Citation: Cong Q, Grishin NV (2014) A new Hermeuptychia (Lepidoptera, Nymphalidae, Satyrinae) is sympatric and synchronic with H. sosybius in southeast US coastal plains, while another new Hermeuptychia species – not hermes – inhabits south Texas and northeast Mexico. ZooKeys 379: 43–91. doi: 10.3897/zookeys.379.6394
Hermeuptychia intricata Grishin, sp. n. is described from the Brazos Bend State Park in Texas, United States, where it flies synchronously with Hermeuptychia sosybius (Fabricius, 1793). The two species differ strongly in both male and female genitalia and exhibit 3.5% difference in the COI barcode sequence of mitochondrial DNA. Setting such significant genitalic and genotypic differences aside, we were not able to find reliable wing pattern characters to tell a difference between the two species. This superficial similarity may explain why H. intricata, only distantly related to H. sosybius, has remained unnoticed until now, despite being widely distributed in the coastal plains from South Carolina to Texas, USA (and possibly to Costa Rica). Obscuring the presence of a cryptic species even further, wing patterns are variable in both butterflies and ventral eyespots vary from large to almost absent. To avoid confusion with the new species, neotype for Papilio sosybius Fabricius, 1793, a common butterfly that occurs across northeast US, is designated from Savannah, Georgia, USA. It secures the universally accepted traditional usage of this name. Furthermore, we find that DNA barcodes of Hermeuptychia specimens from the US, even those from extreme south Texas, are at least 4% different from those of H. hermes (Fabricius, 1775)—type locality Brazil: Rio de Janeiro—and suggest that the name H. hermes should not be used for USA populations, but rather reserved for the South American species. This conclusion is further supported by comparison of male genitalia. However, facies, genitalia and 2.1% different DNA barcodes set Hermeuptychia populations in the lower Rio Grande Valley of Texas apart from H. sosybius. These southern populations, also found in northeastern Mexico, are described here as Hermeuptychia hermybius Grishin, sp. n. (type locality Texas: Cameron County). While being phylogenetically closer to H. sosybius than to any other Hermeuptychia species, H. hermybius can usually be recognized by wing patterns, such as the size of eyespots and the shape of brown lines on hindwing. “Intricate Satyr” and “South Texas Satyr” are proposed as the English names for H. intricata and H. hermybius, respectively.
Biodiversity, cryptic species, DNA barcodes, neotropical, satyr, Hermeuptychia gisella, Hermeuptychia cucullina, Hermeuptychia sosybius kappeli, female genitalia
What could be more exciting than a discovery of a new butterfly species? Perhaps the discovery of a butterfly species in the US that was long overlooked, completely unexpected, and has closest named relatives far away in Bolivia and Brazil. These finds may not be easy to come by, because most of such species are cryptic and appear superficially similar to their more common and well-known relatives. However, DNA-based techniques introduced in taxonomy during the last few decades offer viable tools to facilitate discovery of cryptic species (
The genus Hermeuptychia was proposed by
Data for specimens with DNA sequences used in this study.
| Species | Voucher | GenBank | Locality | Date | Collector |
|---|---|---|---|---|---|
| Hermeuptychia sosybius | NVG-696 | KJ025523 | OK: Atoka Co., 13 air mi E of Atoka, 34.41186, -95.91044, 225 m | 29-Aug-2009 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1632 | KJ025524 | TX: Lamar Co., 11.5 air mi NW of Paris, FM1499 @ Sanders Cr., 140 m | 25-Apr-1998 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1630 | KJ025525 | TX: Marion Co., nr. Carter L., 50 m | 28-Sep-1996 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1633 | KJ025526 | TX: Marion Co., nr. Carter L., 50 m | 29-Sep-1996 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1606 | KJ025527 | TX: Wise Co., LBJ National Grassland, 300 m | 3-Aug-1998 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-783 | KJ025528 | TX: Tyler Co., John H. Kirby SF, 40 m | 19-Mar-2011 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-784 | KJ025529 | TX: Tyler Co., John H. Kirby SF, 40 m | 19-Mar-2011 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-785 | KJ025530 | TX: Tyler Co., John H. Kirby SF, 40 m | 19-Mar-2011 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-786 | KJ025531 | TX: Tyler Co., John H. Kirby SF, 40 m | 19-Mar-2011 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1537 | KJ025532 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1538 | KJ025533 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1539 | KJ025534 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1540 | KJ025535 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1542 | KJ025536 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1543 | KJ025537 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1544 | KJ025538 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1545 | KJ025539 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1546 | KJ025540 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1547 | KJ025541 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1549 | KJ025542 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1550 | KJ025543 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1552 | KJ025544 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1553 | KJ025545 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1557 | KJ025546 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1559 | KJ025547 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1561 | KJ025548 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1562 | KJ025549 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1564 | KJ025550 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1566 | KJ025551 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | NVG-1567 | KJ025552 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia sosybius | 13385H04 | KJ025553 | TX: Comal Co., New Braunfels | 3-Oct-1981 | |
| Hermeuptychia sosybius | 13385H11 | KJ025554 | TX: Williamson Co., Florence | 3-Sep-1974 | J. Parkinson |
| Hermeuptychia sosybius | 13385H03 | KJ025555 | TX: Uvalde Co., Utopia | 10-Jun-1992 | D. E. Gaskin & EAL |
| Hermeuptychia sosybius | 13385H05 | KJ025556 | TX: Uvalde Co., Utopia | {9-23}-Sep-1994 | D. E. Gaskin & EAL |
| Hermeuptychia sosybius | 13385H06 | KJ025557 | TX: Uvalde Co., Utopia | {9-23}-Sep-1994 | D. E. Gaskin & EAL |
| Hermeuptychia sosybius | 13385H07 | KJ025558 | TX: Uvalde Co., Utopia | {13-22}-Apr-1995 | D. E. Gaskin |
| Hermeuptychia sosybius | 13385H08 | KJ025559 | TX: Uvalde Co., Utopia | {13-22}-Apr-1995 | D. E. Gaskin |
| Hermeuptychia sosybius | 13385G12 | KJ025560 | FL: Highlands Co., Lake Placid, Archbold Biological Station | 17-Feb-1985 | D. C. Ferguson |
| Hermeuptychia sosybius* | 13386A07 | KJ025561 | GA: Chatham Co., Savannah | 28-Jul-1958 | Coll. Gordon B. Small |
| Hermeuptychia sosybius** | NVG-1845 | KJ025562 | FL: N of L. Okeechobee | 29-Mar-1983 | Ralf H. Anken |
| Hermeuptychia sosybius | 15609E04 | KJ025563 | FL: Pinellas Co., St. Petersburg | 3-Nov-1938 | H. E. Wilford |
| Hermeuptychia sosybius | 13385G10 | KJ025564 | SC: Clarendon Co. | Aug-1909 | |
| Hermeuptychia sosybius | 13385H09 | KJ025565 | TX: Bastrop Co., Bastrop | prior to 1896 | Collection of O. Meske |
| Hermeuptychia sosybius | 13386A01 | KJ025566 | TX: Guadalupe Co., Seguin | 26-Oct-1905 | F. C. Pratt |
| Hermeuptychia sosybius | 13386A04 | KJ025567 | LA: Jackson Parish, Jonesboro | 4-Jun-1920 | G. W. Rawson |
| Hermeuptychia sosybius | 13386A06 | KJ025568 | LA: Jefferson Parish, Harahan | 11-Aug-1944 | W. D. Field |
| Hermeuptychia hermybius | NVG-1603 | KJ025569 | TX: Cameron Co., E of Brownsville | 17-Mar-2003 | Nick V. Grishin |
| Hermeuptychia hermybius | NVG-1607 | KJ025570 | TX: Cameron Co., E of Brownsville | 18-Jan-2003 | Nick V. Grishin |
| Hermeuptychia hermybius | NVG-1609 | KJ025571 | TX: Cameron Co., E of Brownsville | 30-Mar-2003 | Nick V. Grishin |
| Hermeuptychia hermybius | NVG-1610 | KJ025572 | TX: Cameron Co., E of Brownsville | 9-Mar-2003 | Nick V. Grishin |
| Hermeuptychia hermybius | NVG-1611 | KJ025573 | TX: Cameron Co., E of Brownsville | 14-Mar-2003 | Nick V. Grishin |
| Hermeuptychia hermybius | NVG-1612 | KJ025574 | TX: Cameron Co., E of Brownsville | 16-Mar-2003 | Nick V. Grishin |
| Hermeuptychia hermybius | NVG-1628 | KJ025575 | TX: Cameron Co., E of Brownsville | 19-Oct-1997 | Nick V. Grishin |
| Hermeuptychia hermybius | NVG-1695 | KJ025576 | TX: Hidalgo Co., 1.5 air mi SE of Relampago, Rio Rico Rd., 26.07, -97.891, 21 m | 19-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1698 | KJ025577 | TX: Hidalgo Co., 1.5 air mi SE of Relampago, Rio Rico Rd., 26.07, -97.891, 21 m | 19-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1699 | KJ025578 | TX: Hidalgo Co., 1.5 air mi SE of Relampago, Rio Rico Rd., 26.07, -97.891, 21 m | 19-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1712 | KJ025579 | TX: Starr Co., Rio Grande City, Fort Ringgold, 26.3707, -98.8064, 45 m | 20-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1714 | KJ025580 | TX: Starr Co., Rio Grande City, Fort Ringgold, 26.3707, -98.8064, 45 m | 20-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1726 | KJ025581 | TX: Starr Co., Roma, S of Roma International Bridge, 26.4035, -99.0175, 50 m | 20-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1727 | KJ025582 | TX: Starr Co., Roma, S of Roma International Bridge, 26.4035, -99.0175, 50 m | 20-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1735 | KJ025583 | TX: Starr Co., 0.5 mi S of Fronton, 26.399, -99.085, 50 m | 20-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1737 | KJ025584 | TX: Starr Co., 0.5 mi S of Fronton, 26.399, -99.085, 50 m | 20-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1747 | KJ025585 | TX: Starr Co., Salineno @ Rio Grande, 26.51463, -99.11633, 53 m | 23-Oct-2013 | William R. Dempwolf |
| Hermeuptychia hermybius | NVG-1635 | KJ025586 | TX: Zapata Co., San Ygnacio @ Rio Grande, 92 m | 7-Oct-2007 | Nick V. Grishin |
| Hermeuptychia hermybius | 13385H10 | KJ025587 | TX: Webb Co., Laredo | 15-Apr-1949 | E. L. Todd |
| Hermeuptychia intricata | NVG-1541 | KJ025588 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1548 | KJ025589 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1551 | KJ025590 | TX: Fort Bend Co., Brazos Bend SP, Horseshoe L. tr., 29.38193, -95.61141, 15 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1554 | KJ025591 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1555 | KJ025592 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1556 | KJ025593 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1558 | KJ025594 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata* | NVG-1560 | KJ025595 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1563 | KJ025596 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1565 | KJ025597 | TX: Fort Bend Co., Brazos Bend SP, nr. Hale L., 29.38008, -95.58473, 16 m | 17-Aug-2013 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1629 | KJ025598 | TX: San Jacinto Co., Sam Houston NF, USF217 @ Big Creek, 58 m | 12-Apr-1998 | Nick V. Grishin |
| Hermeuptychia intricata | NVG-1631 | KJ025599 | TX: Brazoria Co., Bar-X Ranch, Rd. 971N, 29.13252, -95.58340, 7 m | 4-Mar-2000 | Nick V. Grishin |
| Hermeuptychia intricata | 13385G07 | KJ025600 | SC: Charleston Co., McClellanville, Wedge Plantation | 6-Apr-1970 | D. C. Ferguson |
| Hermeuptychia intricata | 13385H01 | KJ025601 | FL: Alachua Co., Gainesville | 12-Mar-1983 | Scott W. Gross |
| Hermeuptychia intricata | 13385H02 | KJ025602 | FL: “Putnam Co | Shell Bluff Landing” | 29-Sep-1985 | George Balogh |
| Hermeuptychia intricata | 13386A03 | KJ025603 | LA: Jefferson Parish, Harahan | 28-Jun-1944 | W. D. Field |
| Hermeuptychia intricata | 13385G08 | KJ025604 | SC: Clarendon Co. | 9-Aug-1898 | |
| Hermeuptychia intricata | 13385G09 | KJ025605 | SC: Clarendon Co. | Aug-1910 | |
| Hermeuptychia intricata | 13385G11 | KJ025606 | SC: Clarendon Co. | Aug-1910 | |
| Hermeuptychia intricata | 13386A02 | KJ025607 | “Flatbush LI” | prior to 1941 | G. P. Engelhardt Coll. |
| Hermeuptychia sosybius | DNA-ATBI-0799 | GU089906* | NC: Swain Co., AN9, Smokemont Stables, 35.5504, -83.3084 | 20-Jul-2004 | R. M. Pyle |
| Hermeuptychia sosybius | NSHer-EUA07 | KF466083* | TN: Rutheford Co., 35.70, -86.33 | 2009 | A. V. Z. Brower |
| Hermeuptychia sosybius | NSHer-EUA08 | KF466084* | TN: Rutheford Co., 35.70, -86.33 | 2009 | A. V. Z. Brower |
| Hermeuptychia sosybius | DNA-ATBI-0847 | GU089907* | TN: Blount Co. AN2, Cades Cove, along Forge Cr. Rd. 35.583, -83.838 | 20-Jul-2004 | R. M. Pyle |
| Hermeuptychia sosybius | DNA-ATBI-0848 | GU089908* | TN: Blount Co. AN2, Cades Cove, along Forge Cr. Rd. 35.583, -83.838 | 20-Jul-2004 | R. M. Pyle |
| Hermeuptychia sosybius | DNA-ATBI-0849 | GU089909* | TN: Blount Co. AN2, Cades Cove, along Forge Cr. Rd. 35.583, -83.838 | 20-Jul-2004 | R. M. Pyle |
| Hermeuptychia sosybius | DNA-ATBI-4110 | GU088393* | TN: Sevier Co., Lyon Spring Rd., 35.6, -83.4 | 22-May-2005 | Segebarth |
| Hermeuptychia sosybius | DNA-ATBI-4109 | GU088394* | TN: Sevier Co., Lyon Spring Rd., 35.6, -83.4 | 22-May-2005 | Segebarth |
| Hermeuptychia sosybius | NSHer-EUA02 | KF466080* | FL: Gainesville, 29.65, -82.32 | Apr-2009 | K. R. Willmott |
| Hermeuptychia sosybius | NSHer-EUA03 | KF466081* | FL: Gainesville, 29.65, -82.32 | Apr-2009 | K. R. Willmott |
| Hermeuptychia sosybius | NSHer-EUA06 | KF466082* | FL: Gainesville, 29.65, -82.32 | Apr-2009 | K. R. Willmott |
| Hermeuptychia cucullina | NSHer-PE03 | KF466142* | Peru | C Peña | |
| Hermeuptychia gisella | NSHer-J29 | KF466092* | Brazil: São Paulo, Serra do Japí, Jundiaí, -23.22, -46.92 | 26-Feb-2008 | P. E. C. Peixoto |
| Hermeuptychia atalanta | R10_CA_SP | JN109040* | Brazil: São Paulo, Ribeirão Cachoeira, Campinas | ||
| Hermeuptychia hermes | NSHer-MG08 | KF466108* | Brazil: Minas Gerais, Serra do Cipó, Jaboticatubas, -18.20, -43.50 | Dec-2005 | A. R. M. Silva |
| Hermeuptychia maimoune | NSHer-CO04 | KF466021* | Colombia, Meta, Bosque Bavaria, 4.18, -73.65 | 8-Oct-2006 | M. A. Marín |
| Hermeuptychia pimpla | CP04-10 | GU205843* | Peru: Quebrada Siete Jeringas | ||
| Hermeuptychia harmonia | CP06-93 | GU205842* | Peru: Quebrada Siete Jeringas | ||
| Hermeuptychia fallax | NSHer-J17 | KF466089* | Brazil: São Paulo, Serra do Japí, Jundiaí, -23.22, -46.92 | 26-Feb-2008 | P. E. C. Peixoto |
| Megisto cymela | DNA-ATBI-4114 | GU088434* | TN: Sevier Co., Lyon Spring Rd., 35.6, -83.4 | 22-May-2005 | Segebarth |
| Hermeuptychia intricata? | DNA96-016 | AY508548* | Costa Rica: Puntarenas Province |
Abbreviations: SP State Park; L. Lake; Cr Creek tr. trail; nr. near; Co. County; NF National Forest; SF State Forest Rd. Road
* after the species name indicates primary type specimen, ** is Hermeuptychia hermes kappeli holotype
* after the GenBank number indicates that it was retrieved from GenBank, all other sequences were determined by us in this study
Only DNA ID tags were obtained for the oldest specimens and their dates are shown in bold font.
Here, we show that two distinct species from two different morphogroups as defined by
Specimens used in this study were collected in the field under the permit #08-02Rev from Texas Parks and Wildlife Department to NVG, and inspected in the following collections: Texas A&M University Insect Collection, College Station, TX (TAMU); National Museum of Natural History, Smithsonian Institution, Washington, DC (USNM); Natural History Museum, London, UK (BMNH). Standard entomological techniques were used for dissection (
Two legs (cut with scissors into tiny pieces in lysis buffer) of freshly collected specimens, or two legs that were removed from freshly collected specimens and preserved in alcohol for several years, or an abdomen (dropped into lysis buffer as a whole, and after overnight incubation at 56 °C transferred into 10% KOH for genitalia dissection) of older specimens were used to extract genomic DNA with QIAGEN DNeasy blood and tissue kit complemented with EconoSpin columns from Epoch, or Macherey-Nagel (MN) NucleoSpin® tissue kit following the manufacturer’s protocol. Genomic DNA was eluted in a total volume of 120-150 μl QIAGEN AE buffer (concentration of DNA as measured by Promega QuantiFluor® dsDNA System was from 0.01 to 2.5 ng/μl for legs and from 0.005 to 30 ng/μl for abdomens, depending on specimen age and storage conditions) and was stored at -20 °C.
PCR was performed using Invitrogen AmpliTaq Gold 360 master mix in a 20 μl total volume containing less than 10 ng of template DNA and 0.5 μM of each primer. For legs from freshly collected specimens or those preserved in alcohol, the following primers were used to obtain the complete barcode: LepF: 5’-TGTAAAACGACGGCCAGTATTCAACCAATCATAAAGATATTGG-3'and LepR: 5’-CAGGAAACAGCTATGACCTAAACTTCTGGATGTCCAAAAAATCA-3’. For older specimens the following pairs of primers were used: sCOIF (forward, 5’-ATTCAACCAATCATAAAGATATTGG-3’) – smCOIR (reverse, 5’-CCTGTTCCAGCTCCATTTTC-3’) and bat-smCOIF (forward, 5’-GCTTTTCCTCGTATAAATAATA-3’) – sCOIR (reverse, 5’-TAAACTTCTGGATGTCCAAAAAATCA-3’), to amplify barcode in two overlapping segments (307, 408 bp).
The barcodes of the Hermeuptychia sosybius neotype (designated below) and Hermeuptychia hermes kappeli Anken, 1993 holotype were amplified in four overlapping segments with the following four pairs of Hermeuptychia-specific primers: styr-COIF (forward, 5’-CAACCAATCATAAAGATATTGGAAC-3’) – styr-bCOIR (reverse, 5’-AAAATTATAATAAAAGCATGRGCTGT-3’), styr-bCOIF (forward, 5’-YCCAGGATTTTTAATTGGAGATG-3’) – styr-mCOIR (reverse, 5’-CCTGTYCCACTTCCATTTTCTAC-3’), styr-mCOIF (forward, 5’-TTTTGATTATTACCYCCATCTTT-3’) – styr-eCOIR (reverse, 5’-TTCCTACAGCTCAAATAAATAAAGG-3’), and styr-eCOIF (forward, 5’-TTCATTTAGCTGGAATTTCWTCAA-3’) – sCOIR (reverse, 5’-TAAACTTCTGGATGTCCAAAAAATCA-3’).
For very old specimens (e.g., from 1898 to 1944), amplification of longer DNA segments failed. To obtain their sequences for identification, we developed Hermeuptychia-specific primers for very short, about 100 bp fragments, which we call ID tags. Two regions, in which the three USA Hermeuptychia species differ from each other the most, were selected and the following primers were designed: styr-ID1F (forward, 5’-TTGAGCAGGAATAATTGGWACAT-3’) – styr-ID1R (reverse, 5’-AAAAGCATGRGCTGTAACAA-3’) and styr-ID2F (forward, 5’-TTGGAGGATTTGGTAATTGACTT-3’) – styr-ID2R (reverse, 5’-AAAGATGGRGGTAATAATCAAAAT-3’) to amplify 75 and 56 bp sequence from the specimen (together with both primers, the actual products are 118 and 103 bp).
These primers yielded clear DNA sequence traces (Fig. 65) for 11 out of 12 specimens. The failed traces from DNA voucher 13386A05 showed signs of contamination (i.e., multiple peaks at many positions, probably not even a Hermeuptychia sequence) and were inconclusive. Genitalia, however, offered unambiguous identification of this specimen. We did not pursue re-extraction of DNA from the 13386A05 specimen and were satisfied with higher than 90% success rate (11 out of 12) of this method. The oldest specimens from 1898 and likely prior to 1896 (date not specified on the label of the second specimen, and 1896 is the date of collection donation) yielded excellent traces (e.g., Fig. 65). 6, 1 and 4 specimens of each of the three species were sequenced. For DNA extraction and PCR reactions, they were intermixed and ordered not by species, but as they were placed in USNM collection by curators who did not suspect the presence of more than one species (i.e. semi-randomly, according to DNA voucher numbers assigned to them). Because cross-contamination frequently happens between adjacent specimens, this arrangement alleviates biasing DNA conclusions on the basis of our genitalia and wing pattern-based identification. I.e., if adjacent specimens are the same species (and thus are likely to possess the same DNA barcode), it is more difficult to detect cross-contamination from neighbors. However, if they are different species, disagreement between genitalia-based identification and DNA-based identification would raise suspicions of cross-contamination. All 11 successful DNA identifications were invariably the same as identifications on the basis of genitalia and wing patterns (the voucher 15609E04, Fig. 44, lacked abdomen), and agreed with geographic distribution of these species.
PCR reaction was cleaned up by enzymatic digestion for the whole barcode amplifications of DNA from freshly collected or alcohol preserved specimens and ID tag amplification of old specimens with 4 μl Shrimp Alkaline Phosphatase (20 U/μl) and 1 ul Exonuclease I (1 U/μl) from New England Biolabs. For older specimens that are barcoded in multiple segments, due to the frequent presence of primer dimers and other short non-specific PCR products, Agencourt Ampure XP beads or Invitrogen E-Gel® EX Agarose Gels (followed by Zymo gel DNA recovery kit) were used to select the DNA products of expected length. Sequences were obtained using the M13 primers (for amplification from LepF and LepR primers): 5’-TGTAAAACGACGGCCAGT-3'or 5’-CAGGAAACAGCTATGACC-3'or with primers used in PCR. For the ID tags, PCR products were sequenced in both directions. Sanger sequencing was performed with Applied Biosystems Big Dye Terminator 3.1 kit on ABI capillary instrument in the DNA Sequencing Core Facility of the McDermott Center at UT Southwestern. The resulting sequence traces were proofread in FinchTV <http://www.geospiza.com/Products/finchtv.shtml>. We obtained complete or partial DNA barcode sequences from 85 Hermeuptychia specimens. Sequences and accompanying specimen data were submitted to GenBank and received accession numbers KJ025523–KJ025607. Data about these specimens are provided in Table 1.
Additional DNA sequences were downloaded from GenBank <http://genbank.gov/> using accession numbers provided in
Taxonomic status of various Hermeuptychia populations in Texas has been puzzling (
As a part of a barcoding exercise to shed some light on taxonomy of Hermeuptychia, we obtained DNA sequences from several samples across Texas. The results were not as expected. In fact, populations from extreme south Texas with the small eyespots phenotype characteristic of Hermeuptychia hermes revealed barcodes more similar to those across eastern US. Genitalic examinations showed that even specimens from Tamaulipas and San Luis Potosí, Mexico possessed characters of morphogroup 4 (i.e. the one that includes Hermeuptychia sosybius) from
However, much to our surprise, several specimens from southeast (but not southernmost) Texas, namely from the Brazos Bend State Park in Fort Bend County near Houston, possessed barcodes 3.5% different from those of all other USA populations and, as found by BLAST (
Suspecting DNA introgression, similar to that reported by
Apparently, in Fort Bend County, Texas there exist two sympatric and synchronic Hermeuptychia species (collected on the same day at exactly the same spot!), one from morphogroup 4 and the other one more similar to morphogroups 5, 6 & 7. Interestingly, a possible closest named relative of this second species is either Hermeuptychia gisella or Hermeuptychia cucullina, documented from Bolivia and central to southeastern Brazil. The situation might be analogous to another butterfly recently described from the US, Strymon solitario Grishin & Durden, 2012, whose possible sibling is Strymon jacqueline Nicolay & Robbins, 2005 from Peru (
The two Hermeuptychia species from east Texas are markedly different in genitalia of both sexes and in DNA barcodes. However, upon close inspection of wing patterns, we failed to find strong diagnostic differences that would hold against individual variation. Searching for additional specimens revealed the presence of both species across the eastern US from Texas to Florida and South Carolina, but didn’t reveal obvious wing pattern differences either. This posed a problem with the taxonomic identity of these two species, as it was uncertain which one, if any, is Hermeuptychia sosybius described by
Historical illustrations and specimens of Hermeuptychia sosybius, its original description, and neotype. 1–3 Illustration of Hermeuptychia sosybius syntype(s) by William Jones [1745–1818] from an unpublished book called the “Icones” (
We have taken the following steps to trace the type specimens of Hermeuptychia sosybius. First, we studied relevant publications. For instance,
Second, we consulted knowledgeable historians and scholars of Lepidoptera. John V. Calhoun kindly provided the following information: Drury’s collection was sold at auctions and the catalogs of sales did not list specimens of Hermeuptychia sosybius. However, species names for many sold specimens were not given. It is possible that the types of Hermeuptychia sosybius were acquired by Macleay and are in the Macleay Museum (Sydney, Australia). However, even if Hermeuptychia sosybius specimens could be found in the Macleay collection, it will be nearly impossible to figure out which (if any) served as types. Gerardo Lamas (pers. comm.) was not able to trace Hermeuptychia sosybius syntypes in his comprehensive search for the primary type specimens of all Neotropical butterflies, and expressed an opinion that it would be very difficult to support the status of any found specimens as syntypes.
Nevertheless, as a third step, we contacted the Macleay Museum staff with a request to search for specimens similar to those illustrated by Jones in the Macleay collection. After extensive search of the Macleay holdings (housed in two places), Robert Blackburn, armed with the Jones illustrations and photographs of Hermeuptychia sosybius specimens, was able to find four Hermeuptychia specimens of potential interest. According to Mr. Blackburn (pers. comm.), “the history of these 4 is hard [to determine] due to the absolute lack of labels. Much of the material in these drawers came from a mixture of sources, between William Sharp Macleay’s trading network of entomologists and Alexander Macleay’s purchases at auctions. I think that butterflies like these would be most likely to be Alexander Macleay purchases, and probably came through the purchase of Dru Drury’s collections at auction. I think it’s absolutely possible that they are 1780’s specimens, maybe even through John Abbot, as many of the other butterflies in these drawers are labelled ‘Georgia’.” Two of these (Figs 4–7) would be identifiable as Hermeuptychia sosybius by facies. Unfortunately, neither specimen bears any labels and it will be very difficult to find supporting evidence that these are indeed syntypes. Even if these specimens are from the Drury collection, since Drury exchanged material, it is impossible to know that these are the original specimens, or the ones acquired after the Hermeuptychia sosybius description.
Next, we compared these specimens with the Jones illustrations. The wing pattern and shape of the specimen with abdomen intact (female, Figs 6–7) do not agree closely with the Jones illustrations (Figs 1–3). Most notably, Jones’s illustration of the ventral aspect (Fig. 3) shows two forewing eyespots with strongly developed black rings (near the apex, 2nd and 3rd from the costa), and the specimen has only one (2nd from the costa, the 3rd eyespot entirely lacks black and is more similar to the two posterior eyespots, Fig. 7). The postmedial dark line on ventral hindwing is shaped differently. e.g., it is directed basad near costa in the illustration and is directed distad in the specimen. Other differences in details of placement and shape of eyespots and dark lines are equally obvious, and it is not likely that this specimen was the model for the Jones illustration.
The specimen lacking the abdomen (Figs 4–5) is more similar to the specimen(s) illustrated by Jones, i.e. both 2nd and 3rd eyespots on the forewing are black-ringed and the postmedial hindwing line (slightly) bends basad at costa. However, it seems to be mounted differently than the Jones’s dorsal image shows, i.e. the hindwings that are lowered on the Jones image and touch each other with inner margins, are widely apart in the specimen (Fig. 4). Ventral patterns (in case Jones image Fig. 3 depicts a different specimen from that shown on dorsal image Fig. 2) also differ in detail. In particular, the 3rd hindwing eyespot from the costa lacks black and is more similar to the 4th from costa eyespot in the illustration, but is clearly black-ringed and larger than the 4th eyespot in the specimen (Fig. 5). The submedial and postmedial dark lines on both wings are farther apart in the illustration than in the specimen. The postmedial dark line is strongly bent, directed basad and reaches the hindwing inner margin at an angle in the illustration (more similar to the specimen illustrated in Fig. 47), but is almost perpendicular to the inner margin near the tornus in the specimen. In our opinion, it is not very likely that these obvious pattern differences are caused by inaccuracy of the Jones illustration, in part because we see Hermeuptychia specimens (e.g. Fig. 47) that are more similar in such patterns to the Jones illustration than the specimen in Fig. 5. We see that Hermeuptychia specimens with the characters illustrated by Jones exist, and it seems more likely that their characters were illustrated, rather that invented by Jones. Therefore, we conclude that neither of the specimens from the Macleay collection is the one illustrated by Jones. John V. Calhoun who has vast experience dealing with the analysis of historical illustrations agrees with this opinion (pers. comm.).
To stabilize nomenclature, similarly to
We believe that there is an exceptional need for the neotype to clarify the taxonomic identity of Hermeuptychia sosybius and to define which one of the two USA Hermeuptychia species this name refers to. We hypothesize that it is more likely that the species from morphogroup 4 – i.e. Hermeuptychia sosybius as defined by
In most specimens of eastern US species from a different morphogroup (5, 6, or 7), the forewing postmedial brown line bends basad from vein M1 towards the costa, the hindwing postmedial brown line is more straight near the costa, and it bulges distad around vein M3 (between the two small eyespots in the middle, closer to the posterior eyespot). While the sample of 21 specimens is too small to evaluate the reliability of the wing pattern characters and even this sample already shows variation in these characters (e.g. in some specimens the forewing line is straight towards the costa), morphogroup 4 species seems to be more consistent with Jones’s lectotype drawing in patterns. Combining this albeit rather weak wing pattern evidence with the 20 to 1 ratio of morphogroup 4 specimens found in collections, its possibly wider distribution across eastern US, and the usage of the name “sosybius” in publications to denote this phenotype and DNA barcode (e.g.
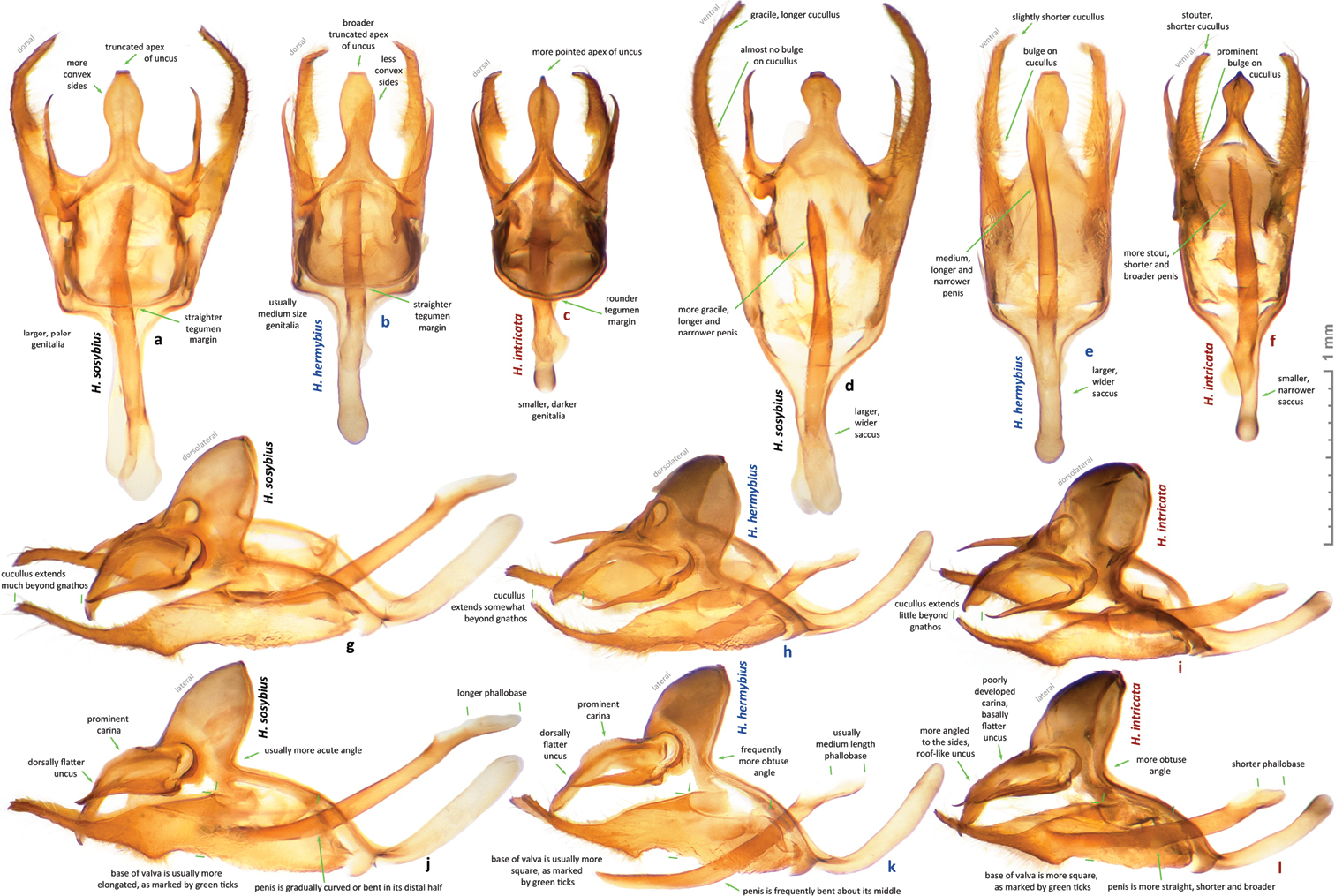
While this species cannot be confidently identified by wing patterns at the moment, it can be differentiated from other Hermeuptychia species by the following combination of male genitalia characters (Figs 60a, d, g, j, 61c, 62o–z2): (1) comparatively large, more gracile and weaker sclerotized (paler) genital capsule (Fig. 60a); (2) medially wider uncus with more prominently convex sides in dorsal (or ventral) view, uncus appears truncated at the apex in dorsal (or ventral) view, but the width of uncus at the apex is generally less than 2/3 of the width of uncus at the narrowest point near the base (Figs 60a, d, 61c); (3) uncus dorsally flatter towards the apex, but convex in lateral view towards the base and with a prominent, thin, membranous carina in basal half (Fig. 60j); (4) valvae elongated, with a saccular lobe, cucullus more gracile, narrower and longer, it projects for close to half of its length farther than the distal end of gnathos (lateral view, Fig. 60g, j); (5) cucullus narrow at the apex, usually with three to five (mostly four) prominent apical teeth (Fig. 60g, j); (6) interior surface of cucullus ventrally without a prominent bulge, best seen in ventral view (Fig. 60d); (7) aedeagus is more gracile, narrower and longer, especially near the distal end, evenly curved or bent distad the middle (Fig. 60d, g, j); (8) longer than wide phallobase (Fig. 60g, j); (9) larger and wider saccus, but shorter than 2/3 of valva length (Fig. 60d). Further analyses and comparisons of genitalia characters between Hermeuptychia species are given in Table 1 of
From the Jones drawing, it is not possible to unambiguously determine the sex of the illustrated specimens because Hermeuptychia are not prominently dimorphic sexually, although the darker color of the specimen shown in dorsal view and wing shape on both illustrations is more consistent with a male. We decided to choose a male specimen as the neotype because male genitalia have been used more widely in Hermeuptychia taxonomy, were illustrated for the majority of known species by
The locality of Hermeuptychia sosybius types was not stated in the original description and currently remains unknown. However, we could attempt to deduce it by comparative analysis of wing patterns on Jones’s drawings. Large eyespots on both wings, some mostly black and pupilled with pale blue are distinctive. Because the size of eyespots is highly variable in Hermeuptychia, it is conceivable that the Drury’s specimens originated in Central or even South America. However, due to very strong development of eyespots and characteristic shape of rusty-brown lines ventrally on both wings, Jones’s drawings are more likely to depict eastern USA Hermeuptychia. Most importantly, the name “sosybius” has been applied to these USA populations historically, and in the interest of stability it is best to secure this name for these populations. If the Hermeuptychia sosybius types were collected in the USA, it is most likely that Drury obtained them from John Abbot and they originated in the eastern coastal US, possibly in Georgia or Virginia (John V. Calhoun, pers. comm.). Populations of the morphogroup 4 species are continuous and widely distributed in east US (
A male specimen (Figs 9–11, genitalia Fig. 62p) bearing three rectangular labels: yellowing white, handprinted on one side - || SAVANNAH, GA. | VII-28-58 ||, grayish, handwritten on the other side - || Coll | G B Small ||; white printed - || DNA sample ID: | 11-BOA-13386A07 | c/o Nick V. Grishin ||; white printed - || NVG131102-61 ||; and a plastic glycerin-filled vial with genitalia on the same pin with the specimen, is hereby designated as the neotype of Papilio sosybius Fabricius, 1793. Upon this publication, red printed label || NEOTYPE ♂ | Papilio sosybius | Fabricius, 1793 | designated by Grishin || will be added. Forewing length of the neotype is 15.5 mm, and this specimen can be recognized by a unique pattern of minor damage to scale cover on wings above, i.e. a longitudinal scratch in the distal half of the left forewing discal cell and a scratch across the discal area of both right wings (Fig. 10). Prior to genitalia dissection, abdomen of the neotype was used to extract total genomic DNA as described in Materials and methods section. The neotype wing pattern mostly agrees with the original description and is similar to Jones illustrations, and the choice of the species is consistent with the usage of this name. The original type locality is not specified in the description (Fig. 8), and the new type locality of Hermeuptychia sosybius according to ICZN Article 76.3 (
Hermeuptychia sosybius. 10–11 neotype designated herein and 12–13 holotype of Hermeuptychia hermes kappeli, data in text 14–15 ♂ USA: Texas, Wise Co., LBJ National Grassland, ex ovum, eclosed 3-Aug-1998, leg. N. V. Grishin 16–17 ♀ ibid, 10-Aug-1998 18–19 ♂ USA: Texas, Brazoria Co., Bar-X Ranch, Rd. 971N, ex ovum, eclosed 18-Apr-2000, leg. N. V. Grishin 20–21 ♀ ibid, 21-Apr-2000. Dorsal/ventral surfaces are in even/odd-numbered figures. Labels are shown for primary types in-line with the specimens and are reduced 2.5-fold compared to specimens as indicated by a smaller scale bar. “F” specifies mirror image (left-right inverted).
Barcode sequence of the neotype: Genbank accession KJ025561, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATAATTGGAACATCATTAAGTTTAATTATCCGAATAGAATTAGGTAACCCAGGATTTTTAATTGGAGATGACCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGTAATTGACTTATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTTCCGCGTATAAATAATATAAGATTTTGATTATTACCTCCATCTTTAATTTTATTAATTTCTAGCAGTATTGTAGAAAATGGAAGTGGAACAGGATGAACTGTTTACCCCCCTCTTTCATCTAATATTGCTCATAGAGGTTCTTCAGTAGATTTAGCAATTTTTTCTCTTCATTTAGCTGGAATTTCATCAATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAATAATATATCTTATGATCAAATACCTTTATTTATTTGAGCTGTAGGAATTACTGCTCTTCTTTTACTTCTCTCATTACCTGTTTTAGCAGGAGCTATTACCATACTTCTTACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCAGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
We believe that our designation of the neotype completely satisfies qualifying conditions of the ICZN Article 75.3 (
The name “Hermeuptychia hermes kappeli” suggested by
No other names have been proposed for North and Central American Hermeuptychia. Now, after the clarification of the morphogroup 4 species identity by the Hermeuptychia sosybius neotype designation and conclusion that Hermeuptychia hermes kappeli is either a subspecies or synonym of Hermeuptychia sosybius, we can proceed with the description of a different morphogroup (5, 6, or 7) species from southeast Texas.
http://zoobank.org/A89BD0A9-9CE9-4DC7-9EFD-42F77A34B2DD
http://species-id.net/wiki/Hermeuptychia_intricata
Figs 22–35, 40–43, 60c, f, i, l, 61a, 62n, 64i–p, 65 part, 66 part, 67 part, 68 partMale (n = 14, Figs 22–23, 28–29, 32, 34–35, 40–43, 68 part) – holotype forewing length = 16.5 mm. Forewing triangular, rounded at apex and tornus, costal and outer margins convex, inner margin almost straight, mildly concave mediad, two discal cell veins bulged at bases, vein 2A thickened basad. Hindwing rounded, almost circular. Wings dorsally dark-brown with sparse olive-beige overscaling and two darker-brown terminal lines. Wings ventrally pale-brown, paler towards inner margin of forewing, with extensive beige overscaling, particularly along veins in distal part in some specimens; submedial and postmedial dark-brown lines and dark-brown end-of-cell streak (smaller on hindwing) between them; forewing postmedial line bent basad near costa in many specimens; hindwing postmedial line almost straight near costa, rarely convex basad and typically convex distad posterior of M3 (between the two small eyespots in the middle, closer to posterior eyespot); two terminal dark-brown evenly curved marginal lines, dark-brown sinuous submarginal line, and row of submarginal eyespots basad of the sinuous line and posteriad of outer discal line, largest eyespots black-centered and pupiled with pale-blue scales: on forewing, largest eyespot in cell M1-M2, eyespot in cell R5-M1 black-centered in some specimens; on hindwing, largest eyespots in cells Cu1-Cu2 and M1-M2, a smaller one in cell Cu2-1A+2A, even smaller, but still black-centered and pale-blue pupilled in cell Rs-M1, and two smallest, usually without black, but in some specimens pale-blue pupilled eyespots in cells M2-M3 and M3-Cu1. Fringes monochrome, a little paler than the ground color of wings. Head, palpi, thorax and abdomen dark-brown above, paler and mostly beige beneath. Antennae dark-brown above with pale scales at segments, orange-brown at the club, beneath beige basad, orange-brown in distal half. Legs brown with beige scales. Male genitalia (n = 14: 12 dissected, 2 inspected in situ, Figs 60c, f, i, l, 61a, 62n) – typical for the genus, smaller and darker in color (more sclerotized) than those of Hermeuptychia sosybius. Tegumen dome-like, rounded at margins. Uncus leaf-shaped in dorsal view, angled to the sides, roof-like, convex distally but almost flat basally in lateral view, without thin, membranous carina in basal half; apex of uncus pointed, not truncated. Gnathos arms thin, wide apart, divergent, about the same length as uncus. Valvae narrow, elongated with thin cuculli extending past gnathos not farther than a third of their length; cucullus more rounded at apex, usually with a couple of small teeth; cucullus ventrally with inner medial bulge. Saccus about the same length as cucullus, narrow. Aedeagus elongated, almost straight, only slightly and evenly curved, not bent, broader and shorter compared to Hermeuptychia sosybius, with a smaller, about as long as wide phallobase. Female (n = 8, Figs 23–27, 30–31, 33, 68 part) – similar to male in facies, with slightly more rounded wings and dorsally paler in color. Female genitalia (n = 8, Fig. 64i–p) with antrum darker in color and smaller than that of Hermeuptychia sosybius. Ostium bursae ellipsoidal, its ventral margin longer than dorsal margin. Antrum narrower anteriad, almost triangular in ventral view, somewhat kidney-shaped in lateral view, mostly symmetric. Ductus and corpus bursae each in length similar to antrum; corpus bursae with two signa, spines in a signum broad, leaf-shaped, usually shingled in two rows.
Hermeuptychia intricata. 22–23 holotype, others are paratypes, data in text and Table 1. Sexes and DNA voucher codes are: 24 ♀ NVG-1554 25 ♀ NVG-1565 26–27 ♀ 13385G09 28–29 ♂ NVG-1631 30–31 ♀ NVG-1629. Dorsal/ventral surfaces are in even/odd-numbered figures, except 24, which is ventral. Labels are shown for the holotype and are reduced 2.5-fold compared to specimens as indicated by a smaller scale bar. “F” specifies mirror image (left-right inverted).
Hermeuptychia intricata paratypes and Hermeuptychia sosybius specimens. 32–35, 40–43 Hermeuptychia intricata 36–39, 44–47 Hermeuptychia sosybius; data in text and Table 1. Sexes and DNA voucher codes are: 32 ♂ 13385G11 33 ♀ 13385G08 34 ♂ 13385G07 35 ♂ 13386A02 36 ♂ 13385G10 37 ♀ 13385G12 38 ♀ 13386A04 39 ♀ 13386A06 40 ♂ 13385H02 41 ♂ 13385H01 42 ♂ 13386A05 43 ♂ 13386A03 44 ♂ 15609E04 45 ♀ 13385H07 46 ♂ 13385H08 47 ♂ 13386A01. All specimens are in USNM collection. Ventral wing surfaces are shown. “F” specifies mirror image (left-right inverted).
Genbank accession KJ025595, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATAATTGGTACATCATTAAGTTTAATTATCCGAATAGAATTAGGTAATCCAGGATTTTTAATTGGAGATGACCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCCATTATAATTGGAGGATTTGGTAATTGACTTGTCCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGTATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAATTTTATTAATTTCTAGTAGTATTGTAGAAAATGGAAGTGGGACAGGATGAACAGTTTACCCCCCCCTCTCATCTAATATTGCTCATAGAGGTTCTTCAGTAGATTTAACAATTTTTTCACTTCATTTAGCTGGAATTTCTTCAATCTTAGGAGCTATTAATTTTATTACAACAATTATTAACATACGAATCAATAATATATCTTATGATCAAATACCTTTATTTATTTGAGCTGTAGGAATTACAGCTCTTCTTTTACTTCTTTCATTACCTGTTTTAGCAGGAGCTATTACTATACTTCTTACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCAGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
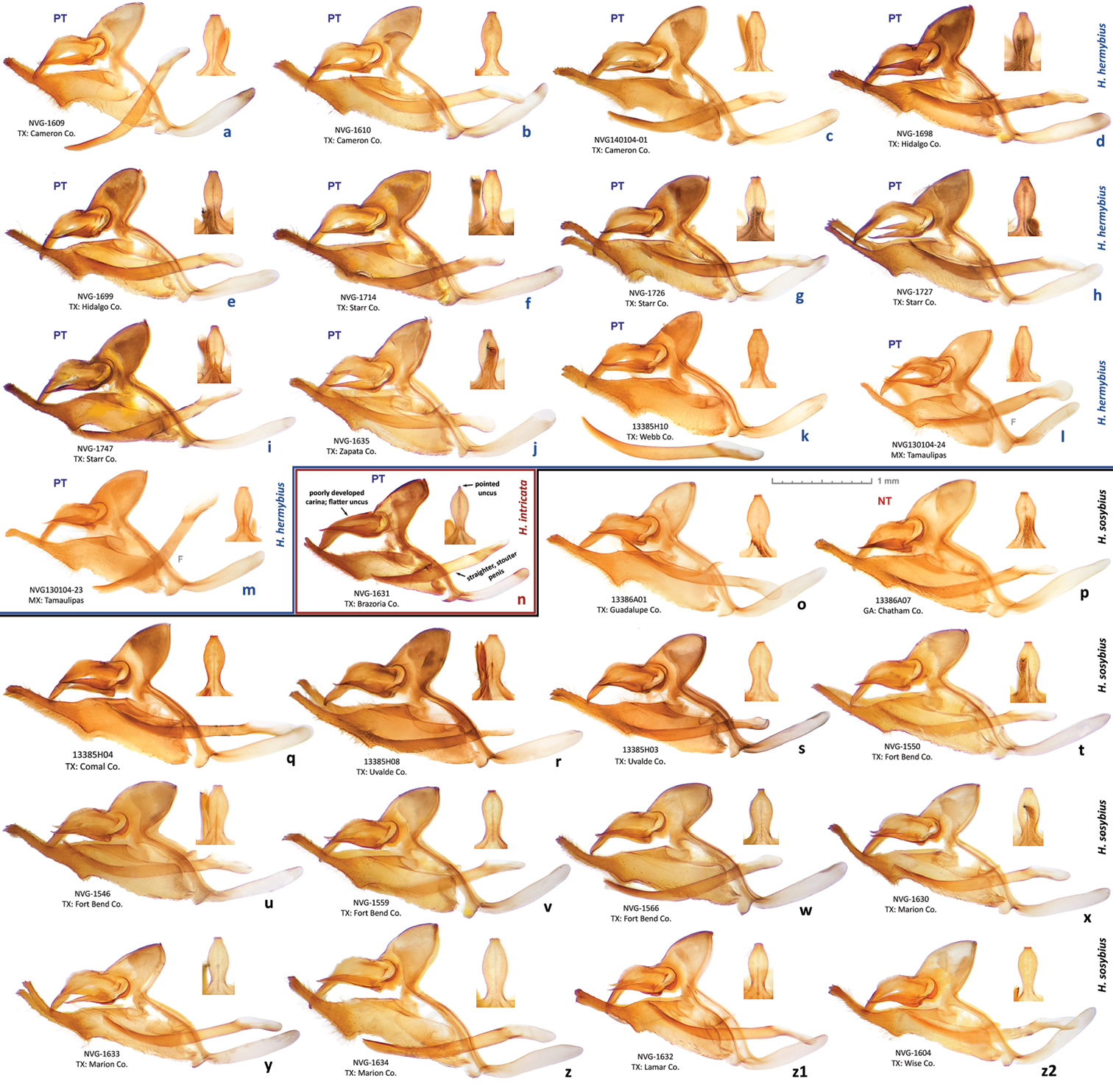
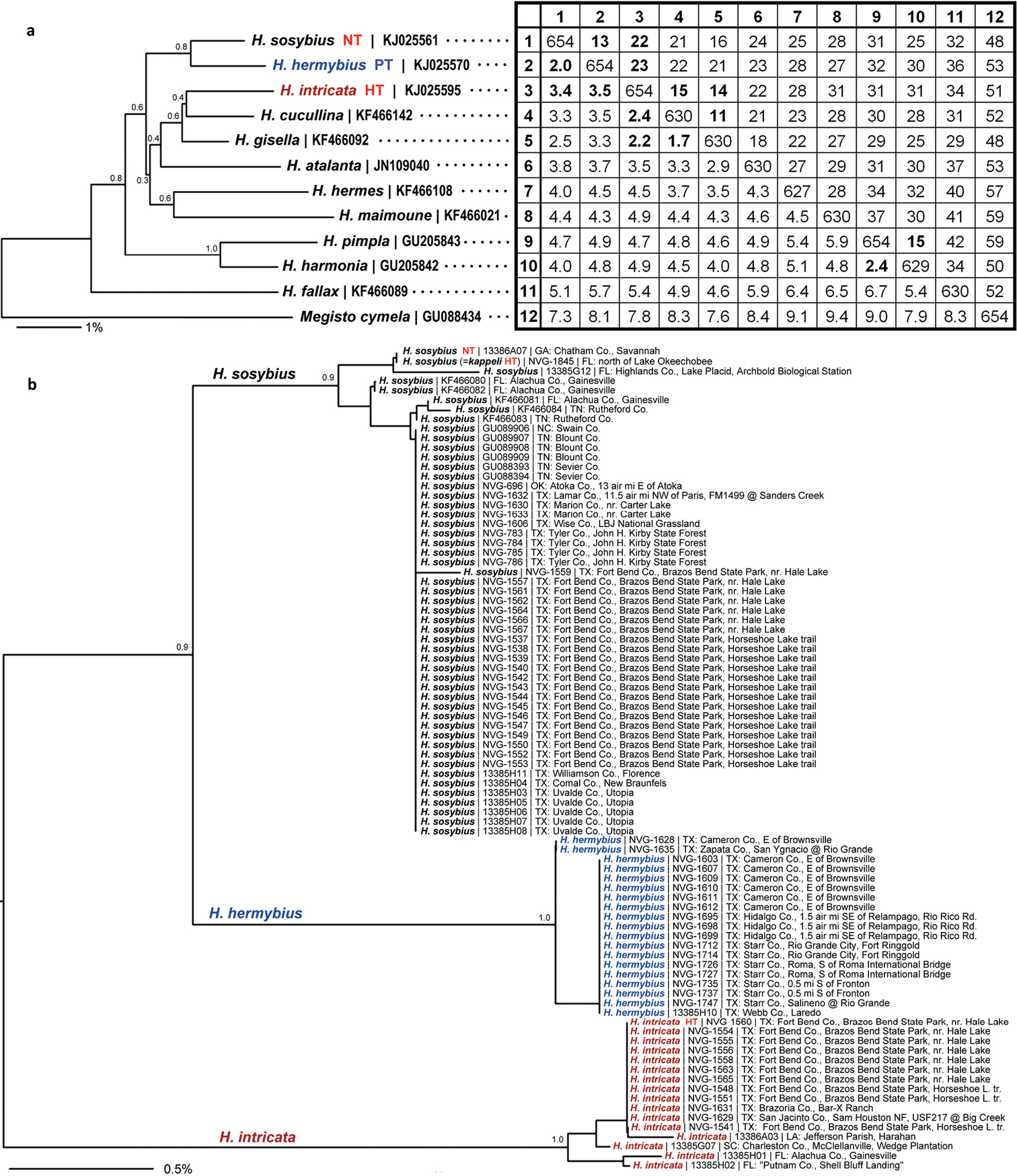
In addition to the holotype, barcodes and ID tags were obtained for 19 paratypes (15 full-length barcodes and 4 ID tags, see Table 1, GenBank accessions: KJ025588–KJ025607, except KJ025595, which is the holotype). Full length barcodes revealed five haplotypes differing from each other by just 1 to 3 base pairs (less than 0.5%). The haplotype of the holotype was more frequently observed (Fig. 66b) and other four haplotypes were confined to a single specimen in the sample.
Holotype: ♂, has the following four rectangular labels: white printed - || USA: TEXAS: Fort Bend Co. | Brazos Bend State Park, | Hale Lake, 29.3801°, -95.5847°| 17-Aug-2013 Grishin N.V. ||; white printed - || DNA extraction | NVG-1560 | 2013-09-05 ||; white printed - || Genitalia vial # | NVG130927-14 | Prep. N. V. Grishin ||; red printed - || HOLOTYPE ♂ | Hermeuptychia | intricata Grishin ||. The holotype is illustrated in Figs 22–23, 60c, f, i, l, & 68 (first image), and the Genbank accession for its DNA COI barcode sequence is KJ025595. Upon publication, the holotype will be deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC (USNM). Paratypes: 13 ♂♂ and 8 ♀♀, all from USA. Of these, 2 ♂♂ and 5 ♀♀ with the same data as the holotype; and 3 ♂♂ (DNA vouchers: NVG-1541, NVG-1548, & NVG-1551) from 2.5 km to the east, i.e. USA: Texas: Fort Bend Co., Brazos Bend State Park, Horseshoe Lake trail, latitude 29°22'54.96", longitude -95°36'41.06", elevation 15 m, 17-Aug-2013, leg. N. V. Grishin. Sexes and GenBank accessions|DNA voucher numbers|genitalia codes (na if not available) for these paratypes (the same format is used below for others) are: ♂ KJ025588|NVG-1541|NVG131003-03, ♂ KJ025589|NVG-1548|na, ♂ KJ025590|NVG-1551|na, ♀ KJ025591|NVG-1554|NVG130927-07, ♂ KJ025592|NVG-1555|NVG131003-04, ♂ KJ025593|NVG-1556|NVG131003-05, ♀ KJ025594|NVG-1558|NVG130927-08, ♀ KJ025596|NVG-1563|NVG130927-11, ♀ KJ025597|NVG-1565|NVG130927-12, ♀ na|na|NVG131003-10. All but one of these paratypes are illustrated in Figs 24, 25, 68 (above the line). 1 ♂ Texas: Brazoria Co., Bar-X Ranch, Rd. 971N, 29.13252, -95.58340, 7 m, 4-Mar-2000, leg. Nick V. Grishin, KJ025599|NVG-1631|NVG131017-08 (Figs 28–29, 62n). 1 ♀ Texas: San Jacinto Co., Sam Houston National Forest, USF217 @ Big Creek, 58 m, 12-Apr-1998, leg. Nick V. Grishin, KJ025598|NVG-1629|NVG131017-06 (Figs 30–31). 1 ♂ South Carolina: Charleston Co., McClellanville, Wedge Plantation, 6-Apr-1970, leg. D. C. Ferguson, KJ025600|13385G07|NVG131102-38 (Fig. 34). 1 ♀ South Carolina: Clarendon Co., 9-Aug-1898, KJ025604|13385G08|NVG131102-39 (Fig. 33). 1 ♀ ibid., Aug-1910, KJ025605|13385G09|NVG131102-40 (Figs 26–27). 1 ♂ ibid., Aug-1910, KJ025606|13385G11|NVG131102-42 (Fig. 32). 1 ♂ Florida: “Putnam Co | Shell Bluff Landing”, 29-Sep-1985, George Balogh, KJ025602|13385H02|NVG131102-45 (Fig. 40). 1 ♂ Florida: Alachua Co., Gainesville, 12-Mar-1983, leg. Scott W. Gross, KJ025601|13385H01|NVG131102-44 (Fig. 41). 1 ♂ Louisiana: Jefferson Parish, Harahan, 28-Jun-1944, W. D. Field, KJ025603|13386A03|NVG131102-57 (Fig. 43). 1 ♂ Louisiana: Jackson Parish, Jonesboro, na|13386A05|NVG131102-59 (Fig. 42). 1 ♂ “Flatbush LI” (specimen curated in the USNM among Hermeuptychia from Louisiana), collected prior to 1941, G. P. Engelhardt Coll., KJ025607|13386A02|NVG131102-56 (Fig. 35).
USA: Texas: Fort Bend Co., Brazos Bend State Park, near Hale Lake, latitude 29°22'48.27", longitude −95°35'05.02", elevation 16 m. This locality is by a wooded, partly open, lowland hiking trail (near and along the park paved road) from a parking lot towards the Big Creek, north of the Hale Lake.
The name refers to the difficulty in recognizing this very distinct species and its intricate ventral wing patterns. The name is an adjective.
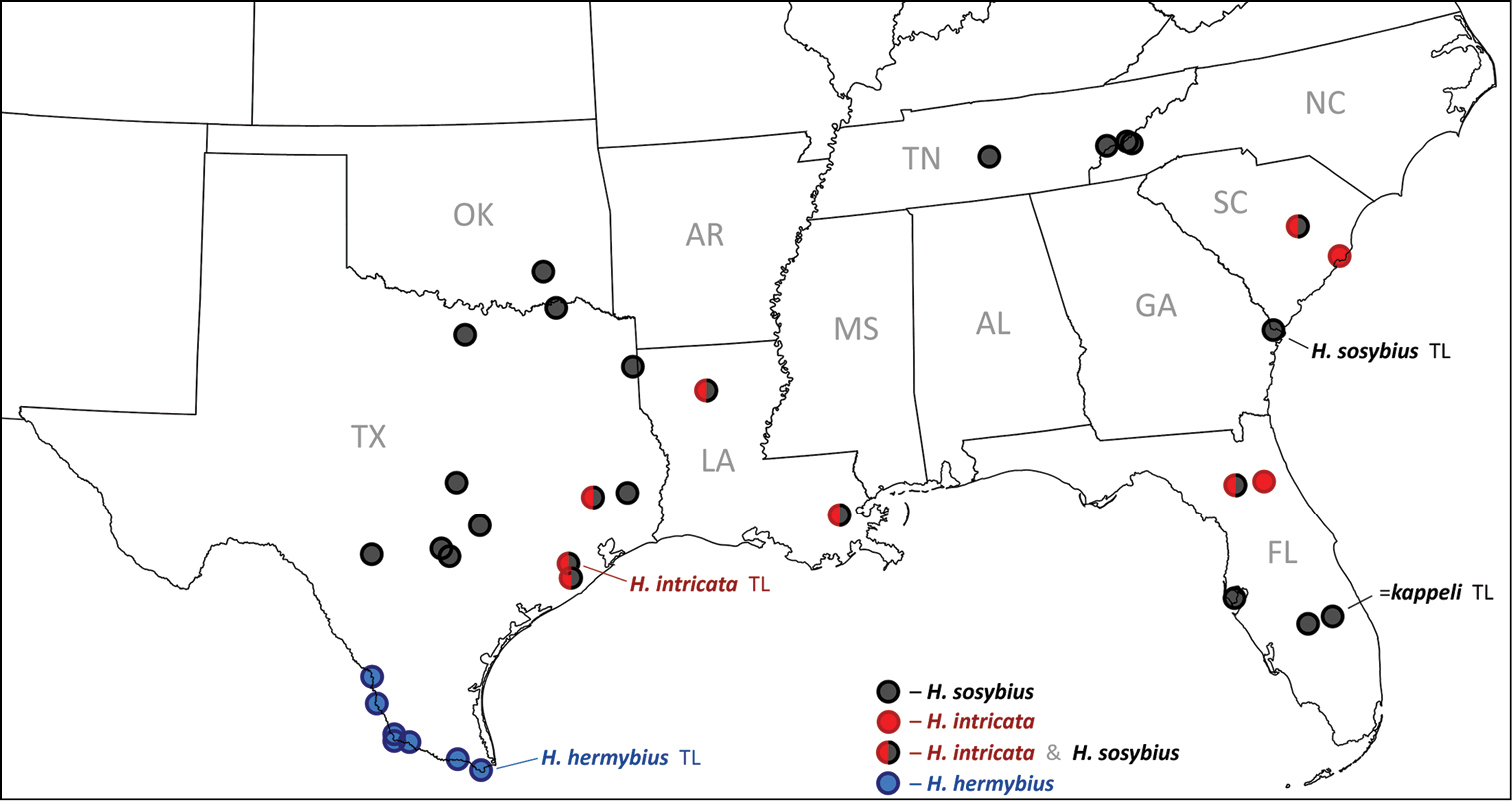
Generally, this is a species of eastern US coastal plains and is currently documented from Texas, Louisiana, Florida, and South Carolina (Fig. 67). It is expected to be more widely distributed in the region and the exact boundaries of the range remain to be investigated. For instance, photographs of live individuals from Alabama: Bibb Co., Blue Girth Creek, 08-VIII-2004 & 18-VI-2005 by Vitaly Charny (
In wing pattern, the new species is very similar to Hermeuptychia sosybius. We were not able to find solid diagnostic characters for the new species, and only hypothetical field marks could be suggested (see discussion). However, it could be easily identified by many distinctive characters of genitalia.
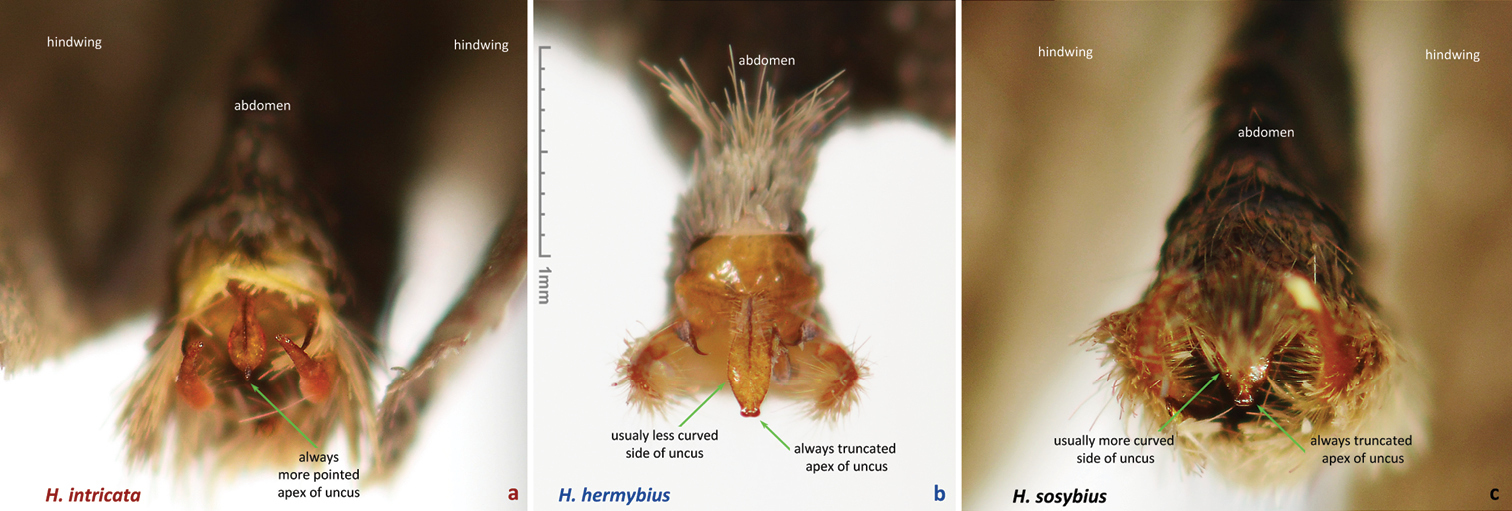
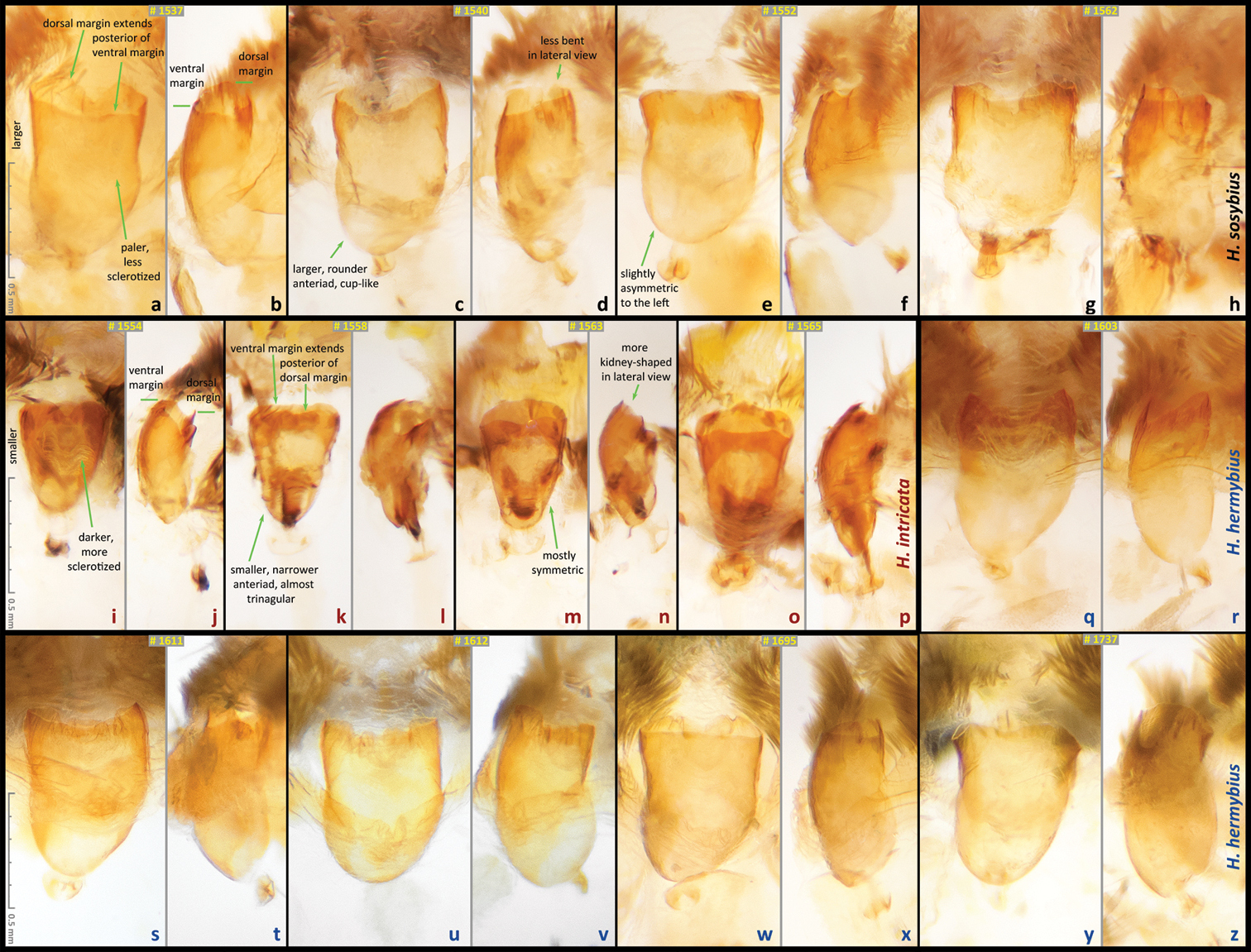
Males of the new species possess: (1) smaller and more robust and darker genital capsule, even in males with larger body size (Fig. 60c)–genitalia of Hermeuptychia sosybius from various parts of the range are larger and look “wider” and are paler (Fig. 60a); (2) narrower and apically pointed uncus (Fig. 60c, f)–uncus of Hermeuptychia sosybius is wider and appears truncated at the apex in dorsal or ventral views (Fig. 60a, d); (3) uncus that is more angled to the sides along the dorsal “rim”, thus appearing “higher” in lateral view (Fig. 60l), but flatter basally due to the lack of prominent carina, vs. a dorsally flatter uncus in distal half, with a well-developed thin, membranous carina in basal half in Hermeuptychia sosybius (Fig. 60j); (4) shorter and stouter cucullus, which projects for less than a third of its length farther than the distal ends of gnathos arms (lateral view, Fig. 60i, l)–cucullus in Hermeuptychia sosybius is more gracile, narrower and longer, it projects for close to half of its length farther than the distal end of gnathos (lateral view, Fig. 60g, j); (5) cucullus more rounded at the apex, usually with a couple of barely defined, very small apical teeth, vs. three to five (mostly four) larger teeth in Hermeuptychia sosybius; (6) interior surface of cucullus ventrally with a more prominent bulge, best seen in ventral view (Fig. 60f vs. 60d); (7) more stout, thicker and shorter penis, best seen in ventral view (Fig. 60f)–penis is more gracile, narrower and longer, especially near the distal end, in Hermeuptychia sosybius (Fig. 60d); (8) shorter phallobase, which is about as long as wide (Fig. 60i, l), vs. phallobase that is much longer than wide in Hermeuptychia sosybius (Fig. 60g, j); (9) smaller and narrower saccus (Fig. 60f), vs. larger and wider one in Hermeuptychia sosybius (Fig. 60d); (10) more obtuse angle formed by the tegumen and vinculum in lateral view (Fig. 60l), vs. typically more acute angle in Hermeuptychia sosybius (Fig. 60j).
Females of the new species possess: (I) narrower ostium bursae and smaller, darker antrum (Fig. 64i, j)–ostium bursae and antrum are larger and antrum is paler in color in Hermeuptychia sosybius (Fig. 64a, b); (II) ventral margin of ostium bursae that extends farther back than its dorsal margin (Fig. 64k, l)–dorsal margin extends posterior of ventral margin in Hermeuptychia sosybius (Fig. 64a, b); (III) antrum that is narrower anteriad, almost triangular in ventral view and symmetric (Fig. 64k, m), vs. rounder, cup-like, slightly asymmetric to the left antrum in Hermeuptychia sosybius (Fig. 64e); (IV) more bent antrum, kidney-shaped in lateral view (Fig. 64n), than that of Hermeuptychia sosybius (Fig. 64j); (V) signa composed of wider, more flattened and rounder spines, mostly in two rows, vs. narrower spines in three to five irregular rows in Hermeuptychia sosybius.
Characters (2) and (3) in males (more pointed apex of uncus and uncus more angled to the sides from the central “rim”) seem to be the easiest to examine without full dissection by brushing the scales off the abdomen tip, even in dry specimens (Fig. 62a, c). Identification of dry females might be more problematic due to abdomen shriveling, however, in freshly caught individuals, ostium bursae and antrum can be easily exposed by squeezing the abdomen in distal third, and the character (II) becomes observable (relative position of ostium bursae margins). Due to these very significant and easily observed differences in genitalia, identification in the field immediately after capture is expected to be straightforward, however, more work remains to be done to discover diagnostic wing pattern characters.
DNA barcodes, consistently with genitalia, set the new species far apart from sympatric Hermeuptychia sosybius, and the difference is about 3.5%, which is significantly higher than “a clear threshold for intra- and interspecific mean distances around 2%”, as quoted from the recent comprehensive analysis of Hermeuptychia (
While the discovery of this second (and new) Hermeuptychia species in eastern USA was very unexpected to us, the next finding is less surprising, although also interesting. Our analysis of DNA barcodes of Texas Hermeuptychia revealed that populations from the lower Rio Grande Valley region of Texas (Webb, Zapata, Starr, Hidalgo, and Cameron Counties) form a tight cluster differing by at least 2% from closely clustered barcodes (divergence average 0.09%, standard deviation 0.19%, maximum below 1%) of over 50 Hermeuptychia sosybius specimens across its range from North Carolina to Texas (south to Uvalde, Comal, Guadalupe and Brazoria Counties, Figs 66–67). These south Texas (and northeast Mexico) Hermeuptychia populations are phenotypically characterized by smaller and more uniformly sized eyespots and more undulated brown lines. This butterfly has been called “Hermeuptychia hermes” in some of the recent literature that advocates the presence of two Hermeuptychia species in the US (
http://zoobank.org/B719B2F8-D0AD-4995-8372-6AA2FC2116E3
http://species-id.net/wiki/Hermeuptychia_hermybius
Figs 48–59, 60b, e, h, k, 61b, 62a–m, 63 part, 64q–z, 66 part, 67 part, 70Male (n = 56, Figs 48–49, 52–56, 58–59) – holotype forewing length = 16 mm. Forewing triangular, rounded at apex and tornus, costal and outer margins convex, inner margin almost straight, mildly concave mediad, two discal cell veins budged at bases, vein 2A thickened basad. Hindwing rounded, almost circular. Wings dorsally dark-brown with sparse olive-beige overscaling and two darker-brown terminal lines. Wings ventrally pale-brown, paler towards inner margin of forewing, with extensive beige overscaling, particularly along veins in distal part in some specimens; submedial and postmedial darker- to rusty- and olive-brown lines and end-of-cell streak (smaller on hindwing) between them; hindwing postmedial line more undulate that in Hermeuptychia sosybius, with a stronger bend in M1-M2 cell; two terminal dark-brown evenly curved marginal lines, dark-brown sinuous submarginal line, more undulate than in Hermeuptychia sosybius, barely touching the eyespot in cell Cu1-Cu2, and row of submarginal eyespots basad of the sinuous line and posteriad of postmedial line, eyespots frequently reduced in size and are more uniformly sized than in Hermeuptychia sosybius; usually largest eyespots black-centered and pupiled with pale-blue scales: on forewing, eyespots about the same size, frequently larger posteriad, but eyespot in cell M1-M2 (usually not the largest in size) and eyespot in cell R5-M1 (in some specimens) black-centered (more eyespots black centered in some specimens); on hindwing, largest eyespots in cells M1-M2 and Cu1-Cu2, a smaller one in cell Cu2-1A+2A, even smaller, but still black-centered and pale-blue pupilled in cell Rs-M1, and two smallest, usually without black, but in some specimens pale-blue pupilled eyespots in cells M2-M3 and M3-Cu1. Fringes monochrome, a little paler than the ground color of wings. Head, palpi, thorax and abdomen dark-brown above, paler and mostly beige beneath. Antennae dark-brown above with pale scales at segments, orange-brown at the club, beneath beige basad, orange-brown in distal half. Legs brown with beige scales. Male genitalia (n = 19, Figs 60b, e, h, k, 61b, 62a–m, 63 part) – typical for the genus, very similar to those of Hermeuptychia sosybius. Tegumen dome-like, rounded at margins. Uncus leaf-shaped in dorsal view, almost flat distally but convex basally in lateral view, with a well-developed thin, membranous carina in basal half; apex of uncus appears truncated in dorsal view and sides usually less concave than in Hermeuptychia sosybius. Gnathos arms thin, wide apart, divergent, about the same length as uncus. Valvae narrow, but typically broader than in Hermeuptychia sosybius, elongated with thin cuculli extending past gnathos usually farther than a quarter of their length; cucullus usually with four apical teeth; cucullus ventrally with inner medial bulge. Saccus about the same length as cucullus, narrow. Aedeagus elongated, bent around its middle, with a medium length phallobase. Female (n = 45, Figs 50–51, 57) – similar to male in facies, with slightly more rounded wings and dorsally paler in color. Female genitalia (n = 9, Fig. 64q–z) as in Hermeuptychia sosybius, with pale, yellowish, weakly sclerotized and broad, rounder anteriad, cup-like antrum slightly asymmetric to the left. Ostium bursae ellipsoidal, its ventral margin shorter or equal to dorsal margin. Ductus and corpus bursae each in length similar to antrum; corpus bursae with two signa, spines in a signum narrow, leaf-shaped, placed in three to five irregular rows.
Hermeuptychia hermybius. 48–49 holotype, others are paratypes, data in text and Table 1. Sexes and DNA or genitalia voucher codes, or data: 50–51 ♀ USA: Texas: Cameron Co., Brownsville, ex ovum, eclosed 2-Apr-2003, leg. N. V. Grishin 52 ♂ NVG-1635 53 ♂ 13385H10 54–55 ♂ NVG-1607 56 ♂ NVG-1699 57 ♀ NVG-1737 58 ♂ NVG130104-23 59 ♂ NVG130104-24. Dorsal wing surfaces are in 48, 50, 54 others are ventral. Labels are shown for the holotype and are reduced 2.5-fold compared to specimens as indicated by a smaller scale bar. “F” specifies mirror image (left-right inverted).
Male genitalia of Hermeuptychia from USA: Texas. a, d, g, j Hermeuptychia sosybius, Fort Bend Co., Brazos Bend State Park, Horseshoe Lake trail, 29°22'54.96", −95°36'41.06", 15 m, 17-Aug-2013, leg. N. V. Grishin, DNA voucher NVG-1542, genitalia NVG130927-03 (forewing length 15 mm) b, e, h, k Hermeuptychia hermybius sp. n. paratype, Cameron Co., E of Brownsville, 18-Jan-2003, leg. N. V. Grishin, DNA voucher NVG-1607, genitalia NVG130927-18 (specimen Figs 54–55, forewing length 15.5 mm) c, f, i, l Hermeuptychia intricata sp. n. holotype, Fort Bend Co., Brazos Bend State Park, near Hale Lake, 29°22'48.27", −95°35'05.02", 16 m, 17-Aug-2013, leg. N. V. Grishin, DNA voucher NVG-1560, genitalia NVG130927-14 [USNM] (specimen Figs 22–23, forewing length 16.5 mm). Views: a–b dorsal, perpendicular to the tegumen-uncus-gnathos plane c–d ventral, perpendicular to the plane of saccus and valvae (appears larger than dorsal view due to different projection axis) e–f right dorsolateral g–h right lateral. All images are to scale. Diagnostic characters are indicated on images. Note that Hermeuptychia intricata with larger than Hermeuptychia sosybius wings has smaller genitalia.
Dorsoposterior view of male abdomens of Hermeuptychia from USA: Texas. a Hermeuptychia intricata, DNA voucher NVG-1548 (mirror image, i.e. left-right inverted) b Hermeuptychia hermybius, DNA voucher NVG-1635 (also shown in Fig. 62j, specimen Fig. 52) c Hermeuptychia sosybius, DNA voucher NVG-1553. Data in Table 1. Scales are brushed off the abdomen tip to expose distal parts of genitalia. The easiest to observe character (the shape of the distal end of uncus) is indicated.
Variation in male genitalia of Hermeuptychia hermybius and Hermeuptychia sosybius. a–m Hermeuptychia hermybius paratypes, DNA (or genitalia, where DNA sequence is not available, and full data for these given) voucher codes: a. NVG-1609 b NVG-1610 c Texas: Cameron Co., Brownsville {10-13}-Mar-1979, T. Friedlander, NVG140104-01 d NVG-1698 e NVG-1699 (specimen Fig. 56) f NVG-1714 g NVG-1726 h NVG-1727 i NVG-1747 j NVG-1635 (also shown in Fig. 61b, specimen Fig. 52) k 13385H10 (specimen Fig. 53) l–m Mexico: Tamaulipas, leg. R. O. & C. A. Kendall: l Quintero cave, 7-Jan-1974, NVG130104-24 (specimen Fig. 59) m Ciudad Mante, Los Arcos Ct., 19-Dec-1973, NVG130104-23 (specimen Fig. 58) n Hermeuptychia intricata paratype, NVG-1631 (specimen Figs 28–29), diagnostic characters are indicated on the image o–z2 Hermeuptychia sosybius: o 13386A01 (specimen Fig. 47) p 13386A07, neotype (specimen Figs 9–11) q 13385H04 r 13385H08 (specimen Fig. 46) s 13385H03 t NVG-1550 u NVG-1546 v NVG-1559 w NVG-1566 x NVG-1630 y NVG-1633 z Texas: Marion Co., W of Caddo Lake, 5-Apr-1997, leg. N. V. Grishin, NVG-1634 z1 NVG-1632 z2 Texas: Wise Co., LBJ National Grassland, ex ovum, eclosed 3-Aug-1998, leg. N. V. Grishin, NVG-1604. c, l, m are in TAMU and o–s are in USNM collections. Data for most specimens are in Table 1, text, or specified above. Complete genitalia are shown in lateral view, and dorsal view of uncus is shown above and to the right of each specimen. Aedeagus is shown below in k DNA (or genitalia, where DNA sequence is not available) voucher codes and general localities are indicated below each genitalia image. “F” specifies mirror image (left-right inverted).
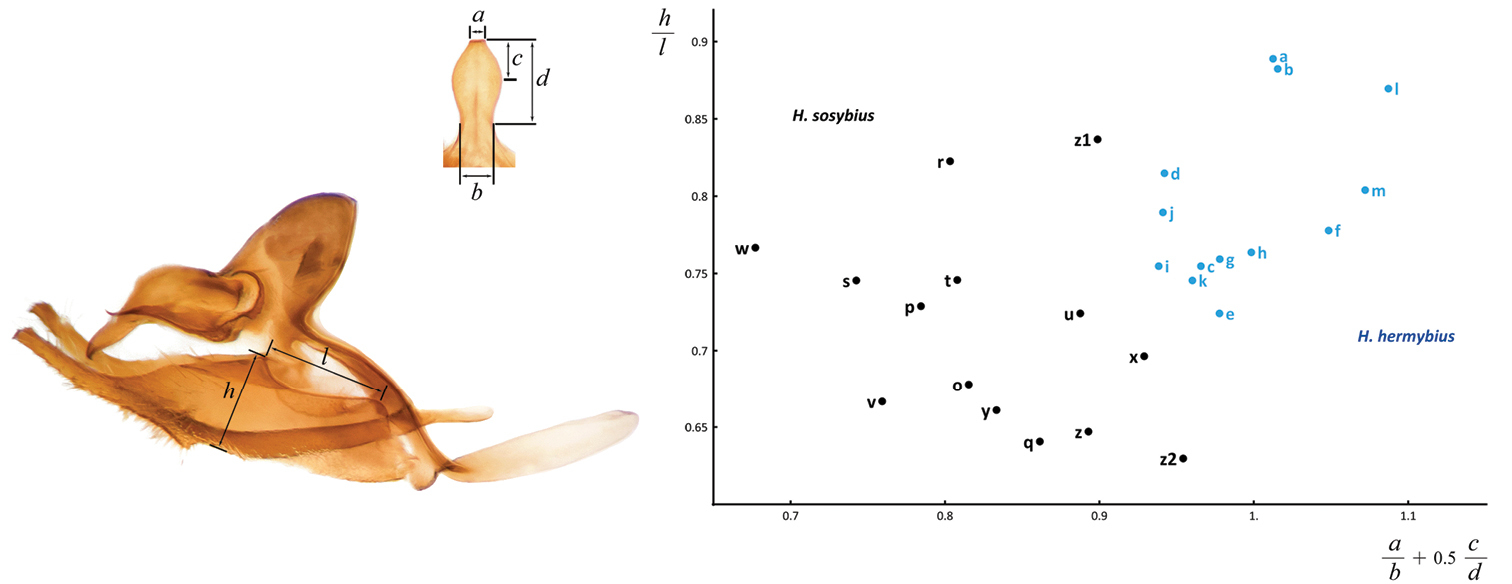
Morphometric differences between male genitalia of Hermeuptychia sosybius (black) and Hermeuptychia hermybius (blue). Measurements used are marked on dorsal view of uncus (top left) and on lateral view of complete genitalia (bottom left): a width of uncus at the apex b width of uncus at the narrowest point near the base (“neck” at the joint with tegumen) c distance from the uncus apex to the cross-section at the widest point d distance from the uncus apex to the cross-section at the narrowest point near the base l length of valval dorsal “window” h height of valva (in lateral view) at the end of the dorsal “window”, direction of height measurement is perpendicular to the direction of length measurement. Measurements of genitalia shown in Fig. 62 are plotted on the right. Horizontal axis combines all uncus measurements into a formula a/b+0.5*c/d and vertical axis shows measurements of valva as h/l. Each point corresponds to a specimen and a letter next to it is the same one that denote its genitalia in Fig. 62.
Antrum in female genitalia of Hermeuptychia from USA: Texas. a–h Hermeuptychia sosybius, Fort Bend Co., Brazos Bend State Park, 17-Aug-2013, leg. N. V. Grishin: a–f is from Horseshoe Lake trail, 29°22'54.96", −95°36'41.06", 15 m and g–h is from near Hale Lake, 29°22'48.27" −95°35'05.02", 16 m; DNA voucher|genitalia dissection codes are: a–b NVG-1537|NVG130927-01 c–d NVG-1540|NVG130927-02 (specimen Fig. 12) e–f NVG-1552|NVG130927-06 g–h NVG-1562|NVG130927-10 i–p Hermeuptychia intricata sp. n. paratypes, Fort Bend Co., Brazos Bend State Park, near Hale Lake, 29°22'48.27", −95°35'05.02", 16 m, 17-Aug-2013, leg. N. V. Grishin; DNA voucher|genitalia dissection codes: i–j NVG-1554|NVG130927-07 (specimen Fig. 24) k–l NVG-1558|NVG130927-08 m–n NVG-1563|NVG130927-11 o–p NVG-1565|NVG130927-12 (specimen Fig. 25) q–z Hermeuptychia hermybius sp. n. paratypes q–r Cameron Co., E of Brownsville, ex ovum ex ♀ captured on 18-Jan-2003, eclosed on 17-Mar-2003, leg. N. V. Grishin, NVG-1603|NVG130927-17 s–t ibid., eclosed on 14-Mar-2003, NVG-1611|NVG131017-03 u–v ibid., eclosed on 16-Mar-2003, NVG-1612|NVG131017-04 w–x TX: Hidalgo Co., 1.5 air mi SE of Relampago, Rio Rico Rd., 26.07, -97.891, 21 m, 19-Oct-2013, leg. W. R. Dempwolf, NVG-1695|NVG131229-03 y–z Starr Co., 0.5 mi S of Fronton, 26.399, -99.085, 50 m 10-Oct-2013, leg. W. R. Dempwolf, NVG-1737|NVG131229-11 (specimen Fig. 57). Additional data for specimens and their DNA barcodes are in Table 1. In all images, posterior end is pointing up (i.e. ostium bursae is closer to the top of each image); a, c, e, g, i, k, m, o, q, s, u, w, y are in lateral view, others are in right ventrolateral view. All images are to scale. Diagnostic characters to tell between Hermeuptychia sosybius and Hermeuptychia intricata are indicated on images, each character was invariantly observed in all inspected samples of a species, but is indicated (for clarity) on a single image only. We failed to find characters distinguishing female genitalia of Hermeuptychia hermybius from Hermeuptychia sosybius and simply illustrate genitalic variation.
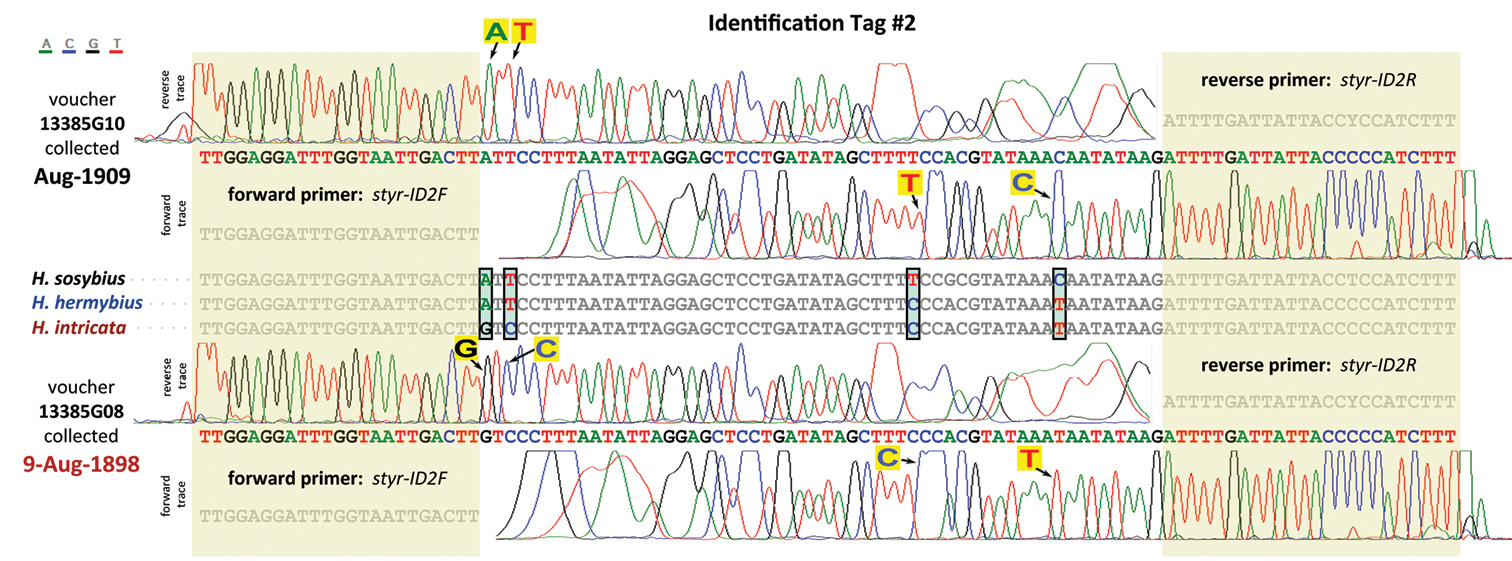
DNA ID tags of specimens that are over 100 years old. ID tag #2 is shown as an example. The tag region sequence alignment of the three species: Hermeuptychia sosybius, Hermeuptychia hermybius, and Hermeuptychia intricata is shown in the middle and positions at which sequences differ are highlighted in cyan and boxed. Each of the three species differs from the other two by at least 2 nucleotides, and Hermeuptychia sosybius is different from Hermeuptychia intricata by 4 nucleotides. Forward and reverse primer regions are shaded. DNA of the tag was amplified and sequenced in both forward and reverse directions from two over-100-years-old specimens from the same locality (SC: Clarendon Co.). Forward and reverse sequences traces for the first specimen are shown above the reference sequences and the two traces for the second specimen are shown below. It is clear from the traces that the specimen above (13385G10, Fig. 36) is Hermeuptychia sosybius, (A, T, T, & C at these 4 positions, no contamination seen) and the one below (13385G08, Fig. 33) is Hermeuptychia intricata (G, C, C, & T at these 4 positions and equally unambiguous traces). Nucleotides that identify each specimen are indicated in large letters on yellow background and arrows point to the trace peaks revealing these nucleotides. This strategy was applied to identify 12 very old specimens of three species in a random order and yielded unambiguous identifications for 11 of them. One sample appeared to be contaminated, and the traces showed the presence of several nucleotides in many positions. All 11 DNA-based identifications agreed with genitalic identifications.
Full length DNA barcodes were obtained for 19 paratypes (GenBank accessions: KJ025569–KJ025587). The most common haplotype present in 17 sequences (including all 5 barcoded siblings of the holotype) is exemplified by the voucher NVG-1603, Genbank accession KJ025569, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATAATTGGAACATCATTAAGTTTAATTATTCGAATAGAGTTAGGTAATCCAGGATTTTTAATTGGAGATGACCAAATTTATAACACTATTGTTACAGCCCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGTAATTGACTTATTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGTATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAATTTTATTAATTTCTAGTAGTATTGTAGAAAATGGAAGTGGAACAGGATGAACTGTTTACCCCCCTCTTTCATCTAATATTGCCCATAGAGGTTCTTCAGTAGATTTAGCAATTTTTTCTCTTCATTTAGCTGGAATTTCATCAATTTTAGGAGCCATTAATTTTATTACAACAATTATTAATATACGAATTAATAATATATCTTATGATCAAATACCTTTATTTATTTGAGCTGTAGGAATTACAGCTCTTCTTTTACTTCTCTCATTACCTGTTTTAGCAGGAGCTATTACCATACTTCTTACTGATCGAAATTTAAATACATCATTTTTTGACCCTGCAGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
The 2 remaining sequences were identical to each other (Fig. 66b) and differed from the sequence shown above by a single base pair (0.15%). Barcode from the oldest and westernmost specimen (TX: Laredo, 15-Apr-1949) was additionally verified with both DNA ID tags as described in Materials and methods section and confirmed to be this species.
DNA-derived data. a Analysis of named Hermeuptychia species b relationships between Hermeuptychia specimens from USA in a form of BioNJ (
Holotype: ♂, has the following two rectangular labels: white printed - || USA: TEXAS: Cameron Co. | E of Brownsville, ex ovum | ex ♀ collected 18-Jan-2003 | ecl. 12-Mar-2003 Grishin N.V. ||; red printed - || HOLOTYPE ♂ | Hermeuptychia | hermybius Grishin ||. The holotype is illustrated in Figs 48–49. Upon publication, the holotype will be deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC (USNM). Paratypes: 55 ♂♂ and 45 ♀♀, from USA: Texas, unless indicated otherwise. Of these, 9 ♂♂ and 12 ♀♀ are siblings of the holotype read from ova, with the same data, their sexes, eclosion dates and GenBank accessions|DNA voucher numbers|genitalia codes (where available, and in this format for other paratypes) are: 1 ♀ 8-Mar-2003; 1 ♂ 9-Mar-2003, KJ025572|NVG-1610|NVG131017-02 (Fig. 62b); 2 ♂♂ and 1 ♀ 9-Mar-2003; 1 ♂ and 1 ♀ 10-Mar-2003; 1 ♂ and 1 ♀ 11-Mar-2003; 3 ♂♂ 12-Mar-2003; 1 ♀ 14-Mar-2003, KJ025573|NVG-1611|NVG131017-03 (Fig. 64s–t); 1 ♀ 15-Mar-2003; 1 ♀ 16-Mar-2003, KJ025574|NVG-1612|NVG131017-04 (Fig. 64u–v); 1 ♀ 17-Mar-2003, KJ025569|NVG-1603|NVG130927-17 (Fig. 64q–r); 2 ♀♀ 17-Mar-2003; 1 ♀ 21-Mar-2003; 1 ♂ 30-Mar-2003, KJ025571|NVG-1609|NVG131017-01 (Fig. 62a); 1 ♀ 2-Apr-2003 (Figs 50–51). Other paratypes are: 1 ♂ ibid., collected on wing 18-Jan-2003, KJ025570|NVG-1607|NVG130927-18 (Figs 54–55, 60b, e, h, k). 1 ♀ Cameron Co., E of Brownsville, 19-Oct-1997, leg. N. V. Grishin, KJ025575|NVG-1628|NVG131017-05. 1 ♂ Cameron Co., Brownsville, {10-13}-Mar-1979, leg. T. Friedlander, NVG140104-01 [TAMU] (Fig. 62c). 1 ♂ (06-Jun-2007) 1 ♀ (07-Jun-2007) Cameron Co., Los Fresnos, Ted Hunt & Loop Rd., leg. William R. Dempwolf. 4 ♀♀ Hidalgo Co., 1.5 air mi SE of Relampago, Rio Rico Rd., 26.07, -97.891, 21 m, 13-Jun-2013, leg. W. R. Dempwolf; 2 ♂♂ ibid., 19-Oct-2013, KJ025577|NVG-1698|NVG131229-04 (Fig. 62d) and KJ025578|NVG-1699|NVG131229-05 (Figs 56, 62e); 1 ♀ ibid., 19-Oct-2013, KJ025576|NVG-1695|NVG131229-03 (Fig. 64w–x); 3 ♂♂ 4 ♀♀ ibid., 19-Oct-2013; 2 ♂♂ 4 ♀♀ ibid., 21-Oct-2013; 3 ♂♂ ibid., 24-Oct-2013. 1 ♀ TX: Starr Co., Rio Grande City, Fort Ringgold, 26.3707, -98.8064, 45 m, 12-Nov-2010, leg. W. R. Dempwolf; 1 ♀ ibid., 13-Jun-2013; 1 ♂ ibid., 20-Oct-2013, KJ025580|NVG-1714|NVG131229-07 (Fig. 62f); 1 ♀ ibid., 20-Oct-2013, KJ025579|NVG-1712|NVG131229-06; 2 ♂♂ ibid., 20-Oct-2013; 1 ♂ ibid., 23-Oct-2013; 2 ♂♂ 1 ♀ ibid., 9-Nov-2013. 2 ♂♂ Starr Co., Roma, S of Roma International Bridge, 26.4035, -99.0175, 50 m, 20-Oct-2013, leg. W. R. Dempwolf, KJ025581|NVG-1726|NVG131229-08 (Fig. 62g) and KJ025582|NVG-1727|NVG131229-09 (Fig. 62h); 8 ♂♂ 7 ♀♀ ibid., 20-Oct-2013. 1 ♀ Starr Co., Roma Creek, Hwy 650/Hwy 83, 29-Oct-2007, leg. W. R. Dempwolf. 2 ♀♀ Starr Co., 0.5 mi S of Fronton, 26.399, -99.085, 50 m, 20-Oct-2013, leg. W. R. Dempwolf, KJ025583|NVG-1735|NVG131229-10 and KJ025584|NVG-1737|NVG131229-11 (Figs 57, 64y–z); 7 ♂♂ 3 ♀♀ ibid., 20-Oct-2013. 1 ♂ Starr Co., Salineno @ Rio Grande, 26.51463, -99.11633, 53 m, 23-Oct-2013, leg. W. R. Dempwolf, KJ025585|NVG-1747|NVG131229-12 (Fig. 62i). 1 ♂ Zapata Co., San Ygnacio @ Rio Grande, 92 m, 7-Oct-2007, leg. N. V. Grishin, KJ025586|NVG-1635|NVG131017-12 (Figs 52, 61b, 62j). 1 ♂ Webb Co., Laredo, 15-Apr-1949, leg. E. L. Todd KJ025587|13385H10|NVG131102-53 [USNM] (Figs 53, 62k). 1 ♂ Mexico: Tamaulipas: Rt. 101 at Rio Corona, 1-Jan-1980, leg. P. W. Kovarik & D. S. Bogar, NVG140104-04 [TAMU]. 1 ♂ Mexico: Tamaulipas: El Canindo, nr. Ejido San José, 7.5 km W Gómez Farías, 1400 m, {19-21}-Jul-1994, leg. C. Cate & T. Riley, NVG140104-67 [TAMU]. 2 ♂♂ Mexico: Tamaulipas: Ciudad Mante, Los Arcos Ct., 19-Dec-1973, leg. R. O. & C. A. Kendall, NVG140104-22 and NVG130104-23 [TAMU] (Figs 58, 62m); 1 ♂ ibid., 28-Jan-1995, ex larva, foodplant Panicum maximus Jacq., NVG140104-24 [TAMU]. 1 ♂ Mexico: Tamaulipas: Quintero cave [22.6333, -99.0333], 7-Jan-1974, leg. R. O. & C. A. Kendall, NVG130104-24 [TAMU] (Figs 59, 62l). 1 ♂ 1 ♀ Mexico: San Luis Potosí: El Salto Falls, 30-Dec-1979, leg. P. W. Kovarik & D. S. Bogar, NVG140104-03 and NVG140104-02 [TAMU].
USA: Texas: Cameron County, east of Brownsville. It is a shaded area covered in Guinea grass (Panicum maximus), situated near a ravine and overgrown with taller trees.
The name is a fusion of two words: herm[es] beginning and [sos]ybius ending. It symbolizes that this species traditionally and previously regarded as Hermeuptychia hermes is phylogenetically closer to Hermeuptychia sosybius, and yet is distinct from it. The resulting word is unique and currently unknown to internet search engines, which is expected to ease its searches. The name is a noun in apposition.
This species is currently recorded from the lower Rio Grande Valley region of Texas along the Rio Grande from Laredo to the Gulf coast (Webb, Zapata, Starr, Hidalgo, and Cameron Counties, Fig. 67) and in neighboring Mexico (Tamaulipas, San Luis Potosí).
USA localities of Hermeuptychia specimens with available DNA barcode information. Color of circles corresponds to species: Hermeuptychia sosybius – black; Hermeuptychia hermybius – blue, Hermeuptychia intricata – red, split red/black circles mark localities where both Hermeuptychia sosybius and Hermeuptychia intricata were recorded. Type localities are indicated with a corresponding name followed by “TL”. Hermeuptychia hermes kappeli was treated as a junior subjective synonym of Hermeuptychia sosybius by
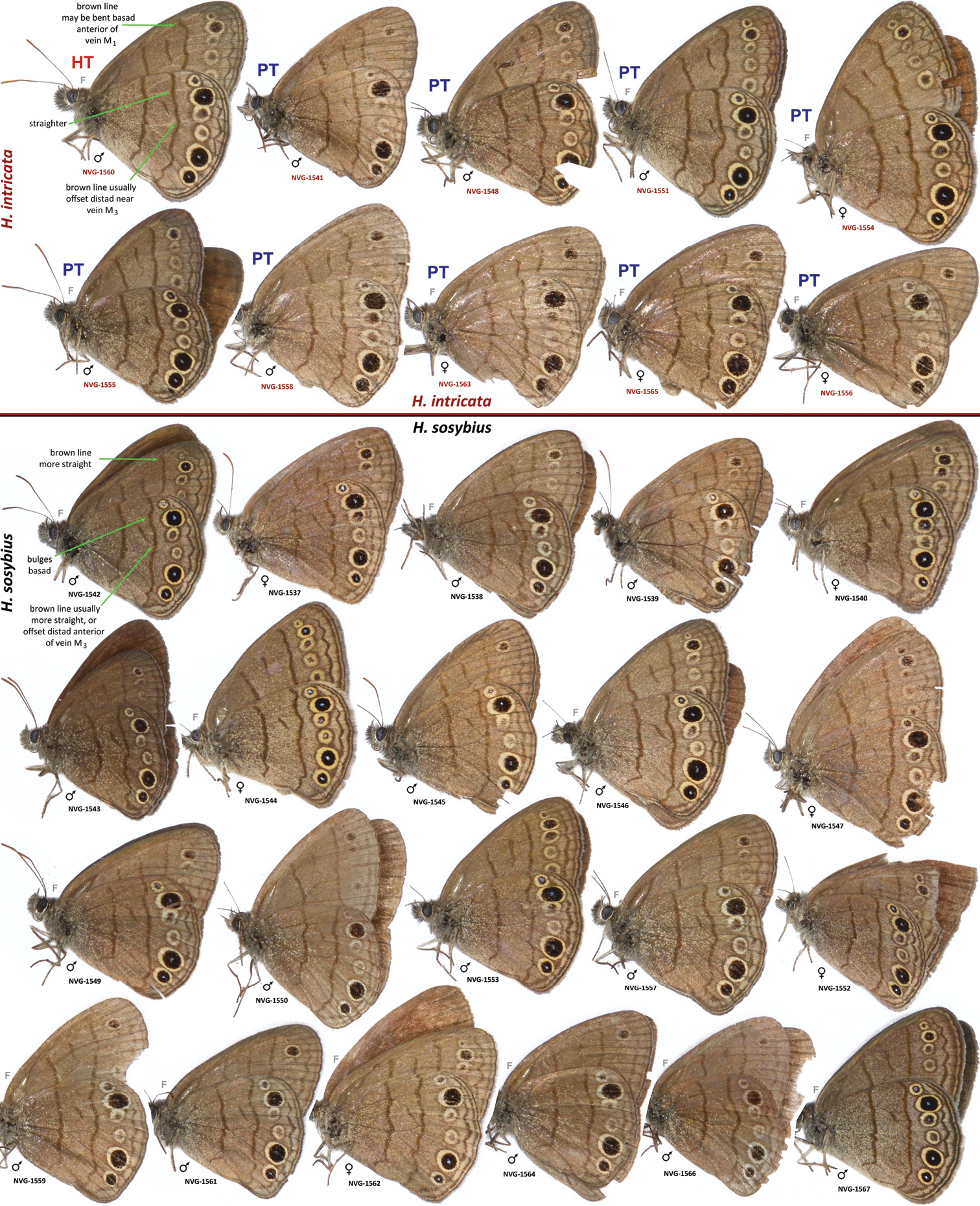
DNA-barcoded Hermeuptychia specimens from USA: Texas: Fort Bend Co., Brazos Bend State Park. Hermeuptychia intricata is above the line and Hermeuptychia sosybius is below the line, photographed prior to removal of body parts for DNA extraction. DNA voucher codes (see Table 1 for data) are shown below each specimen. Hypothetical field marks are indicated on the first specimen of each species. NVG-1537–NVG-1553 are from Horseshoe Lake trail, 29°22'54.96", −95°36'41.06", 15 m; and NVG-1554–NVG-1567 are from near Hale Lake, 29°22'48.27", −95°35'05.02", 16 m, all collected on 17-Aug-2013. Both species are present in each locality. Images are scaled approximately. “F” specifies mirror image (left-right inverted).
In wing pattern, the new species is most similar to Hermeuptychia sosybius, but typically can be differentiated from it by: (a) eyespots that are not only smaller, but also more uniform in size, i.e. out of 5 forewing eyespots, 4 (except the one near costa) are usually about the same size, and the eyespot that is black-ringed in most specimens (second from costa) is typically not the largest (this eyespot is frequently the largest in Hermeuptychia sosybius), but the next-to-last eyespot (4th from the costa) is usually the largest one; (b) more undulate postmedial line on ventral hindwing, that frequently strongly bulges basad by the largest eyespot near apex (in cell M1-M2); (c) more undulate submarginal sinuous line, which on ventral hindwing barely touches the largest eyespot near the tornus (in cell Cu1-Cu2, second eyespot from tornus, indicated in Fig. 57)–this line is usually fully merged with this eyespot border for some distance in Hermeuptychia sosybius. Wing-based identification is not absolute due to extensive pattern variation in both species.
In male genitalia, the new species is also closest to Hermeuptychia sosybius and should be attributed to the same morphogroup 4 of
Finally, the most confident identification is provided by DNA barcode sequences (Fig. 66) that show little variation within each species (most sequences are identical across the range, maximum difference below 1% in Hermeuptychia sosybius), but reveal a definitive 2% hiatus between central and south Texas populations (Figs 66–67). We selected all positions that were invariant in the barcode sample of each species but different between the two species as characters to differentiate Hermeuptychia hermybius from Hermeuptychia sosybius. The resulting 11 positions are listed in the format “k X (not Y)”, where k is a sequential number of the position (numbering is from 1 to 658 for the barcode sequence shown above as a reference), X is a nucleotide in Hermeuptychia hermybius barcodes and Y is a nucleotide in Hermeuptychia sosybius barcodes: 64 T (not C), 73 G (not A), 82 T (not C), 118 C (not T), 133 C (not T), 235 C (not T), 238 A (not G), 364 C (not T), 436 C (not T), 526 A (not T), 616 C (not T). These positions distinguish the two species; however, some of the positions are expected to show variation when a larger sample of sequence is accumulated.
The holotype of the new species, along with 21 paratypes are specimen reared in the lab from ova obtained from a captive female. All life history stages are illustrated in Fig. 70, and could be compared to the images of Hermeuptychia sosybius life history (Fig. 69). Immature stages of both species are very similar and without larger sample it is difficult to derive solid conclusions about the differences. Nevertheless, the following observations were made. Natural foodplants seems to be Panicum maximus (Guinea grass) per R. O. Kendall & C. A. Kendall, who reared caterpillars found on this grass in Mexico: Tamaulipas [TAMU collection]. This plant is also common in the lower Rio Grande Valley and is ubiquitously present where Hermeuptychia adults were encountered. Caterpillars hatched from eggs in captivity readily accepted Cynodon dactylon (L.) Pers. (Bermuda grass) and were successfully reared on it. Both Hermeuptychia sosybius and Hermeuptychia hermybius caterpillars go through four instars prior to pupation, and the first instar has black head capsule (Figs 69b–d, 70b–c). In subsequent instars, head capsule is green and round, without horns and projections (Figs 69e–m, 70d–m). Caterpillars of both species typically rest below leaves on loosely made silk pads, frequently in pairs, when two caterpillars face each other “head-to-head” (Figs 69h, 70c). When disturbed, caterpillars first curl into a C head-to-tail while legs being attached to the leaf (Figs 69f, j, 70e), then to a full O, head-to-legs (Fig. 70g). White dorsolateral spots in ultimate instar seem to be more pronounced in Hermeuptychia hermybius than in Hermeuptychia sosybius (compare Fig. 70g–k with 69k). Pupae of Hermeuptychia hermybius were stronger patterned with brown on the sides (Fig. 70o) than those of Hermeuptychia sosybius from two distant-from-each-other Texas localities (Fig. 69n–o), and some Hermeuptychia hermybius pupae were brown in color (Fig. 70n).
Life history of Hermeuptychia sosybius. USA: TX: Brazoria County, Bar-X Ranch, Rd. 971N, 29.13252, -95.58340, ex ovum ex ♀ collected on 4-Mar-2000, except o, which is TX: Wise Co., LBJ National Grassland. a ovum, 6-Mar-2000 b–d 1st instars, photographed on 14- 14- & 16-Mar-2000, respectively e–g 2nd instars photographed on 21- 19- & 21-Mar-2000 e, f are just after molt, shed larval skins are behind and 1st instar head capsule (black) is on the left in e, f is in a curled position adopted when disturbed h pre-molt quiescent 2nd instar larvae in a typical “head-to-head” resting position, 24-Mar-2000 i–j 3rd instars, 24- & 27-Mar-2000 k–l 4th (ultimate) instars, ♂♂, 3- & 6-Apr-2000 l close to pupation, note the color and shape change m prepupa, 6-Apr-2000 n–p pupae, 9-Apr-2000, 8-Aug-1998, & 17-Apr-2000 o is from Wise Co., wing color is starting to develop p near eclosion, dark adult is seen through semi-transparent pupal cuticle. Most images show different individuals. Images a–g are enlarged 2 times (scale on f) compared to the rest (scale on l).
Life history of Hermeuptychia hermybius. USA: TX: Cameron County, E of Brownsville, ex ovum ex ♀ collected on 18-Jan-2003. a ovum, 23-Jan-2003 b–c 1st instars, photographed on 30-Jan & 1-Feb-2003, respectively b prior to feeding, thus is white in color, c shows two caterpillars in a typical “head-to-head” resting position d 2nd instar, 10-Feb-2003 e–f 3rd instars 14- & 15-Feb-2003 g–k 4th (ultimate) instars, 25- 28- 26- 26- & 25-Feb-2003 g is in a curled position adopted when disturbed; l–m prepupae, 26- & 23-Feb-2003 n–p pupae, 10-Mar 25-Feb & 4-Mar-2003 n is a brown form, shed larval skin is still attached near cremaster in o and p and is hanging behind the pupa in n, p near eclosion, dark adult is seen through semi-transparent pupal cuticle. Most images show different individuals, those that eclosed are paratypes. Images a–d are enlarged 2 times (scale on d) compared to the rest (scale on k).
We pose and answer some questions that are likely to arise regarding our description of Hermeuptychia intricata and Hermeuptychia hermybius.
Can Hermeuptychia intricata be identified by wing patterns alone? The sample of 20 barcoded and dissected Hermeuptychia intricata specimens from the type series (plus 2 more dissected paratypes without barcode sequences) is too small to judge with confidence. At the moment, it seems risky to accept wings-only identification. However, comparative analysis of wing patterns suggests the following three characters that should be investigated further (marked on representative specimens in Fig. 68). First, the postmedial dark-brown line on the ventral hindwing in Hermeuptychia intricata bulges distad near the vein M3, in between the two smaller eyespots, closer to the posterior eyespot (e.g., compare Figs 32–47, 68). The line is more straight, or sinuous in Hermeuptychia sosybius, but frequently with a bulge anterior of the vein M3 (closer to large eyespot that is nearer the apex). Second, this line is relatively straight anterior of vein M2 in Hermeuptychia intricata, but is frequently bulged basad in Hermeuptychia sosybius. Third, in some Hermeuptychia intricata specimens, the postmedial dark-brown line on ventral forewing bends basad toward the costa (Fig. 68). More precisely, this line bends slightly distad towards the largest eyespot (in cell M1-M2) and then bends basad from near vein M1 (just anteriad of the largest eyespot) to costal margin. In Hermeuptychia sosybius, this line is typically straight or even bends distad towards costa. If the line bends basad, the bend is more gradual and begins posterior of M1 vein (posteriad of the largest eyespot). However, in some Hermeuptychia intricata specimens this bend is not present (Figs 29, 31, 34, 41). If these hypothesized field marks are indeed meaningful, then an individual photographed by
Are DNA barcode sequences necessary for confident identification ofHermeuptychia intricata? Although the DNA barcoding study has been instrumental in this project, we believe that its conclusions would hold without the knowledge of barcode sequences. Although we first noticed the difference in DNA barcodes, had we dissected the specimens prior to that, the presence of the two species (Hermeuptychia sosybius and Hermeuptychia intricata) and distinction between them would have become equally clear. We think male and female genitalia offer solid diagnostic characters that are sufficient for confident identification. These characters are numerous, are listed in the diagnosis above, and are illustrated in Figs 60–62 & 64. Therefore, DNA barcode sequences are not required for confident identification of this new species. Nevertheless, barcodes were very valuable to suggest that south Texas Hermeuptychia sosybius-like populations with typically smaller, more uniformly-sized eyespots and more undulate ventral hindwing lines are not conspecific with eastern USA populations, but represent a new species, Hermeuptychia hermybius. DNA barcodes were equally valuable to confirm that the name Hermeuptychia hermes should be best reserved for a South American species (
Should Hermeuptychia intricata be described now from a small sample? We believe that a description of a new species is an invitation to study it further. The discovery of a new butterfly species in the USA, especially rather distant evolutionarily from other members of the fauna (closest relatives in Bolivia and Brazil), is very exciting and should be made public without delays. It is interesting that the new species is cryptic in wing patterns, and its cryptic nature allowed it to remain unnoticed for over 200 years of research in butterfly taxonomy. Despite the small sample (type series of 22 specimens), we think that our taxonomic conclusions are solid, and genitalic differences are so pronounced that the species status of this taxon is fully justified. However, much remains to be studied, with the most obvious question being the distribution range of Hermeuptychia intricata. We hope that our description will stimulate its future studies, including those by citizen scientists and butterfly enthusiasts. Thus we think it is beneficial to describe this new species right away.
Could Hermeuptychia intricata be an extreme variation or a subspecies of Hermeuptychia sosybius? These two taxa are sympatric and synchronic. They could be found flying together at exactly the same spot, with two individuals of different species landing on the same leaf. We think that prominent and easily observable differences in both male and female genitalia are sufficient to strongly support distinction between Hermeuptychia intricata and Hermeuptychia sosybius as species under essentially any species concept (
Could Hermeuptychia intricata be a northern subspecies of Hermeuptychia gisella or Hermeuptychia cucullina? DNA barcode distance tree (Fig. 66a) shows that Hermeuptychia intricata is in the same clade with Hermeuptychia cucullina and Hermeuptychia gisella (bootstrap support for the clade is about 60%) and is more distant from sympatric Hermeuptychia sosybius. Even without the reconstructed tree, analysis of pairwise differences in DNA barcodes (Fig. 66a right, row and column 3 in the matrix) leads to the same conclusion: species closest to Hermeuptychia intricata are Hermeuptychia gisella (2.2% difference) and Hermeuptychia cucullina (2.4% difference). Hermeuptychia sosybius appears to be more distant at 3.4% difference. By genitalia, Hermeuptychia cucullina is quite different from the other species in its very short, thick and curved penis (
Should the neotype for Hermeuptychia sosybius be designated now? It is difficult to derive firm conclusions about taxa without firm identity. We think that it is not prudent to describe a new species sympatric with and hardly separable by wing patterns from Hermeuptychia sosybius without clarity about what the name Hermeuptychia sosybius stands for. Although the nature of Fabricius types of Hermeuptychia sosybius remains unclear and might never come to light, essentially everyone used and uses this name to refer to a common Hermeuptychia species widely distributed in the eastern US. Therefore it is sensible to stabilize this meaning by neotype designation. Data we gathered suggests which of the two US species should be Hermeuptychia sosybius (in the traditional use of the name) and a specimen of this species was selected as neotype.
What happened to Hermeuptychia hermes for USA populations? We agree with
Can Hermeuptychia hermybius be identified by wing patterns alone? The Hermeuptychia hermybius type series of 101 specimens from all 5 Texas Counties bordering the Rio Grande from Laredo to Brownsville and northeastern Mexico (Tamaulipas & San Luis Potosí) offers excellent material to study variation. This sample suggests that pattern-based identification using the three characters described in the diagnosis above is rather reliable, and Hermeuptychia hermybius (in contrast to Hermeuptychia intricata) could be mostly identified by wing patterns. Interestingly, while Hermeuptychia hermybius is much closer to Hermeuptychia sosybius phylogenetically than Hermeuptychia intricata, it seems easier to identify by wing patterns. Nevertheless, due to extreme variability of Hermeuptychia patterns, some specimens, especially those with smaller, underdeveloped eyespots and other elements of ventral hindwing pattern will not be identifiable by facies.
Are DNA barcode sequences necessary for confident identification of Hermeuptychia hermybius? Hermeuptychia hermybius is a species very close to Hermeuptychia sosybius. Our analysis shows that the best characters to tell between the two species are indeed DNA barcodes (Fig. 66). However, we think that wing-based and male genitalia-based (using measurements and graph in Fig. 63) identification will be unambiguous in most cases without knowing the locality of a specimen. Nevertheless, it is likely that some specimens would not be identifiable with confidence in the absence of DNA barcodes.
Could Hermeuptychia hermybius be a southern subspecies of Hermeuptychia sosybius? In our opinion, consistency between the differences in DNA barcodes, wing patterns, male genitalia and historic treatment of south Texas populations as a distinct species by several authors (
Is there a specimen age limit for successful DNA barcoding? The oldest specimens we succeeded with were about 120 years old. These were the oldest specimens we tried. We see that DNA fragments into smaller pieces with age. Therefore, for successful amplification, primers for shorter DNA segments should be designed. Per Materials and methods section, we developed primers for two very short (75 bp and shorter) segments of DNA that contain the highest density of differences between the three US Hermeuptychia species. We called these regions ID tags. These tags were successfully and consistently amplified from over 100 years old specimens. Example of the results is shown in Fig. 65. The ID tags allowed us to identify these specimens by DNA and these identifications always matched genitalic identifications. This method could be applied to older specimens and is expected to yield similar success. Potentially, even the oldest preserved butterfly specimen could contain usable DNA that can be extracted and amplified with current methods.
What could be the English name for Hermeuptychia intricata and Hermeuptychia hermybius? Although some object to using “common” names for butterflies (i.e. the names in the native language of a country the insect inhabits), especially in research publications, we believe that common names are beneficial to attract public interest to butterflies and to disseminate knowledge about them more efficiently, thus possibly aiding their conservation. We suggest “Intricate Satyr” as the English name for Hermeuptychia intricata to indicate its delicate wing patterns, cryptic nature, difficulty of identification, and the fact that it has remained overlooked in over 200 years of exploration of North American butterflies despite very significant differences in genitalia. We propose the English name “South Texas Satyr” for Hermeuptychia hermybius to emphasize its type locality and distribution in the US, and to highlight the importance of South Texas in the studies of butterfly fauna.
Qian Cong is a Howard Hughes Medical Institute International Student Research fellow. We acknowledge Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the permit #08-02Rev making research based on material collected in Texas State Parks possible. We are grateful to Edward G. Riley (Texas A & M University insect collection, College Station, TX), Robert K. Robbins, John M. Burns and Brian Harris (National Museum of Natural History, Smithsonian Institution, Washington DC), David Lees and Blanca Huertas (Natural History Museum, London, UK), for facilitating access to the collections under their care and stimulating discussions; to Bill Dempwolf for collecting part of the Hermeuptychia hermybius type series; to André V. L. Freitas (Instituto de Biologia, Universidade Estadual de Campinas, Campinas, SP Brazil) and Jonathan P. Pelham for fruitful discussions; to John V. Calhoun for information about historic facts and literature; to Jude Philp and Robert Blackburn (Macleay Museum, The University of Sydney, Sydney, Australia) for discussions of Macleay collection and photographs of possible Hermeuptychia sosybius syntypes; to Kathleen Santry (Oxford University Museum of Natural History, Oxford, UK) for the images of Hermeuptychia sosybius illustrations by Jones; Peter N. Grishin for assistance with translations from Latin; Ralf H. Anken for sending the holotypes of Hermeuptychia taxa described by him and many useful discussions; to Gerardo Lamas (Museo de Historia Natural, Universidad Nacional Mayor de San Marcos, Lima, Peru) for most helpful discussions, critical review of the manuscript and insightful suggestions that lead to significant improvement of this work, to Kim Garwood, Bryan Reynolds and Brian Banker for reading the manuscript and corrections, and to the reviewers for helpful comments and suggestions.