(C) 2013 Eric Alfonsi. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Alfonsi E, Méheust E, Fuchs S, Carpentier F-G, Quillivic Y, Viricel A, Hassani S, Jung J-L (2013) The use of DNA barcoding to monitor the marine mammal biodiversity along the French Atlantic coast. In: Nagy ZT, Backeljau T, De Meyer M, Jordaens K (Eds) DNA barcoding: a practical tool for fundamental and applied biodiversity research. ZooKeys 365: 5–24. doi: 10.3897/zookeys.365.5873
In the last ten years, 14 species of cetaceans and five species of pinnipeds stranded along the Atlantic coast of Brittany in the North West of France. All species included, an average of 150 animals strand each year in this area. Based on reports from the stranding network operating along this coast, the most common stranding events comprise six cetacean species (Delphinus delphis, Tursiops truncatus, Stenella coeruleoalba, Globicephala melas, Grampus griseus, Phocoena phocoena)and one pinniped species (Halichoerus grypus). Rare stranding events include deep-diving or exotic species, such as arctic seals. In this study, our aim was to determine the potential contribution of DNA barcoding to the monitoring of marine mammal biodiversity as performed by the stranding network.
We sequenced more than 500 bp of the 5’ end of the mitochondrial COI gene of 89 animals of 15 different species (12 cetaceans, and three pinnipeds). Except for members of the Delphininae, all species were unambiguously discriminated on the basis of their COI sequences. We then applied DNA barcoding to identify some “undetermined” samples. With again the exception of the Delphininae, this was successful using the BOLD identification engine. For samples of the Delphininae, we sequenced a portion of the mitochondrial control region (MCR), and using a non-metric multidimentional scaling plot and posterior probability calculations we were able to determine putatively each species. We then showed, in the case of the harbour porpoise, that COI polymorphisms, although being lower than MCR ones, could also be used to assess intraspecific variability. All these results show that the use of DNA barcoding in conjunction with a stranding network could clearly increase the accuracy of the monitoring of marine mammal biodiversity.
DNA barcoding, COI, control region, marine mammals, cetaceans, pinnipeds, biodiversity monitoring, stranding network
The aim of DNA barcoding is to concentrate the efforts of molecular taxonomists on a single part of the mitochondrial genome, chosen because it presents portions conserved across taxa that are appropriate for primer design, while including polymorphism among and within species (
DNA barcoding also possesses some inherent limitations (
In the present study, we assess the contributions that DNA barcoding could provide to the monitoring of the marine mammal biodiversity along the coasts of Brittany, in the North West of France. For almost 20 years, the stranding network has been collecting data and, when possible, sampling, each time a marine mammal stranding is reported. Field correspondents are organized in a geographical area covering the entire Brittany coasts. The network is coordinated regionally by Océanopolis (Brest, France), and nationally by Pelagis (La Rochelle, France).
DNA barcoding could be useful for the monitoring of marine mammal strandings at different levels. First, by confirming the quality and the reproducibility of a species identification made by the field correspondents. Beside common species, which are often encountered and easily identified, exotic or deep living species represent rare stranding events. In such cases, DNA barcoding could provide a confirmation or an additional degree of precision of taxonomic determination (
We evaluated the usefulness of DNA barcoding in the monitoring of marine mammal biodiversity along the coasts of Brittany at three levels: by confirming the taxonomic identification performed by field correspondents, by identifying degraded carcasses or parts of carcasses, and by determining intraspecific variations for two species commonly found off Brittany, the harbour porpoise and the grey seal. For this last part of our study, we also compared COI and the mitochondrial control region in terms of their effectiveness in species identification.
The CRMM (Centre de Recherche sur les Mammifères Marins, La Rochelle, France), presently the Joint Service Unit PELAGIS, UMS 3462, University of La Rochelle-CNRS has created the French marine mammal stranding recording program at the beginning of the 70s. The network comprises about 260 field correspondents, members of organizations or volunteers (
Since 1995, the LEMM (Laboratoire d’Etude des Mammifères Marins, Océanopolis, Brest, France) has coordinated this network at a regional scale in Brittany, North West of France. Data are collected from the Brittany coastlines, analyzed, and then added to the central database maintained in La Rochelle. The Brittany coasts have been divided into 18 sections covering the whole coastline (
Genomic DNA was extracted from blood samples or from muscle and skin tissues using a standardized protocol and the DNeasy Blood and Tissue kit (Qiagen), following the instructions of the manufacturer. The quality and the concentration of all the DNA extracts were estimated by agarose gel electrophoresis and by spectrophotometry using a Nanodrop 1000 (Thermo Scientific).
A 736 base-pair (bp) fragment of the 5’ region of the COI fragment (position 5352 to 6087 of the complete mitochondrial genome of the harbour porpoise, GenBank acc. no. AJ554063), was amplified using two newly designed primers, LCOIea (5’-tcggccattttacctatgttcata-3’) and HBCUem (5’-ggtggccgaagaatcagaata-3’). The 50 µl PCR final volume included approximately 50 ng of genomic DNA, and 25 pmole of each primer in the Hotgoldstar master mix x 1 (Eurogentec) with a final concentration of MgCl2 of 2.5 mM. After an initial denaturation step of 10 min at 95 °C, the thermocycle profile consisted of 32 cycles for cetaceans or 35 cycles for pinnipeds at 95 °C for 30 s, 53 °C for 30 s and 72 °C for 60 s, with a final extension at 72 °C for 10 min.
For some animals, we also amplified and sequenced another part of the mitochondrial genome including the control region (MCR). For cetaceans, the primers and reaction conditions are described in (
Electropherograms were analyzed and edited manually using the Sequence scanner software (Applied Biosystems), and alignments were produced using CLUSTAL W (
DNA sequences and specimen information have been added to two BOLD projects. The first project includes specimens for which the species had been identified without doubt using classical morphological identification, and is referred to as IMMB (Identified Marine Mammals in Brittany). The IMMB project is a part of the campaign “barcoding mammals of the world”. The second project, UMMB (Unidentified Marine Mammals in Brittany), includes specimens only identified to the genus or to higher taxonomic levels. This second project is a part of the campaign “barcoding application”.
Genetic distances (intraspecific, interspecific and minimal distance to the nearest neighbour) were calculated using the Kimura 2-parameter (K2P) model (
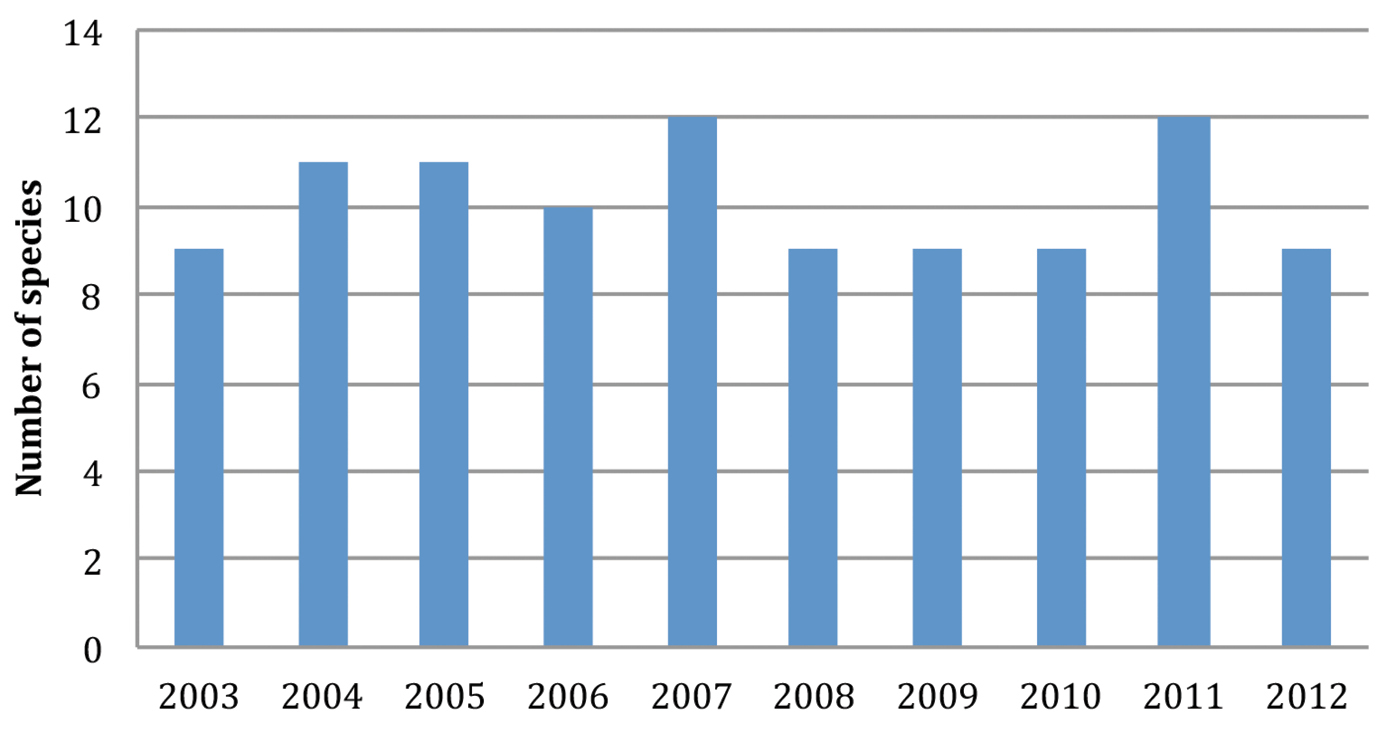
From 2003 to 2012, 1530 marine mammal strandings were recorded along the coastline of Brittany (Table 1). Fourteen species of cetaceans and five species of pinnipeds were identified. The most frequent cetaceans were six indigenous species of the Brittany waters, viz. five members of the Delphinidae (Delphinus delphis, Tursiops truncatus, Stenella coeruleoalba, Globicephala melas, Grampus griseus), and the harbour porpoise (Phocoena phocoena). Two members of the Zyphiidae (Hyperoodon ampullatus and Ziphius cavirostris), three other species of Delphinidae (Lagenorhynchus acutus, Orcinus orca and Stenella frontalis), one species of Physeteridae (Physeter macrocephalus) and two mysticete species (Balaenoptera acutorostrata and Balaenoptera physalus) were rare stranding events. Halichoerus grypus was by far the most commonly encountered pinniped, far before Phoca vitulina, and some uncommon arctic seals (Phoca hispida, Cystophora cristata and Phoca groenlandica). Between 9 and 12 different marine mammal species stranded each year (Figure 1).
Numbers of different species of marine mammals stranded along the coasts of Brittany (North West of France) in the period 2003–2012.
Strandings of marine mammals along the coasts of Brittany, North West of France (2003–2012)
| 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cetaceans | |||||||||||
| Balaenoptera acutorostrata | 1 | 1 | 2 | ||||||||
| Balaenoptera physalus | 1 | 2 | 2 | 3 | 4 | 2 | 14 | ||||
| Delphinidae (undetermined) | 40 | 30 | 36 | 22 | 15 | 9 | 9 | 6 | 16 | 8 | 191 |
| Delphinus delphis | 56 | 61 | 109 | 53 | 51 | 56 | 40 | 39 | 72 | 57 | 594 |
| Globicephala melas | 6 | 5 | 7 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 28 |
| Grampus griseus | 2 | 1 | 7 | 3 | 1 | 7 | 2 | 1 | 2 | 4 | 30 |
| Hyperoodon ampullatus | 1 | 1 | |||||||||
| Lagenorhynchus acutus | 1 | 2 | 1 | 1 | 5 | ||||||
| Orcinus orca | 1 | 1 | |||||||||
| Phocoena phocoena | 18 | 13 | 12 | 15 | 20 | 23 | 9 | 10 | 15 | 11 | 146 |
| Physeter macrocephalus | 2 | 1 | 3 | ||||||||
| Stenella coeruleoalba | 1 | 7 | 9 | 8 | 4 | 5 | 9 | 6 | 3 | 52 | |
| Stenella frontalis | 1 | 1 | |||||||||
| Tursiops truncatus | 6 | 2 | 7 | 6 | 4 | 5 | 3 | 8 | 3 | 3 | 47 |
| Ziphius cavirostris | 1 | 1 | 2 | ||||||||
| Mysticeti (undetermined) | 1 | 4 | 5 | ||||||||
| Odontoceti (undetermined) | 5 | 1 | 1 | 3 | 1 | 11 | |||||
| Cetacea (undetermined) | 3 | 3 | 2 | 1 | 9 | ||||||
| Pinnipeds | |||||||||||
| Cystophora cristata | 1 | 3 | 4 | ||||||||
| Halichoerus grypus | 20 | 29 | 41 | 37 | 51 | 41 | 37 | 13 | 34 | 24 | 327 |
| Phoca groenlandica | 1 | 1 | |||||||||
| Phoca vitulina | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 12 | ||
| Pusa hispida | 1 | 1 | 2 | ||||||||
| Phocidae (undetermined) | 5 | 7 | 4 | 13 | 4 | 5 | 2 | 1 | 41 | ||
| Unknown | 1 | 1 | |||||||||
| Total | 161 | 149 | 240 | 165 | 175 | 157 | 113 | 91 | 162 | 117 | 1530 |
Members of the stranding network are trained to identify the stranded animals. Nevertheless, 258 animals (16.8% of the strandings) were not characterized to the species level, generally because of an advanced state of decomposition of the animal body, sometimes in conjunction with bad field-work conditions.
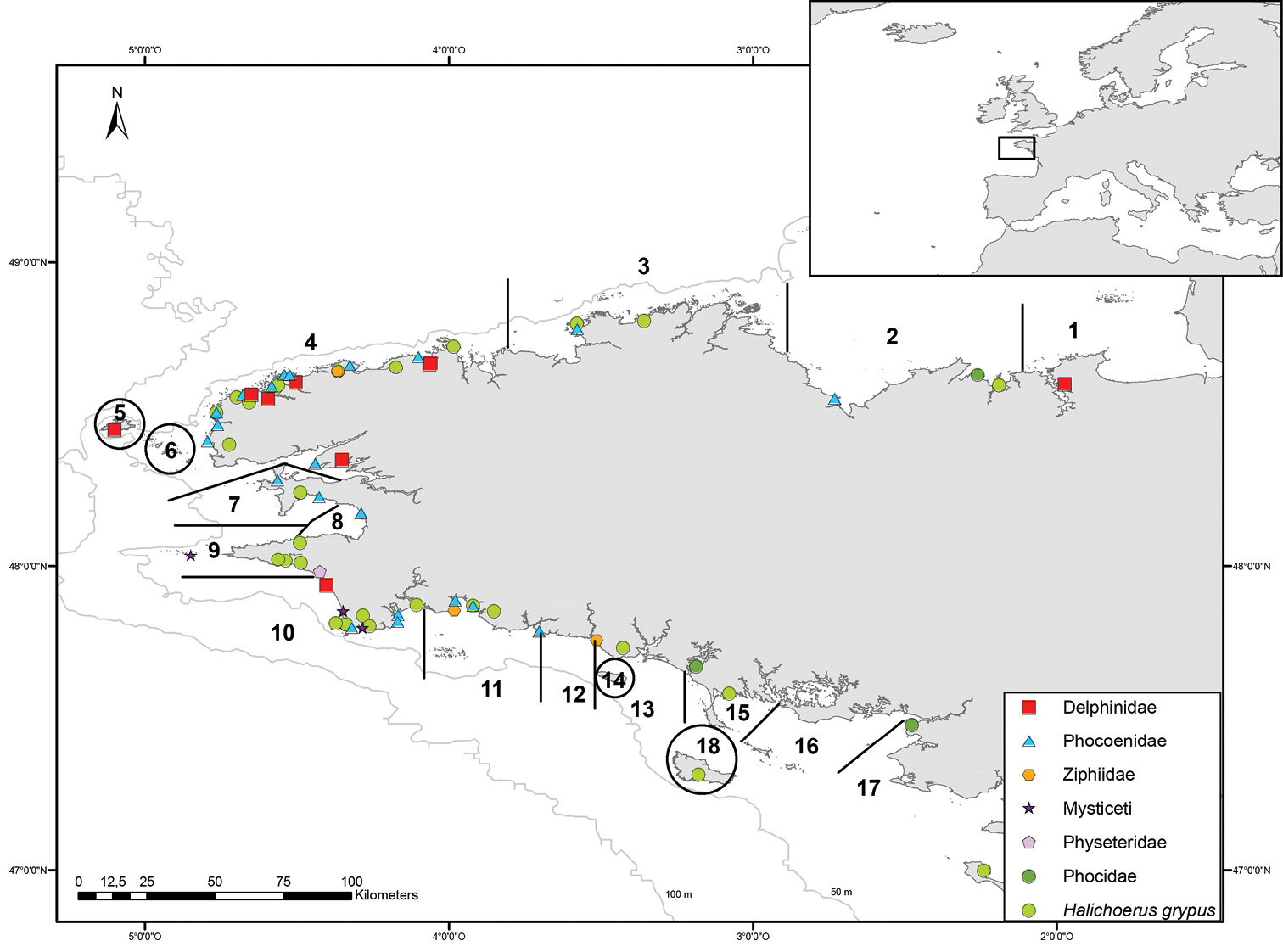
DNA was extracted from 92 stranded animals, i.e. from dead cetaceans and pinnipeds, but also from 40 grey seals stranded alive, which were treated in the care center of Océanopolis (Brest, France) and from which a small blood sample was taken and kept at -20 °C. All the samples came from animals stranded at the coasts of Brittany, except for one grey seal (Hgc406), which stranded alive in Spain in 2009 and which was transported to the care center (Figure 2). Our sampling included 12 species of cetaceans, and three species of pinnipeds (Table 2). Two species were very common, the harbour porpoise (29 samples) and the grey seal (44 samples), thus allowing intraspecific distance analyses.
Organization of the stranding network in Brittany (North West of France) and localization of the stranded specimens used in this study. Numbers indicate the 18 geographic sections of the stranding network in this area. The map was drawn using ArcGIS Desktop: Release 9.3.1 (Environmental Systems Research Institute, Redlands, CA, USA) with WGS 84 coordinates.
Numbers of samples included in the IMMB project.
| Cetaceans (12 species) | |
|---|---|
| Balaenoptera acutorostrata | 1 |
| Balaenoptera physalus | 1 |
| Delphinus delphis | 1 |
| Grampus griseus | 3 |
| Hyperoodon ampullatus | 2 |
| Lagenorhynchus acutus | 2 |
| Phocoena phocoena | 29 |
| Physeter macrocephalus | 1 |
| Stenella coeruleoalba | 1 |
| Stenella frontalis | 1 |
| Tursiops truncatus | 1 |
| Ziphius cavirostris | 1 |
| Pinnipeds (3 species) | |
| Cystophora cristata | 2 |
| Halichoerus grypus | 44 |
| Phoca vitulina | 2 |
| Total (15 species) | 92 |
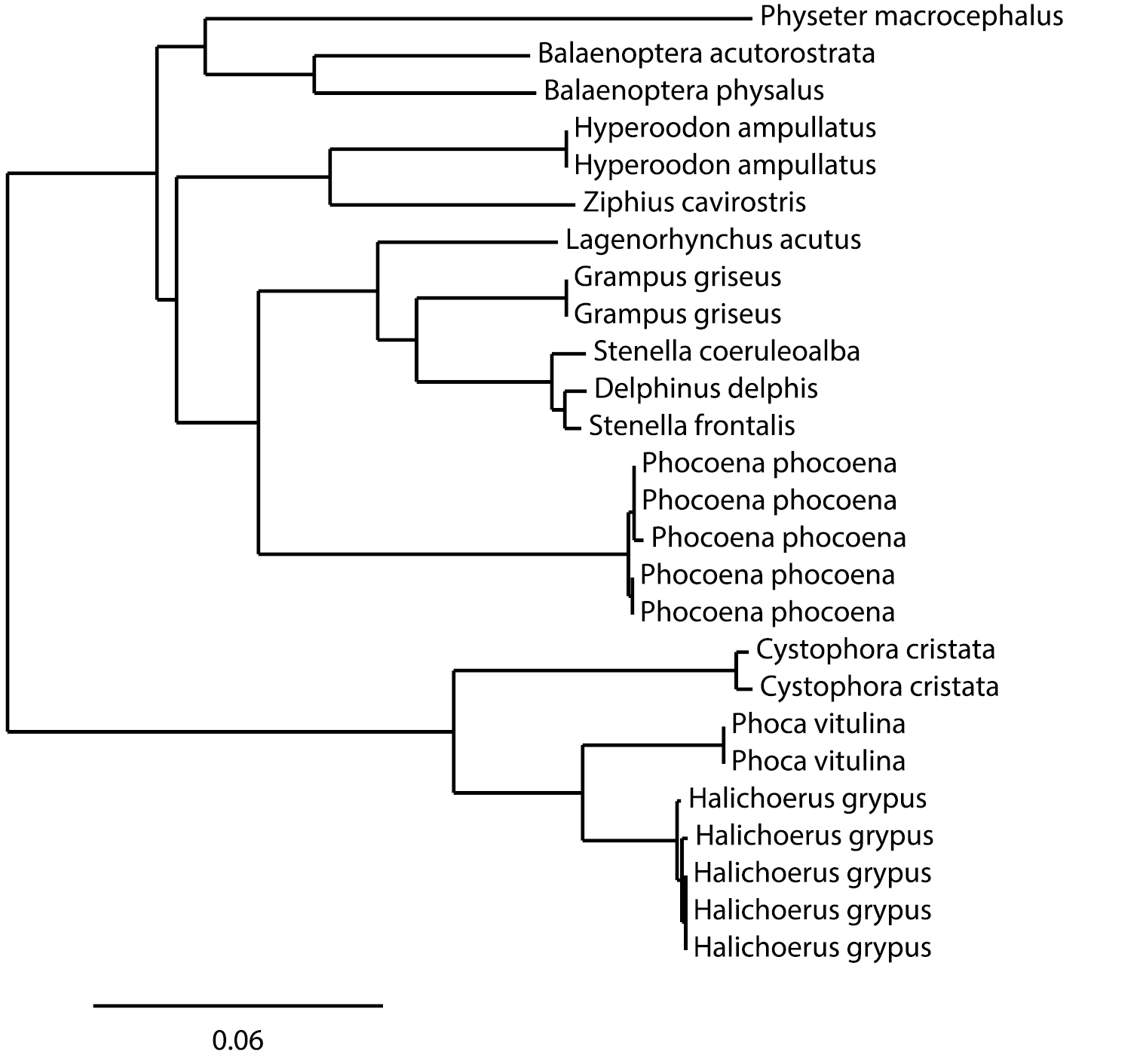
A COI amplicon was recovered from 89 samples, and good quality sequences of more than 500 bp were obtained for all samples (GenBank accession numbers KF281608–KF281697). The sequence alignment used in the analyses was 507 bp long. About 32% of the positions were polymorphic in the cetaceans and 13.1% in the pinnipeds (Table 3). The maximal intraspecific distance was 0.46% for the grey seal and 0.83% for the harbour porpoise. The COI sequences of three species of the Delphininae (Stenella frontalis, Stenella coeruleoalba and Delphinus delphis) showed very low interspecific distances (0.84% between Delphinus delphis and the nearest species Stenella frontalis, and 1.18% between the two Stenella species). All other interspecific distances were above 3.9% for pinnipeds and above 6% for cetaceans. The Neighbour-Joining (NJ) tree built on the BOLD interface using K2P-distances (Figure 3) confirms that, except for of the Delphininae, all the cetacean and pinniped species analyzed are distinguished unambiguously.
Neighbour-Joining tree of major species of marine mammals, based on K2P-distances calculated from 507 bp of COI. All sequences come from the IMMB project on BOLD, and only 5 harbour porpoise and 5 grey seal samples among those of the IMMB project have been included in the analysis.
Polymorphism levels of COI between 12 species of marine mammals stranded in Brittany, and comparison with intra-species variation for harbour porpoise and grey seal.
| Total | Cetaceans (12 species) | Pinnipeds (3 species) | Harbour porpoises | Grey seals | |
|---|---|---|---|---|---|
| Number of species | 14 | 11 | 3 | 1 | 1 |
| Number of sequences | 89 | 41 | 48 | 45* | 44 |
| Length (bp) | 507 | 508 | 656 | 610 | 658 |
| Polymorphic sites | 186 | 163 | 86 | 8 | 7 |
| Polymorphism (%) | 36.7 | 32.1 | 13.1 | 1.3 | 1.06 |
| Minimal distance to NN | - | 0.84 | 3.9 | 13.46 | 3.9 |
| Maximal distance to NN | - | 17.3 | 11.2 | - | - |
| Maximal intraspecific distance | - | - | - | 0.83 | 0.46 |
*This sampling includes 28 harbour porpoises stranded along the coasts of Brittany, and 17 more samples, stranded or by-caught in the Bay of Biscay, included to better characterize intraspecific variation. NN: nearest neighbour.
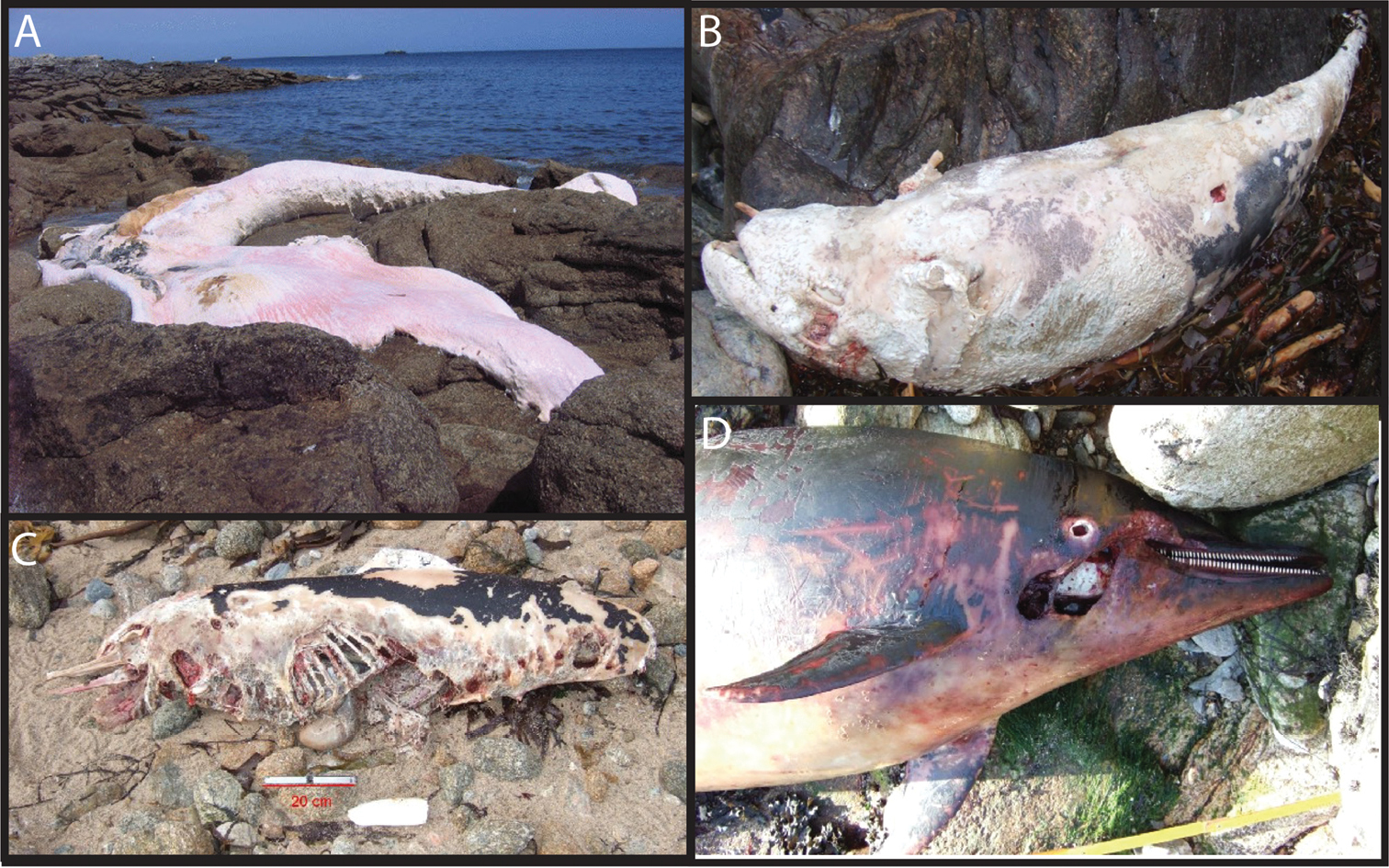
We then determined COI sequences from 10 cetacean samples whose species could not be determined accurately using morphological characters (Figure 4), either because only parts of the animal were recovered (Figure 4A) or because of the highly degraded state of the carcasses (Figure 4C). COI sequences of good qualities were obtained from all these samples, and three of them were identified unambiguously using the BOLD identification engine: Ms250511 was identified as a Balaenoptera physalus, Ds160111 as a Grampus griseus and Ds290811 as a Phocoena phocoena. The other seven samples were Delphininae, as confirmed by COI sequences. Yet, neither the BOLD identification engine, nor a BLAST search on GenBank allowed a more precise determination. We therefore sequenced MCR, which is more variable than COI, from six unidentified samples. BLAST searches on GenBank confirmed the COI results: all these samples were Delphininae, but a more precise identification could not be achieved.
Examples of marine mammals stranded along the coasts of Brittany and the species-level identifications of which were determined or confirmed thanks to DNA barcoding. A Sample Ms250511, stranded on the “Île de Sein” during May 2011, and identified as a Balaenoptera physalus B Sample Ds160111, stranded on the Ushant Island during January 2011, and identified as a Grampus griseus C Sample Ds130211, stranded on the Ushant Island in February 2011, and identified as belonging to the Delphininae subfamily (putatively identified as a Delphinus delphis on the nMDS plot in Figure 5) D Sample Ds080410 stranded on the Ushant Island during April 2010, and identified as belonging to the Delphininae (putatively identified as a Stenella coeruleoalba on the nMDS plot in Figure 5).
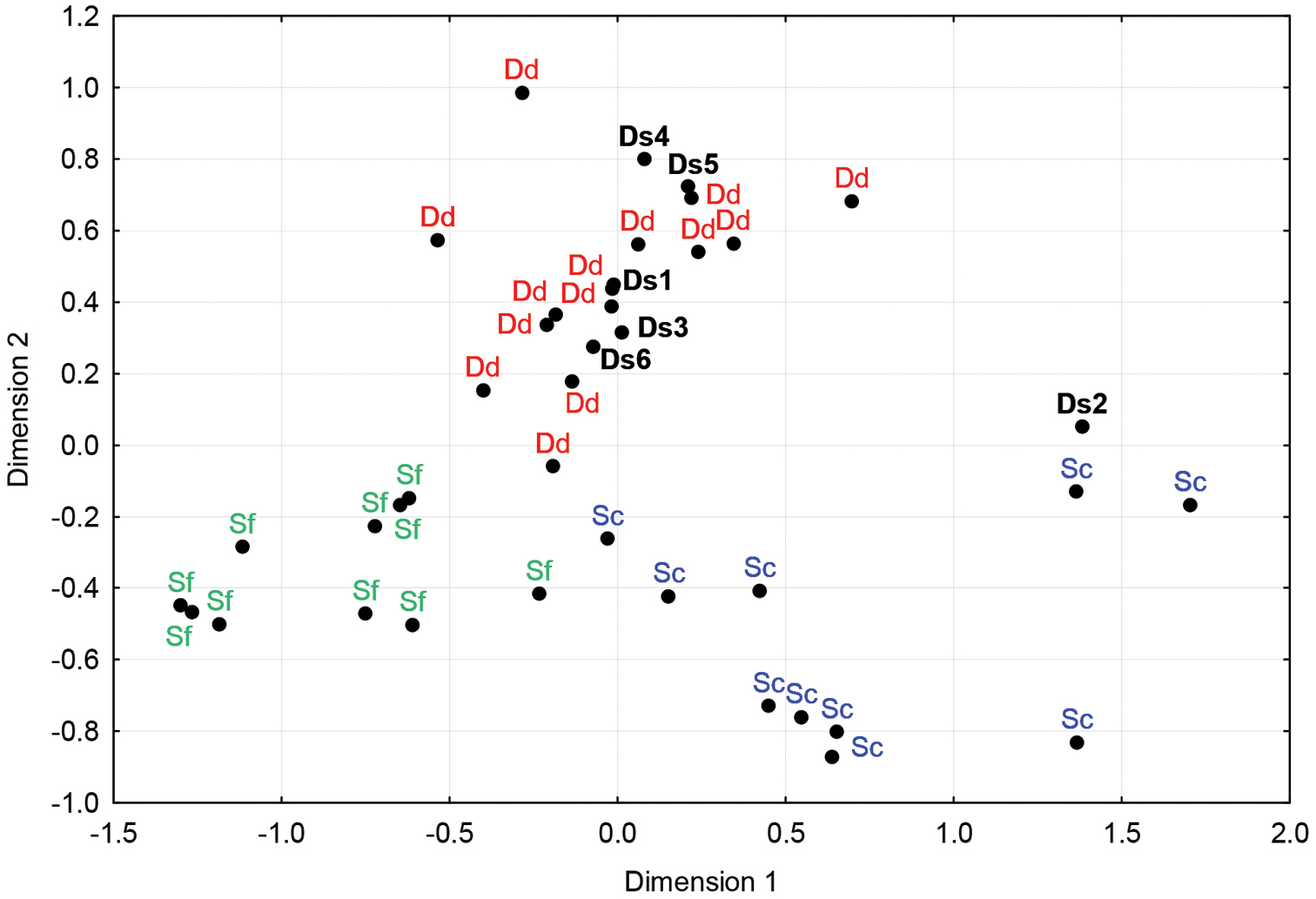
We constructed a nMDS plot of the distances between MCR sequences of Stenella coeruleoalba, Stenella frontalis and Delphinus delphis taken from GenBank: for Stenella coeruleoalba, we used sequences AM498725, AM498723, AM498721, AM498719, AM498717, AM498715, AM498713, AM498711, AM498709, AM498707 (Mace et al. unpublished), for Delphinus delphis FM211560, FM211553, FM211545, FM211535, FM211527, FM211519, FM211511, FM211503, FM211495 (
The three species were clearly discriminated by the nMDS (Figure 5). The posterior probabilities are given in Appendix 2. This analysis suggests that five of our unidentified samples could belong to Delphinus delphis, and one to Stenella coeruleoalba.
Non-metric Multidimensional Scaling plot of K2P-distance between MCR sequences of Stenella coeruleoalba (in blue), Delphinus delphis (in red) and Stenella frontalis (in green). Individuals of each species are clearly clustered together, and unidentified samples (in black) stranded along the coasts of Brittany group with one of the three species. Dd280211A (Ds1), Ds130210 (Ds3), Ds230409 (Ds4), Ds250412 (Ds5) and Sc210910 (Ds6) are putatively identified as Delphinus delphis, whereas Ds080410 (Ds2) would more likely belong to Stenella coeruloalba.
For the intraspecific analysis of the harbour porpoise, we included 17 additional samples of animals stranded or by-caught from the Bay of Biscay (Appendix 1,
Comparison of intraspecific COI and MCR polymorphisms for grey seal and harbour porpoise
| Harbour porpoise (Phocoena phocoena) | Grey seal (Halichoerus grypus) | |||
|---|---|---|---|---|
| Markers | COI | MCR | COI | MCR |
| Number of sequences | 45 | 45 | 35 | 35 |
| Sequence length (bp) | 610 | 579 | 658 | 482 |
| Haplotypes | 9 | 14 | 6 | 14 |
| Polymorphic sites | 8 | 22 | 5 | 23 |
| Polymorphism | 1.30% | 3.80% | 0.76% | 4.77% |
| Haplotype diversity | 0.695 | 0.832 | 0.553 | 0.935 |
| Nucleotide diversity | 0.00242 | 0.00632 | 0.00098 | 0.00945 |
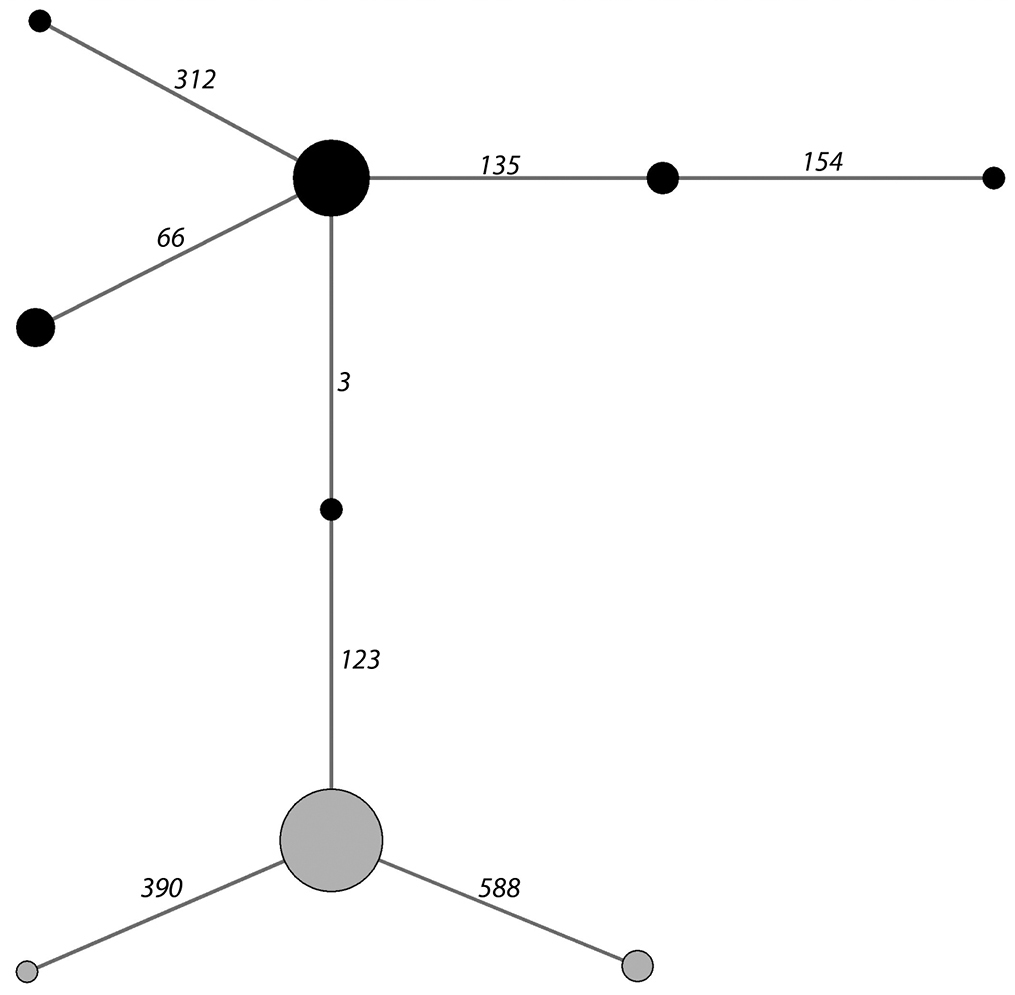
The haplotype network of the COI sequences in harbour porpoises clearly differentiated two haplogroups (Figure 6), that correspond perfectly to those described for MCR in
Haplotype network established from the COI sequences of 45 harbour porpoises stranded along the Atlantic coast of France (Appendix 1). Numbers on a line connecting two haplotypes correspond to the sequence position of the mutation differentiating these haplotypes. Two mitochondrial haplogroups appear (black circles - grey circles), that group the same individuals as the haplogroups alpha and beta determined using MCR polymorphisms and described in
Stranding networks collect opportunistic data that are ecologically significant (
The aim of this study was to evaluate the feasibility of a routine use of DNA barcoding in a stranding network; and to determine which gains this use could bring in terms of data relevance. The Brittany stranding network is a part of the French stranding network, and has to analyze an average of around 150 marine mammal strandings per year, with a high species biodiversity (19 species during 2003–2012).
We obtained DNA sequences of good quality for almost all the samples studied, whatever their origin, their collectors, or even their state of degradation. This is consistent with the numerous molecular genetic studies that have used samples taken on stranded cetaceans or pinnipeds (e.g.
The quality of the whole functioning and organization of the stranding network, from the field-work achieved by the correspondents to the preservation of the samples is therefore confirmed by our study. All the samples analyzed by DNA barcoding led to correct identification of the expected species with no exceptions.
We obtained COI good quality sequences for 10 unidentified animals, some of which were in a highly degraded body state. This showed that DNA barcoding can help to identify such specimens, which represent more than 16% of the stranded animals in the period 2003–2012. Hence, a routine use of DNA barcoding would noticeably decrease the proportion of unidentified animals.
Within the Delphininae, species are difficult to discriminate (
DNA barcoding is informative for animals that belong to species that infrequently strand along the coasts of Brittany, which can involve either species living far off the coasts or living in deep water, but also exotic species. Such species can be more difficult to identify by the field correspondents simply because of their scarcity. Along the coast of Brittany, we observed a Stenella frontalis, a temperate to tropical Atlantic Ocean inhabitant, and three species of arctic seals (Phoca hispida, Cystophora cristata and Phoca groenlandica). It is likely that other members of such rare species are listed among the “undetermined” species, just because their morphological characteristics are less well known by field correspondents. Additionally, a species that rarely strands along the French coast may be mistakenly identified as its more common sister-species. This issue can be illustrated by the case of the two pilot whale species: Globicephala melas, the long-finned pilot whale, commonly strands along the French Atlantic coast, while only a few stranding events of Globicephala macrorhynchus, the short-finned pilot whale, have been reported (the Bay of Biscay is the northern limit of the geographical range of Globicephala macrorhynchus). The two species have overlaping morphological characters, which adds to the difficulty of detecting rare stranding events of Globicephala macrorhynchus based on morphological data only (Viricel and Sabatier unpublished data). A systematic use of DNA barcoding when morphological taxonomic characteristics are not straightforward, would clearly lower the percentage of exotic animals not listed. The existence of natural interspecific hybrids between the two Globicephala sister-species (
It is important to note that a main limitation of DNA barcoding is the use of a single locus, leading to some problematic species identification such as within the Delphininae, but also to an inability to detect hybrids without complementary genetic studies. This limitation may well be removed in the near future thanks to next-generation sequencing, allowing the accumulation of large amount of DNA sequence data in a cost-effective manner. Multi-locus barcoding, including mitochondrial and nuclear polymorphic loci, will certainly represent a next step for the barcoding community.
A routine use of DNA barcoding could also allow monitoring the marine mammal biodiversity at intraspecific levels. For instance, global climate change has some effects on genetic diversity that must be studied and quantified (
This project is part of the collaboration between the Laboratory BioGeMME of the “Université de Bretagne Occidentale” (Brest, France), Océanopolis, a public private company (http://www.oceanopolis.com), the “Parc naturel marin d’Iroise” (http://www.parc-marin-iroise.com) and the French Stranding Network, coordinated by Pelagis, Université de La Rochelle, France. All the specimens and sequence data described in this manuscript are deposited in BOLD under the institution called “Oceanopolis-BioGeMME” in two projects, UMMB and IMMB. Our mixed institution became the first contributor to BOLD for the Cetacea, as well as for the Phocidae, and these two BOLD projects will be publicly available, and all the sequences published on GenBank.
The samples in this study were collected by the French Stranding Network and stored at Océanopolis (Brest) and at Pelagis (La Rochelle). We especially thank all the members of the LEMM, Christine Dumas who is in charge of the care center at Océanopolis and Willly Dabin and Olivier Van Canneyt from Pelagis. This work is part of a collaboration with ‘‘The Parc Naturel Marin d’Iroise’’, and especially with Phillipe Le Niliot, Cécile Lefeuvre and all the technicians working on the field.
We are indebted to Paul Marec, who collected the samples and took the photographs of the stranded animals on Ushant Island. Loriane Mendez, Anne Moulinet, Sandrine Quemener participated to some experiments during their internships in BioGeMME. Special thanks go to Chantal Hily-Mazé.
This manuscript benefited from useful comments during the reviewing and editing process.
List of the 46 Phocoena phocoena analyzed. (doi: 10.3897/zookeys.365.5873.app1) File format: Microsoft Word file (doc).
Posterior probabilities for species identification determined by the nMDS analysis. (doi: 10.3897/zookeys.365.5873.app2) File format: Microsoft Word file (doc).