(C) 2012 Yun Bu. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Littoral dipluran Parajapyx pauliani Pagés, 1959 was redescribed based on the specimens collected in Hainan Island, South China. The littoral habitat was confirmed for the species, as the first report of arenicolous dipluran in China. DNA barcoding fragment was sequenced for five Parajapyx species (18 individuals) from China, and this is the first report on DNA barcodes used for dipluran identification. The mean intra- and interspecific divergencesare 1.9% and 19.1% respectively. Synonymy of Parajapyx paucidentis and Parajapyx isabellae was confirmed.
Diplura, Parajapyx, littoral, DNA barcodes analysis, China
The genus Parajapyx was erected by
Later, Parajapyx was divided into two subgenera (Grassjapyx and Parajapyx) according to the shape of cerci (inner margin of cerci straight, tooth 1 not separated from others by a sinus in Grassjapyx vs. teeth 2-5 or 3-5 on a convexity of the internal margin, tooth 1 is separated from others by a sinus in Parajapyx) (
In April 2011, during the research of the diversity of basal hexapods in littoral of Asia-Pacific coast, seven specimens of Parajapyx were collected from intertidal zone of several beaches of Hainan Island, South China. Those specimens were identified as Parajapyx pauliani Pagés, 1959, which was firstly described based on only specimen from intertidal zone of Nosy Be, Madagascar Island, and Pagés doubted about the habitat where the species was collected (
In this study, we provided a detailed redescription of this species based on our specimens, and more discussion on its littoral habitat. We analyzed the DNA barcoding sequences (
With flotation method, the specimensof Parajapyx pauliani were collected directly from the water surface in Hainan, China, and stored in 80% ethanol. Specimens of other species were extracted by the Tullgren funnels from soil samples (Table 1). For Parajapyx isabellae, two individuals of its synonym Parajapyx paucidentis identified from the morphology were also sampled.
Parajapyx species and outgroups used in the study.
| Classification | Species | Locality | Number of individuals | GenBank Accession Numbers |
|---|---|---|---|---|
| Diplura | ||||
| Parajapygidae | ||||
| Parajapyx | Parajapyx pauliani | Hainan | 2 | JQ692327, JQ796634 |
| Parajapyx emeryanus | Shanghai | 6 | JQ796635-JQ796640 | |
| Parajapyx isabellae | Shanghai | 5 | JQ796641-JQ796645 | |
| Parajapyx isabellae (Syn. Parajapyx paucidentis) | Shanghai | 2 | JQ796646, JQ796647 | |
| Parajapyx hwashanensis | Qinghai | 1 | JQ796648 | |
| Parajapyx yangi | Gansu | 2 | JQ796649, JQ796650 | |
| Japygidae | Occasjapyx japonicus | Shanghai | 1 | HQ882833 |
| Campdeidae | Lepidocampa weberi | Shanghai | 1 | HQ882832 |
Seven specimens of Parajapyx pauliani were collected: four of which were mounted in Hoyer’s solution for identification, two were morphological identified in the alcohol first and then used for DNA extraction, and one was reserved in pure alcohol. Measurements and photos were taken by the help of a phase contrast microscope NIKON E600. The species was identified by the comparison of characters of all known species of the genus. For the name of chaetotaxy, we used the nomenclature proposed by
Abbreviations. Ant. I-XXI= antenna segments I-XXI; BS= baculiform sensillum; M = macroseta; the position on dorsal of body as: ma = medial anterior, la= lateral anterior, mp = medial posterior, lp = lateral posterior; ms= microsensillum; m = microseta, n* = normal seta; s=sensillum; t1-t5= teeth of cercus.
* including all “s” setae named by
Eighteen individuals from five Parajapyx species were used for DNA barcoding analyses (Table 1), and two dipluran specimens from Japygidae and Campodeidae were used as the outgroups. All specimens were morphological identified in the alcohol first and then used for DNA extraction. We followed the experimental procedure for Collembola described in
DNA sequences were analyzed with the software DNASTAR (
4 females, South China, Hainan Island, Sanya city, shingly beach of Ximaozhou island (samples No. 6 and 8), 18°14'N, 109°22'E, 5-IV-2011; 1 female, from sand beach of the Ximaozhou Island (sample No. 17), 6-IV-2011; 2 female, South China, Hainan Island, Changjiang County, Changhua town, from sand beach of Qizi Bay (sample No. 54), 19°21'N, 108°40'E, 7-IV-2011, coll. Y. Bu, C. W. Huang, M. B. Potapov and N. A. Kuznetsova. All specimens are kept at Institute of Plant Physiology & Ecology, CAS.
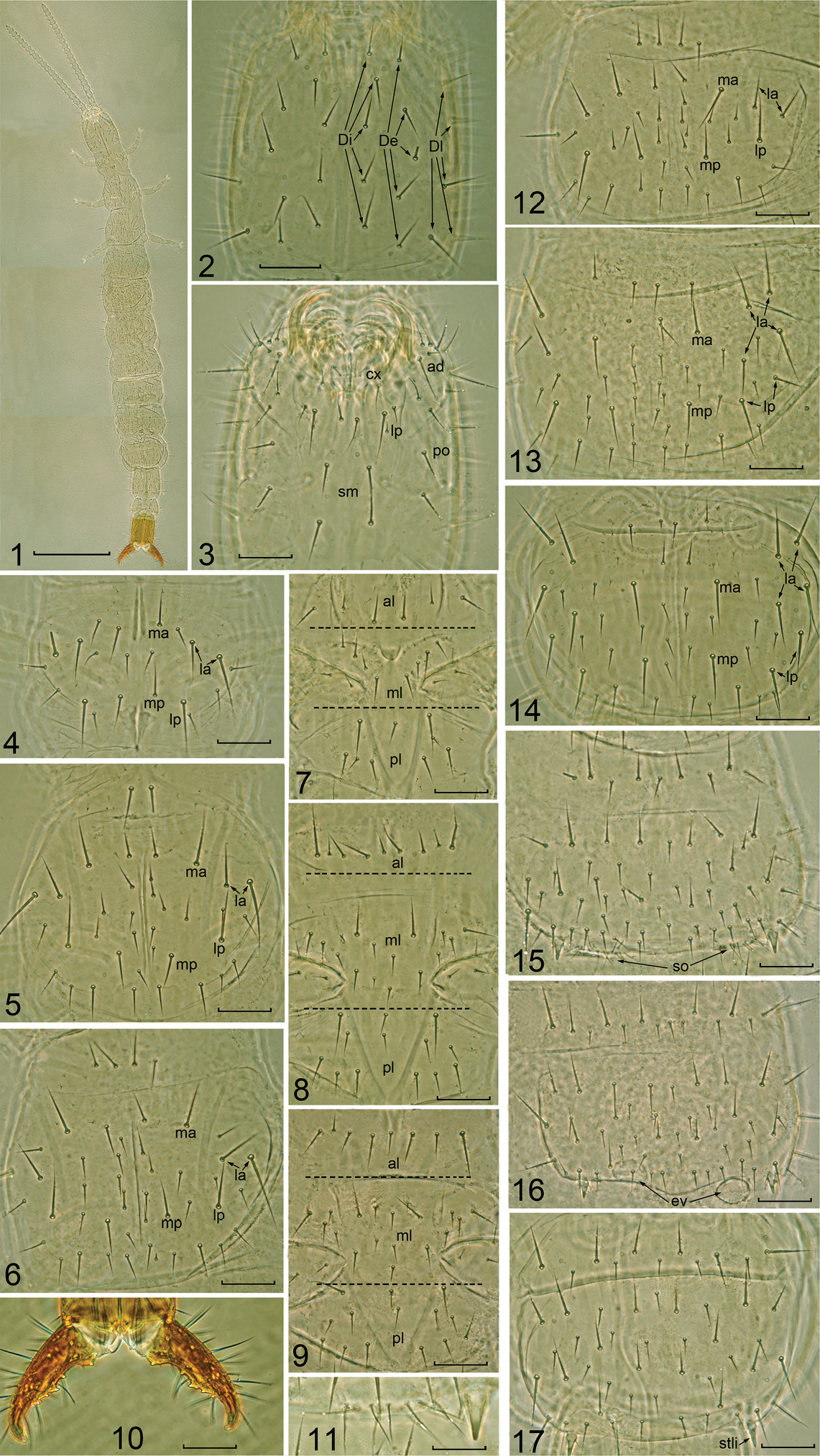
Body length and width of adult female 2.8–3.0 mm, and 0.3–0.35 mm, respectively (four specimens, antenna and cerci not included). Tegument smooth, without ornamentation (Fig. 1).
Length 0.23–0.25 mm, width 0.23–0.25 mm. Dorsal side with 5+5 interior (Di), 5+5 exterior (De), and 10+10 lateral setae (Dl) (only show five on the picture), without front setae (Fig. 2). Labrum with two pairs of medial setae (1+1 M and 1+1 n), 6+6 m. On ventral side internal lobe (li) with 1+1 m; external lobe (le) with 9 +9 setae; coxae (cx) with 1 M and 3n; labial palpus absent, replaced with 1 M accompanied by two normal seate; admentum with 11 setae, 3 M and 8 n; pli oral region with 4–5 setae; submentum with 2+2 setae (Fig. 3).
Mouthparts. Lacinia composed by five lobes, the first lobe (distal) is very acute and smooth, and the following four larger and pectinate. Mandible with five teeth and three denticles between them. Maxillary palpus with 10 n and 2 m setae.
Antenna with 21 segments, length 0.8 mm. Antenna segment I with seven microsetae dorsally and 5 setae, Ant. II and III each with 9 setae, Ant. IV with 11 setae, Ant. I-IV without sensilla and trichobothria, Ant. V with two bacilliform sensilla (BS) and 14–17 setae, Ant. VI with three BS and 16–18 setae, Ant. VII with three BS and 17–20 setae; Ant. VIII-XIX each with 4 BS and 18–28 setae, Ant. XX with 6 BS and 34–35 setae, Ant. XXI with eight BS and four placoid sensilla, and 55–60 seate. Single microsensillum asymmetrically present on Ant. IX-XIII, XVII, and XIX.
Thorax. Chaetotaxy of thorax as show in Table 2, 3. Pro-, meso- and metanotum each with 5+5 M setae and 6-17 n setae (Figs 4–6). Pro-, meso- and metasternum as show in Figs 7–9. Leg III length 0.3 mm, coxa with 1 M, 3 n and 2 m; trochanter with 1 M and 2 n dorsally, 1 m ventrally; femur with 10 n and 3 m setae; tibia with 8 n; tarsus with 10 n; claw symmetrical and with single medial unguis.
Abdomen. Chaetotaxy of the abdomen as shown in Table 2 and 3. Urotergite I (Fig. 12): prescutum with 4+4 m and 2+2 n, scutum with 6+6 m, 5+5 M and (10-14)+(9-14) n. Urotergites II-VII (Figs 13-14): prescutum with 4+4 m and 2+2 n, scutum with (6-8)+(6-8) m, 8+8 M and (9-18)+(9-19) n. Urotergite VIII with 12+12 m, 8+8 M and (9-10)+2(3)+(7-13) n. Urotergite IX with 7+7 m, 3+3 M and 2+1+2 n. Urotergite X with 4+4 m, 6+1+6 M and (6-8)+(6-8) n.
Urosternite I (Fig. 15): prescutum with 2+2 m and 5 +5 n, scutum with 5+5 m, 10+10 M and (10-18)+1(2)(3)+(11-18) n. Subcoxal organ composed by 10-13 setae, including 6-8 slender glandular setae and 4-5 sensory setae, without medial glandular organ (Fig. 11). Urosternites II to VII (Figs 16-17): prescutum with 4 +4 m and (5-7)+1+(5-7) n setae, scutum with 5 +5 m, 12 +12 M, (7-12) + 2(3)+(6-11) n. Urosternite VIII with 4+4 m, 2 +2 M, (4-5)+(4-5) n. Urosternite IX with 4+4 m, 2 +2 M and 3+3 n. Urosternite X with 4 +4 m, 6+6 M and 3+1+3 n. Eversible vesicles present on the urosternites II-III, diameter 32-37 μm. Styli on urosternites I-III each with one short sensilla and one m seta, on urosternites IV-VII with single seta m. Female genital papilla with 10+2+10 n.
Cerci (Fig. 10) singly segmented, symmetrical, with five distinct internal teeth, crooked; t3 larger than others; interval between t1-t2 as two times as t2-t3 and t3-t4; t2-t4 with shoulder, dorsal side with 9 M, 5 n and 5 m, ventral side with7 M, 3 n and 3 m; each cercus with 7–8 evaporation plates.
Chaetotaxy of dorsal side of body in adult Parajapyx pauliani
| segments | m | M | n | ||||
|---|---|---|---|---|---|---|---|
| ma | mp | la | lp | ||||
| Pronotum | 6+6 | 1+1 | 1+1 | 2+2 | 1+1 | 6+6 | |
| Mesonotum | Prescutum | 6+6 | 1+1 | ||||
| Scutum | 3+3 | 1+1 | 1+1 | 2+2 | 1+1 | (8-13)+(8-12) | |
| Metanotum | Prescutum | 7+7 | 2+2 | ||||
| Scutum | 3+3 | 1+1 | 1+1 | 2+2 | 1+1 | (13-17)+(10-16) | |
| Abd. I | Prescutum | 3+3 | 2+2 | ||||
| Scutum | 5+5 | 1+1 | 1+1 | 2+2 | 1+1 | (10-14)+(9-14) | |
| II | Prescutum | 4+4 | 2+2 | ||||
| Scutum | 6+6 | 1+1 | 1+1 | 4+4 | 2+2 | (11-17)+(11-17) | |
| III | Prescutum | 4+4 | 2+2 | ||||
| Scutum | 7+7 | 1+1 | 1+1 | 4+4 | 2+2 | (14-17)+(11+17) | |
| IV | Prescutum | 4+4 | 2+2 | ||||
| Scutum | 7+7 | 1+1 | 1+1 | 4+4 | 2+2 | (11-17)+(11-18) | |
| V | Prescutum | 4+4 | 2+2 | ||||
| Scutum | 7+7 | 1+1 | 1+1 | 4+4 | 2+2 | (12-17)+(13-18) | |
| VI | Prescutum | 4+4 | 2+2 | ||||
| Scutum | 7+7 | 1+1 | 1+1 | 4+4 | 2+2 | (12-14)+(10-16) | |
| VII | Prescutum | 4+4 | 2+2 | ||||
| Scutum | 8+8 | 1+1 | 1+1 | 4+4 | 2+2 | (9-14)+(9-14) | |
| VIII | Scutum | 12+12 | 1+1 | 1+1 | 4+4 | 2+2 | (9-10)+2(3)+(7-13) |
| IX | Scutum | 7+7 | 3+3 | 2+1+2 | |||
| X | Scutum | 4+4 | 6+1+6 | (6-8)+(6-8) | |||
Chaetotaxy of ventral side of body in adult Parajapyx pauliani
| segments | m | M | n | |
|---|---|---|---|---|
| Prosternum | Anterior lobe | 3+7+3 | 2+2 | 1+1 |
| Middle lobe | 7+7 | 2+2 | 2+2 | |
| Posterior lobe | 6+6 | 1+1 | 2(3)+1+2(3) | |
| Mesosternum | Anterior lobe | 3+3 | 2+2 | 2+2 |
| Middle lobe | 7+7 | 4+4 | 2+2+2 | |
| Posterior lobe | 6+6 | 3+3 | 3+2+3 | |
| Metasternum | Anterior lobe | 4+4 | 2+2 | 2+1+2 |
| Middle lobe | 5+5 | 4+4 | 3+2+3 | |
| Posterior lobe | 5+5 | 3+3 | 3+2+3 | |
| Abd. I | Prescutum | 2+2 | 5+5 | |
| Scutum | 5+5 | 10+10 | (10-18)+2(3)+(11-18) | |
| II | Prescutum | 4+4 | (5-7)+1+(5-7) | |
| Scutum | 5+5 | 12+12 | (8-11)+2(3)+(8-11) | |
| III | Prescutum | 4+4 | 6(7)+1+6(7) | |
| Scutum | 5+5 | 12+12 | (8-9)+2(3)+(8-11) | |
| IV | Prescutum | 4+4 | 6(7)+1+6(7) | |
| Scutum | 5+5 | 12+12 | (8-11)+2(3)+(8-11) | |
| V | Prescutum | 4+4 | (5-7)+1+(5-7) | |
| Scutum | 5+5 | 12+12 | (7-12)+2+(7-10) | |
| VI | Prescutum | 4+4 | 6+1+6 | |
| Scutum | 5+5 | 12+12 | (8-12)+2+(7-10) | |
| VII | Prescutum | 4+4 | 5+1+5 | |
| Scutum | 5+5 | 12+12 | (9-10)+2+(6-9) | |
| VIII | Scutum | 4+4 | 2+2 | (4-5)+(4-5) |
| IX | Scutum | 4+4 | 2+2 | 3+3 |
| X | Scutum | 4+4 | 6+6 | 3+1+3 |
Parajapyx pauliani 1 Habitus 2 head, dorsal view (Di= dorsal interior setae; De= dorsal exterior setae; Dl= dorsal lateral setae) 3 head, ventral view (ad= admentum cx= coxae lp= labial palps area sm= submentum po= pli oral region) 4 pronotum 5 mesonotum 6 metanotum 7 prosternum (al= anterior lobe ml= middlelobe pl= posterior lobe, same for figs 8–9) 8 mesosternum 9 metasternum 10 cerci 11 subcoxal organ of urosternite I, right side 12 urotergite I 13 urotergite II 14 urotergite VII 15 urosternite I (so= subcoxal organ) 16 urosternite II (ev= eversible vesicles) 17 urosternite VII. Scale bar: 0.5 mm in Fig. 1; 0.1 mm in Figs 2–17.
So far, the species is known only from two localities: Hainan, China and Madagascar.
Parajapyx pauliani is characterized by the antenna with 21 segments, nota each with 5+5 M setae and numerous normal setae, urotergites II-VII each with 8+8 M setae and numerous normal setae, and prescutum of urotergites II-V each with 2+2 normal setae. Ithas more normal setae than in other congeners. The numbers of M and m setae are relatively stable, but the numbers of normal setae are quite variable in different individuals.
Littoral habitat of Parajapyx pauliani.Three intertidal locations where Parajapyx pauliani was found are shown in Figs 18–20. All habitats are positioned lower than supralittoral, devoid of halophytes, and are directly influenced by sea water. The animal lives in shingly or sand beaches (Figs 18–20), between particles of different size: from 9 mm (with the whole variation from 5 to 16 mm, n=100) to 1.5 mm (1.0–2.3 mm, n=150) in diameter. Parajapyx pauliani appears to be a dipluran member of a genuine littoral community and is often associated with collembolan species like Yuukianura sp., Isotogastrura trichaetosa
Habitats of Parajapyx pauliani in Hainan (S China). 18 shingly beach of Ximaozhou Island, inset shows the size of stone 19 sand beach of Ximaozhou Island, inset show the size of sand granules 20 sand beach of Qizi Bay, inset show the size of sand granules ✱ indicates the sample site.
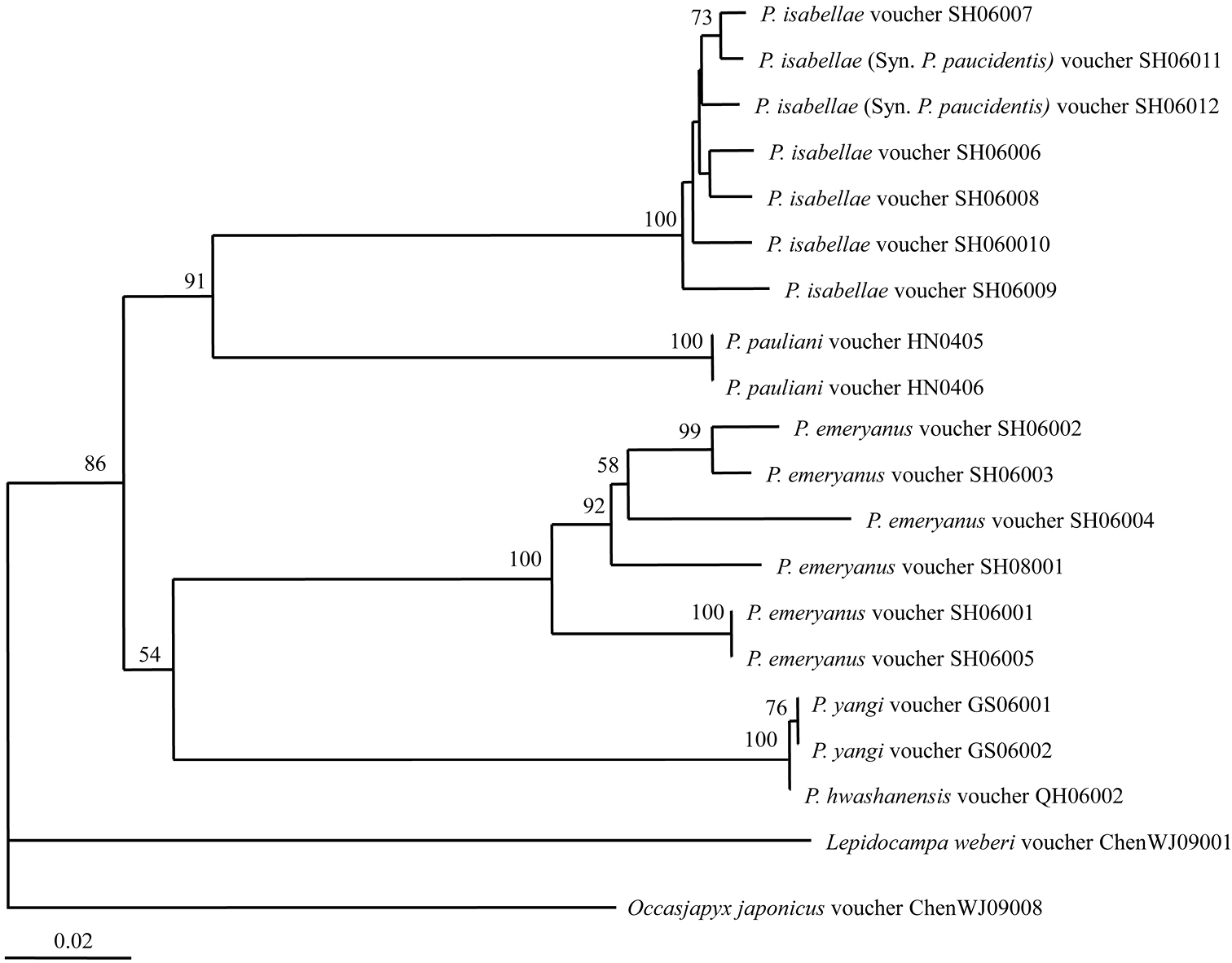
Neighbour-joining tree (p-distance, Bootstrap 1000 replicates) of Chinese Parajapyx inferred from COI gene sequences. Numbers on the nodes show the bootstrap values (> 50%).
The DNA barcoding of 18 individuals from five Parajapyx species from China were sequenced, and deposited in GenBank (the accession numbers showed in Table 1). The genetic divergence between individuals of the same species is 1.9% in average, with span 1.5–5.3%, and it is 19.1% in average, with span 16.3–21.3% between different Parajapyx species.
The Neighbour-joining tree was constructed based on the barcoding sequences (Fig. 21). Parajapyx pauliani is clustered with Parajapyx isabellae. Parajapyx isabellae and Parajapyx emeryanus are valid species respectively well supported by barcoding analyses. Two individuals of Parajapyx isabellae (Syn. Parajapyx paucidentis) (teeth absent on the cerci) clustered together with five individuals of Parajapyx isabellae (teeth present on the cerci). The genetic divergence between Parajapyx isabellae (Syn. Parajapyx paucidentis) and Parajapyx isabellae is only 1.7% in average (with span 0.8–2.6%). In addition, individuals of Parajapyx yangi and Parajapyx hwashanensis clustered together with high support value, and the genetic divergence between them is low (0.2%).
This is the first record of littoral dipluran in China. When Parajapyx pauliani was first found in intertidal zone in 1959, Pagés supposed that it is “purely fortuitous, and the single specimen collected was, in fact, might be pulled far away from its normal habitat by runoff” (
Numerous normal setae on body of Parajapyx pauliani are shared with Parajapyx botosaneanui Pagés, 1975, described from intertidal zone of Caribbean coast of Cuba (
The DNA barcodes have been widely used in identification of microarthropod species, for instance, collembolans (
The formal morphological difference in second problematic couple, Parajapyx yangi and Parajapyx hwashanensis, is the number of teeth on the cerci: the former species has four teeth, while the latter has five. Our DNA barcoding data showed only one nucleotide difference between examined individuals of Parajapyx yangi and Parajapyx hwashanensis. All individuals, identified formally by us as Parajapyx yangi were, however, immature that indicated the possible age nature of this differences. The type materials of the two species call for study to make the final conclusions.
We sincerely thank Natalya A. Kuznetsova who taken the photos for the size of granules, and Cheng-Wang Huang for their help during the collection. Special thanks are given to Rong-Dong Xie for his suggestion on species identification. We also owe our deepest gratitude to Markus Koch (Germany), Alberto Sendra (Spain) and Arturo García-Gómez (Mexico) for generously providing good suggestions and references of Parajapyx. The study was supported by the National Natural Sciences Foundation of China (30870282, 31071911, 31071887), NSFC-RFBR Cooperative Research Project (31111120077 / 11-04-91179-GFENa), and Bureau of International Co-operation Chinese Academy of Sciences.