(C) 2013 Marco Ballardini. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Ballardini M, Mercuri A, Littardi C, Abbas S, Couderc M, Ludeña B, Pintaud JC (2013) The chloroplast DNA locus psbZ-trnfM as a potential barcode marker in Phoenix L. (Arecaceae). In: Nagy ZT, Backeljau T, De Meyer M, Jordaens K (Eds) DNA barcoding: a practical tool for fundamental and applied biodiversity research. ZooKeys 365: 71–82. doi: 10.3897/zookeys.365.5725
The genus Phoenix (Arecaceae) comprises 14 species distributed from Cape Verde Islands to SE Asia. It includes the economically important species Phoenix dactylifera. The paucity of differential morphological and anatomical useful characters, and interspecific hybridization, make identification of Phoenix species difficult. In this context, the development of reliable DNA markers for species and hybrid identification would be of great utility. Previous studies identified a 12 bp polymorphic chloroplast minisatellite in the trnG (GCC)-trnfM (CAU) spacer, and showed its potential for species identification in Phoenix. In this work, in order to develop an efficient DNA barcode marker for Phoenix, a longer cpDNA region (700 bp) comprising the mentioned minisatellite, and located between the psbZ and trnfM (CAU) genes, was sequenced. One hundred and thirty-six individuals, representing all Phoenix species except P. andamanensis, were analysed. The minisatellite showed 2-7 repetitions of the 12 bp motif, with 1-3 out of seven haplotypes per species. Phoenix reclinata and P. canariensis had species-specific haplotypes. Additional polymorphisms were found in the flanking regions of the minisatellite, including substitutions, indels and homopolymers. All this information allowed us to identify unambiguously eight out of the 13 species, and overall 80% of the individuals sampled. Phoenix rupicola and P. theophrasti had the same haplotype, and so had P. atlantica, P. dactylifera, and P. sylvestris (the “date palm complex” sensu Pintaud et al. 2013). For these species, additional molecular markers will be required for their unambiguous identification. The psbZ-trnfM (CAU) region therefore could be considered as a good basis for the establishment of a DNA barcoding system in Phoenix, and is potentially useful for the identification of the female parent in Phoenix hybrids.
Chloroplast psbZ-trnfM (CAU) region , DNA barcode, minisatellite, palms
The genus Phoenix L. (Arecaceae) comprises 14 species (
The taxonomy, phylogeny and evolution of the genus itself have been assessed using morphological and molecular approaches. According to
The cultivated date palm Phoenix dactylifera L. is the most important fruit crop in the Middle East and North African countries. This species was probably domesticated around 4 000 B.C. in the Mesopotamia-Arabic Gulf area (
Phoenix species are largely interfertile and many interspecific hybrids have been recognized or suspected (
Added to the common hybridization process between Phoenix species, the paucity of systematically useful morphological and anatomical characters within the genus (
However, despite all efforts, no locus (alone or in combination), has proven to be 100% efficient as universal DNA barcode in plants at the species level.
The first DNA barcoding analysis in palms (
Investigating the taxonomic status of Phoenix atlantica, in comparison with its close relatives Phoenix dactylifera, Phoenix canariensis and Phoenix sylvestris,
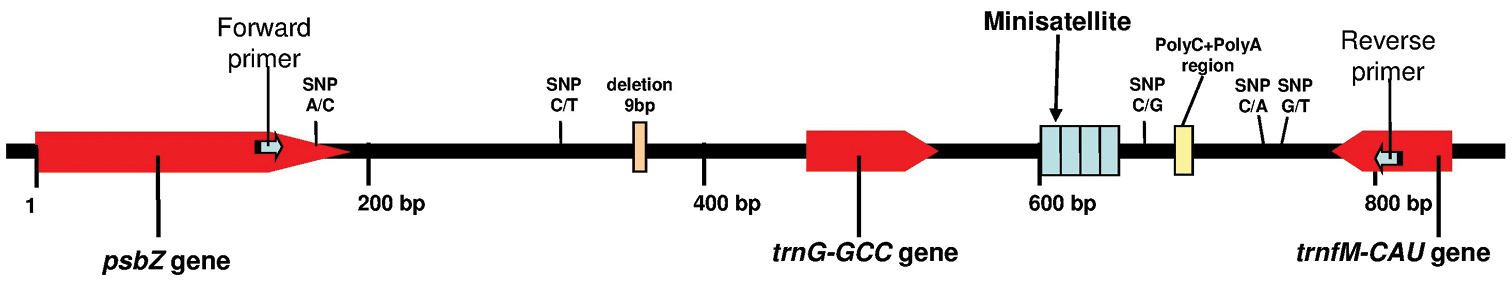
Given the potential of the trnG (GCC)-trnfM (CAU) spacer for barcoding in Phoenix, we examined a wider cpDNA region, viz. a ~700 bp sequence psbZ-trnfM (CAU) (Figure 1), in search of an efficient DNA barcode locus for species delimitation and identification of female parents in hybrids in the genus Phoenix.
The sequenced cpDNA psbZ-trnfM region.The location of PCR primers used and polymorphisms found in this study are shown. DNA fragment length refers to the Phoenix dactylifera cv. Khalas cpDNA sequence (
One hundred and thirty-six individuals, belonging to 13 Phoenix species, with emphasis on Phoenix dactylifera, were analysed in this work (Appendix). Phoenix andamanensis was not included in the analysis due to a lack of material.
For each sample, genomic DNA was extracted from 40 mg of freeze-dried leaf tissue which was first grinded using a bead-mill homogenizer Tissuelyser (Qiagen, France). Extraction was performed using the DNeasy Plant Mini Kit protocol along with the QIAcube robotic workstation for DNA automated purification (Qiagen, France). Extracted DNA was quantified by means of a Nanodrop ND1000 spectrophotometer (Thermo Fisher Scientific Inc., USA) and visualized on 1% agarose gels stained with ethidium bromide.
The PCR amplification was carried out using the monocotyledoneous universal primers psbZ-IGS-F: GGTACMTCATTATGGATTGG, and trnfM-IGS-R: GCGGAGTAGAGCAGTTTGGT (
The chromatograms obtained with the forward and reverse primers were combined and edited with SeqMan II 5.00 software (DNASTAR Inc., USA), to generate consensus sequences, which were aligned in BioEdit (
To assess the potential of the psbZ-trnfM region as a barcode for accurate species identification, we evaluated the proportion of correct identifications using TaxonDNA (
The amplification of the plastid target region psbZ-trnfM (CAU) was successful for all samples, and the sequencing with both primers was achieved for 123 individuals, while a single read (forward or reverse) was retrieved for the other 13 individuals, whose sequences were approximately 20% shorter.
The analysis of the intra- and interspecific variation within the sequenced region by direct observation of the sequence alignment showed four mutation types that contributed to the separation of Phoenix species: single nucleotide polymorphisms (SNPs), indels, length variation at the 12 bp minisatellite locus, and in homopolymers, allowing in total to identify unambiguously eight out of the 13 species (Table 1).
Distribution of observed polymorphisms in the region psbZ-trnfM (CAU)
| Substitutions |
9 bp deletion |
Minisatellite |
Homo-polymer |
Species |
||||
|---|---|---|---|---|---|---|---|---|
| 36607 | 36754 | 37099 | 37183 | 37190 | 36795-36803 | 37050-37098 | 37128-37139 | |
| Haplotypes recorded in a single species | ||||||||
| C | T | G | A | T | absent | 5M1+1M2 |
7 C + 5 A | Phoenix canariensis (7) |
| C | T | C | A | T | absent | 2M1+5bp+1M2 |
6 C + 5 A | Phoenix reclinata (4) |
| C | T | G | A | T | absent | 1M1+2M2 |
7 C + 5 A | Phoenix reclinata (6) |
| C | T | G | C | T | absent | 6M1+1M2 |
7 C + 5 A | Phoenix caespitosa (2) |
| C | C | G | A | G | absent | 2M1+1M2 |
7 C + 5 A | Phoenix loureiroi (1) |
| A | C | G | A | T | absent | 3M1+1M2 |
6 C + 6 A | Phoenix loureiroi (1) |
| A | C | G | A | T | absent | 2M1+1M2 |
7 C + 5 A | Phoenix acaulis (1) |
| C | C | G | A | T | absent | 2M1+1M2 |
6 C + 6 A | Phoenix pusilla (2) |
| C | T | G | A | T | present | 2M1+1M2 |
6 C + 6 A | Phoenix paludosa (2) |
| C | T | G | A | T | present | 4M1+1M2 |
5 C + 7 A | Phoenix roebelenii (3) |
| C | T | G | A | T | present | 3M1+1M2 |
5 C + 7 A | Phoenix roebelenii (1) |
| C | T | G | A | T | absent | 4M1+1M2 |
7 C + 5 A | Phoenix dactylifera (78) |
| C | T | G | A | T | absent | 2M1+1M2 |
8 C + 5 A | Phoenix sylvestris (1) |
| Haplotypes shared by two species (5.1%) | ||||||||
| C | T | G | A | T | absent | 6M1+1M2 |
7 C + 5 A | Phoenix rupicola (3) |
| Phoenix theophrasti (4) | ||||||||
| Haplotypes shared by three species (14.8%) | ||||||||
| C | T | G | A | T | absent | 3M1+1M2 |
7 C + 5 A | Phoenix atlantica (1) |
| Phoenix dactylifera (16) | ||||||||
| Phoenix sylvestris (3) | ||||||||
a Position in the complete chloroplast genome of Phoenix dactylifera ‘Khalas’ accession NC_013991.2.
b Number of individuals analysed for each species in parentheses (total sampling of 136 specimens).
c Number of repetitions of the 12 bp minisatellite units, including number of units of motif 1 (M1) and motif 2 (M2) as represented in Figure 2.
d Species-specific mutations in bold.
(1–7) Minisatellites haplotypes as reported in Figure 2 (1 to 7).
The minisatellite located in the trnG (GCC)-trnfM (CAU) intergenic spacer showed seven haplotypes. Most haplotypes corresponded to a Variable Number Tandem Repeat (VNTR) stepwise mutational pattern of 12 bp units. These units corresponded to two motifs: CTAACTACTATA (motif 1) and GTAGTTAGTATA (motif 2), which form between themselves a pattern of 12 bp inverted repeats shifted with respect to the boundaries of the mutational units (Figure 2). One haplotype, found in four out of ten Phoenix reclinata individuals, departed from this pattern, with two complete units of motif 1 plus an incomplete third unit with a 7 bp-deletion (CTAACTA) (haplotype 6; Figure 2). These four specimens were further characterized by a SNP (C instead of G) at position 37099. The other six Phoenix reclinata samples were also unique in having two repeats of motif 2 instead of one as in the six other haplotypes (haplotype 7; Figure 2). Phoenix canariensis was characterised by a private haplotype with five repeats of motif 1 (haplotype 4; Figure 2). The maximum number of haplotypes per species was three; in that case, one or two of them were shared by different species (Table 1).
Structure and variation of the minisatellite in the trnG-trnfM intergenic spacer. The repeats of the two mutational motifs (1 and 2) are indicated above the sequence alignment of the 7 haplotypes recorded. The pattern of inverted repeats generated by the two motifs and their reverse complements (RC) is shown below the alignment. See Table 1 for haplotype distribution among species.
Additional deletions and SNPs were detected in some of the analysed species, both upstream and downstream of the minisatellite (Table 1). Phoenix roebelenii and Phoenix paludosa shared a 9 bp-deletion (GTACTTTAC, upstream to the minisatellite, in position 36795-36803), Phoenix acaulis, Phoenix loureiroi, and Phoenix pusilla shared a SNP at position 36754 (C instead of T), while Phoenix caespitosa had a SNP at position 37183 (C instead of A). One of the three samples of Phoenix loureiroi showed a SNP in position 37190 (G instead of T), and another sample shared a SNP with the single specimen of Phoenix acaulis (A instead of C in position 36607). Some more differences among species and/or individuals were found in an homopolymer region (Cn + An), located at position 37128-37139, downstream to the minisatellite (Table 1).
Phoenix theophrasti and Phoenix rupicola shared the 6 repetitions of motif 1 minisatellite haplotype (haplotype 5; Figure 2) and could not be distinguished from each other (Phoenix caespitosa had also the 6 repetitions haplotype but was further differentiated by a species-specific SNP). Within the “date palm-complex” (
The TaxonDNA pairwise comparison analysis of the 121 samples retained resulted in 115 sequences with a closest match at 0%. There were 18 allospecific matches at 0% (15.65%). At the individual level, the Best Match test, and the Best Close Match test with a threshold of 3%, resulted in 82.64% correct identifications, 14.87% ambiguous identifications and 2.47% incorrect identifications. At the species level, however, only Phoenix caespitosa could be unambiguously identified, since it was the only species in the sampling with an autapomorphic SNP.
The haplotype sequences used, and the new ones obtained during this study, are deposited in GenBank under accessions: JF745571, EU043486, EU043484, EU043485, JX970915–JX970936.
In this study, we tested the usefulness of the psbZ-trnfM (CAU) region as a barcode locus in Phoenix. The successful amplification and sequencing of this marker within all of the analysed species confirms its value in terms of universality. Moreover, its high performance should allow the acquisition of barcode information even with partially degraded DNA samples.
TaxonDNA unambiguously identified a single species, Phoenix caespitosa, due to the scarcity of SNPs, most of them shared by two or more species, or on the contrary restricted to a subset of individuals within species. Therefore, it is important to take into account the other polymorphisms (indels, minisatellites and homopolymers) which usually represent half or more of the mutations in non-coding chloroplast DNA (
The 9 bp-deletion, shared by Phoenix roebelenii and Phoenix paludosa, supports Barrow’s conclusions (
Regarding the 12 bp minisatellite, our results revealed much more complexity than previously reported (
In total, considering all mutation types, our results allowed us to efficiently identify eight out of 13 species. This indicates that the locus psbZ-trnfM (CAU) has some potential to yield DNA barcodes that can be used for species identification within the genus Phoenix. This locus could also be useful to identify the female parent in many interspecific crosses, such as Phoenix dactylifera × Phoenix canariensis. Hybrids involving Phoenix canariensis as female parents are particularly easy to track because this species is monomorphic with a private haplotype at the locus studied. Hybrids between these two species are a concern for the genetic integrity of native populations of Phoenix canariensis in the Canary Islands (
Nevertheless, in order to increase resolution, other DNA regions should be examined, in search of characters allowing the identification of all taxa. Given their proven utility in palms, the psbA-trnH locus (
We wish to thank the following persons and institutions for their valuable help during the various phases of our study: Rita Bregliano†, Paolo Curir, Laura De Benedetti, Federica Nicoletti (CRA-FSO, Sanremo, Italy); Frédérique Aberlenc-Bertossi, Natalie Chabrillange, Nora Scarcelli (IRD Montpellier, France); Muriel Latreille, Sylvain Santoni (AMM at INRA SupAgro, Montpellier, France); Patrizia Martini (IRF, Sanremo, Italy); Mauro Roggero (Cooperativa “Il Cammino”, Sanremo, Italy); Giancarlo Pignatta (“U Risveiu Burdigotu” Association, Bordighera, Italy); the Natta family (Bordighera, Italy); Robert Castellana (CRP, Nice, France), Salwa Zehdi (University of Tunis, Tunisia). We also thank the three anonymous reviewers and the scientific editor for their useful comments and suggestions.
List of samples. DNA bank reference: IRD = Institut de Recherche pour le Développement, 911 Av. Agropolis, F-34394 Montpellier Cedex 5, France. Tissue bank reference: CRA-FSO = Consiglio per la Ricerca e la sperimentazione in Agricoltura - Unità di Ricerca per la Floricoltura e le Specie Ornamentali, Corso degli Inglesi 508, I-18038 Sanremo (IM), Italy.
Phoenix acaulis Roxb: MWC5559, Kew, UK (IRD). Phoenix atlantica A. Chev.: SH25, Cape Verde (IRD). Phoenix caespitosa Chiov.: MWC1195, MWC1802, Kew, UK (IRD). Phoenix canariensis Chabaud: 93.100, 93.101, 93.103, 93.107, Sanremo, Italy (IRD, CRA-FSO); MWC1396, Kew, UK (IRD); JCP169, JCP210, Canary Isl., Spain (IRD). Phoenix dactylifera L.: 93.003, 93.004, 93.005, 93.025, 93.027, 93.030, 93.037, 93.043, 93.045, 93.047, 93.048, 93.049, 93.052, 93.054, 93.055, 93.056, 93.059, 93.060, 93.061, 93.065, 93.066, 93.067, 93.070, 93.071, 93.072, 93.073, 93.076, 93.077, 93.080, 93.085, 90.002, 90.003, 90.004, 90.005, 90.006, 90.007, 90.008, 90.009, 90.010, 90.011, 90.012, 90.013, 90.014, 90.015, 90.025, 90.026, 90.027, 90.028, 90.029, 91.005, Sanremo, Italy (IRD, CRA-FSO); 00.01, 00.02, 00.03, 00.04, 00.05, 00.06, 00.07, 00.08, 00.09, 00.10, 00.11, 00.13, 00.14, 00.83, 00.85, 00.88, 46.02, 46.04, 46.05, 46.06, 46.08, 46.09, 46.14, 46.15, 46.16, 46.17, 46.18, 46.19, 46.20, 46.21, 46.23, JCP413, JCP414, JCP415, JCP416, JCP417, Bordighera, Italy (IRD, CRA-FSO); DAT077-365, DAT079-366, Oman (IRD); JCP260, Murcia, Spain; JCP426, Elche, Spain; SZ1, SZ2, SZ5, SZ10, Tunisia (IRD). Phoenix loureiroi Kunth var. loureiroi: JCP409, Montgomery Botanical Garden, Miami, USA (IRD); MWC1187, Kew, UK (IRD). Phoenix paludosa Roxb.: MWC1190, MWC1877, Kew, UK (IRD). Phoenix pusilla Gaertn.: JCP213_5, Sri Lanka (IRD); MWC1806, Kew, UK (IRD). Phoenix reclinata Jacq.: ECH3-A, ECH4-A, ECH5-B, 91.001, 91.007, 91.008, 91.009, 91.033, 92.003, Sanremo, Italy (IRD, CRA-FSO); MWC1397, Kew, UK (IRD). Phoenix roebelenii O’Brien: ECH1-A, ECH2-A, Sanremo, Italy (IRD, CRA-FSO); MWC1400, MWC1805, Kew, UK (IRD). Phoenix rupicola T. Anderson: ECH6-A, ECH8-A, Sanremo, Italy (IRD, CRA-FSO); MWC1399, Kew, UK (IRD). Phoenix sylvestris (L.) Roxb.: DAT057-345, Elche, Spain (IRD); JCP214, The Palm Center, UK, (IRD); JCP405-388, Thuret, France (IRD); MWC1876, Kew, UK (IRD). Phoenix theophrasti Greuter: ECH7-A, ECH9-A, Sanremo, Italy (IRD, CRA-FSO); JCP215, The Palm Center, UK (IRD); MWC1163, Kew, UK.