heteropteran 28S rRNAs are distributed in 10 regions (D2–D11). Only D3 region of most superfamilies has corresponding data in GenBank, therefore only this region was comparatively analyzed and the results may serve as an inspiring case for the other LVRs of 28S rRNA. Alignment results suggested that in the D3 region, LVRs can be further divided into three sections (D3-1, D3-2 and D3-3), which are interspaced by several short length-conservative regions. Among these three separate LVRs, D3-2 is the most variable one in length.
The 5’-half part of secondary structure model of 28S rRNA of Eurydema maracandica. The numbers D2 to D7 represent six LVRs. PM, GO and AL represent monophyletic groups Paraphrynoveliidae+Macroveliidae, Gelastocoridae+Ochteridae and Acanthosomatidae+Lestoniidae, respectively.
The 3’-half part of secondary structure model of 28S rRNA of Eurydema maracandica. The numbers D8 to D11 represent four LVRs.
According to the above results, the local length variations of heteropteran rRNAs are moderate in comparison with the length variations of eukaryotic rRNAs (
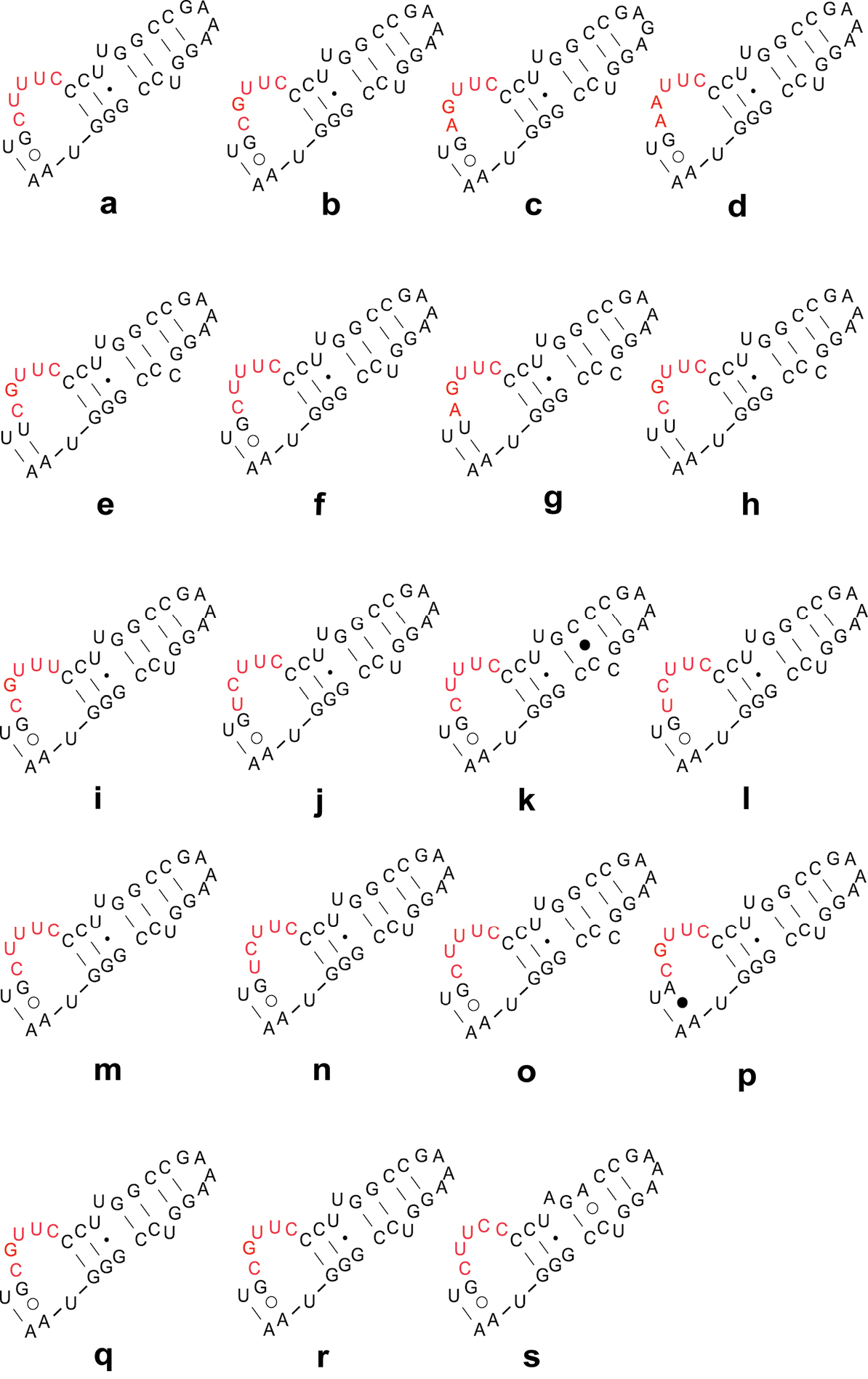
Secondary structure models of LVR W of Naboidea and Cimicoidea. These sequences are from 15 genera, 19 species of Naboidea and Cimicoidea. The species names and GenBank Accession numbers are as follow: Nabidae (a) Nabis ferus EF487300 (b) Nabis flavomarginatus GQ258424 (c) Himacerus apterus GQ258425; Lyctocoridae (d) Lyctocoris beneficus EF487298; Anthocoridae (e) Anthocoris sp. AY252319 (f) Anthocoris confusus EF487297 (g) Anthocoris montanus EF487307 (h) Tetraphleps aterrimus EF487295 (i) Amphiareus obscuriceps EF487301 (j) Orius agilis EF487296 (k) Physopleurella armata EF487308 (l) Montandoniola moraguesi EF487310 (m) Xylocoris cerealis GQ258395 (n) Bilia sp. GQ258406 (o) Buchananiella crassicornis GQ258407 (p) Lasiochilus japonicus GQ258410 (q) Lasiochilus luceonotatus GQ258411; Cimicidae (r) Cimex lectularius GQ258396; Curaliidae (s) Curalium cronini EU683128.
Monophyletic groups within Heteroptera with some LVRs serving as potential synapomorphies.
| Monophyletic group | Reference | LVR (number of sequences examined) | |
| 18S rDNA | 28S rDNA | ||
| Neoheteroptera | 9nt U (131) | ||
| Paraphrynoveliidae+Macroveliidae | 4nt D3-1 (3) | ||
| 6nt D3-2 (3) | |||
| 15nt D3-3 (3) | |||
| Gelastocoridae+Ochteridae | 4nt D3-1 (4) | ||
| Cimicomorpha+Pentatomomorpha | 10nt M (117) | ||
| Naboidea+Cimicoidea | 5nt W (36) | ||
| Pentatomomorpha | |||
| 11nt B (39) | |||
| 4nt T (37) | |||
| Acanthosomatidae+Lestoniidae | 5nt D3-1 (8) | ||
| 19nt D3-2 (8) | |||
Note: Detailed information see Supplementary Files 4 and 5.
We are grateful to Dr. Petr Kment (National Museum, Prague) for confirming the identity of Eurydema maracandicum. We also thank Dr. Zhong-Hua Fan (Nankai University) for helpful advices. This work was supported by the National Science Foundation of China (grant numbers 31222051, J1210005, J0930005, 30970350), China Postdoctoral Science Foundation (grant number 20110490769) and Science Foundation of Tianjin (grant number 11JCYBJC08100).
Primer pairs and the annealing temperatures for amplifying rDNAs of Eurydema maracandica. (doi: 10.3897/zookeys.319.4178.app1) File format: Mircrosoft Word Document (doc).
Explanation note: List of primer pairs and annealing temperatures.
Alignment of 18S rDNA sequence of Heteroptera. (doi: 10.3897/zookeys.319.4178.app2) File format: DNA and Protein Sequence Alignment (fas).
Explanation note: The alignment of Eurydema maracandica 18S rDNA sequence with other 140 orthologous sequences of Heteroptera.
Alignment of D3 region in 28S rDNA sequence of Heteroptera. (doi: 10.3897/zookeys.319.4178.app3) File format: DNA and Protein Sequence Alignment (fas).
Explanation note: The alignment of Eurydema maracandica D3 region in 28S rDNA sequence with other 84 orthologous sequences of Heteroptera.
The summarization of length variation in 18S rDNA sequence of Heteroptera. (doi: 10.3897/zookeys.319.4178.app4) File format: Microsoft Excel Worksheed (xls).
Explanation note: The length of 12 LVRs in all currently known 18S rDNA sequences of Heteroptera.
The summarization of length variation in D3 region of 28S rDNA sequence of Heteroptera. (doi: 10.3897/zookeys.319.4178.app5) File format: Microsoft Excel Worksheed (xls).
Explanation note: The length of 3 LVRs in all currently known sequences of D3 region of 28S rDNA of Heteroptera.