(C) 2013 Karin Breugelmans. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Breugelmans K, Jordaens K, Adriaens E, Remon JP, Cardona JQ, Backeljau T (2013) DNA barcodes and phylogenetic affinities of the terrestrial slugs Arion gilvus and A. ponsi (Gastropoda, Pulmonata, Arionidae). In: Nagy ZT, Backeljau T, De Meyer M, Jordaens K (Eds) DNA barcoding: a practical tool for fundamental and applied biodiversity research. ZooKeys 365: 83–104. doi: 10.3897/zookeys.365.6104
The Iberian Peninsula is a region with a high endemicity of species of the terrestrial slug subgenus Mesarion. Many of these species have been described mainly on subtle differences in their proximal genitalia. It therefore remains to be investigated 1) whether these locally diverged taxa also represent different species under a phylogenetic species concept as has been shown for other Mesarion species outside the Iberian Peninsula, and 2) how these taxa are phylogenetically related. Here, we analysed DNA sequence data of two mitochondrial (COI and 16S) genes, and of the nuclear ITS1 region, to explore the phylogenetic affinities of two of these endemic taxa, viz. Arion gilvus Torres Mínguez, 1925 and A. ponsi Quintana Cardona, 2007. We also evaluated the use of these DNA sequence data as DNA barcodes for both species. Our results showed that ITS did not allow to differentiate among most of the Mesarion molecular operational taxonomic units (MOTUs) / morphospecies in Mesarion. Yet, the overall mean p-distance among the Mesarion MOTUs / morphospecies for both mtDNA fragments (16.7% for COI, 13% for 16S) was comparable to that between A. ponsi and its closest relative A. molinae (COI: 14.2%; 16S: 16.2%) and to that between A. gilvus and its closest relative A. urbiae (COI: 14.4%; 16S: 13.4%). Hence, with respect to mtDNA divergence, both A. ponsi and A. gilvus, behave as other Mesarion species or putative species-level MOTUs and thus are confirmed as distinct ‘species’.
DNA barcoding, terrestrial slugs, Gastropoda, taxonomy, Iberian Peninsula, Arion ponsi, Arion gilvus
The genus Arion Férussac, 1819 is the most species rich genus of the terrestrial slug family Arionidae (Mollusca, Pulmonata, Gastropoda). It comprizes approximately 40 species, grouped into four subgenera, viz. Arion s.s. Férussac, 1819, Kobeltia Seibert, 1873, Carinarion Hesse, 1926 and Mesarion Hesse, 1926. Species of the subgenus Mesarion (type species: Limax subfuscus Draparnaud, 1805) are characterized by 1) a medium body-size (up to 75 mm when extended), 2) an orange to dark brown dorsum, 3) two dark bands on the sides of the mantle, 4) (usually) yellow to orange body mucus, and 5) an enlarged free-oviduct with a long and V-shaped ligula (
Arion ponsi (Figure 1) was described from Menorca (Balearic Islands, type locality: Barranc d’Algendar). The species has a medium body size (range: 54–66 mm), an orange to beige dorsal body colour with dark lateral bands that can be blurry in the posterior parts, a foot sole that is cream coloured with a greyish hue, and a transparent body mucus (
Arion ponsi Quintana Cardona, 2007 from Menorca (Balearic Islands, Spain).
Arion gilvus (Figure 2) was described from ‘Mandol’ in the Spanish Province of Tarragona. However, the toponym ‘Mandol’ seems to be erroneous (e.g.
Arion gilvus Torres Mínguez, 1925 from Serra de Pandóls (Valencia, Spain). A dorsal view B lateral view C ventral view.
As illustrated by Arion ponsi and Arion gilvus, the alleged species-specific genital differences among the Iberian species of the Arion subfuscus complex are very subtle and little is known about their intraspecific variation. Moreover, genital differences among arionid taxa do not necessarily imply reproductive isolation (
Because DNA sequence data do not only provide phylogenetic information, but can also serve as DNA barcodes for species identification (
Information on the species and specimens included here is provided in Table 1. In total, we screened 45 specimens (Table 1). DNA was extracted from small parts of the foot using a NucleoSpin Tissue Kit (Macherey-Nagel, Düren) following the manufacturer’s instructions. PCR reactions were done in 25 µl reaction volumes that contained 1.5 mM MgCl2 in 1 × PCR buffer (Qiagen), 0.2 mM of each dNTP, 0.2 µM of each primer and 0.5 units of Taq polymerase (Qiagen). A fragment of the mitochondrial COI and 16S genes was amplified using primer pairs LCO1490 and HCO2198 (
List of specimens used in this study with specimen ID, sampling locality, GenBank accession numbers for the COI, 16S and ITS1 sequences, and collection number at the museums (if available). Neo-, para- and topotypes have been indicated. Specimen codes with an asterisk are data taken from
| Species/ID | Locality, country | COI | 16S | ITS1 | Collection number |
|---|---|---|---|---|---|
| Subgenus Mesarion Hesse, 1926 | |||||
| Arion anguloi Martín and Gómez, 1988 | |||||
| ang-SU2777 | Torralba del Rio, Spain | AY987869 | AY947348 | AY947386 | RBINS Brussels, I.G. 32471 |
| ang-115 (topotype) | Osma, Álava, Spain | KF305196 | KF356212 | AJ509055 | RBINS Brussels, I.G. 32471 |
| AANG 73A* | Burgos, Spain | NA | NA | AY316291 | |
| Arion baeticus Garrido, Castillejo and Iglesias, 1995 | |||||
| bae-556 (paratype) | Malaga, Spain | AY987871 | AY947350 | AJ509054 | MNCN Madrid 15.05/6969 |
| Arion flagellus Colligne, 1893 | |||||
| fla-130 | Glasgow, UK | AY987880 | AY947359 | AJ509053 | RBINS Brussels, I.G. 32471 |
| fla-161 | Glasgow, UK | AY987881 | AY947360 | AJ509052 | RBINS Brussels, I.G. 32471 |
| fla-SU672 | Salamir, Spain | AY987882 | AY947361 | AY947388 | RBINS Brussels, I.G. 32471 |
| AFLA 44A* | Croydon, UK | NA | NA | AY316278 | |
| Arion fuligineus Morelet, 1845 | |||||
| AFUL 43A* | São Silvestre, Portugal | NA | NA | AY316277 | |
| Arion fuscus (Müller, 1774) | |||||
| fus-SU155 | Grudki, Poland | AY987885 | AJ786721 | AY947390 | RBINS Brussels, I.G. 32471 |
| fus-2320 | Predel, Bulgaria | AY987886 | AJ786722 | AY947391 | RBINS Brussels, I.G. 32471 |
| fus-SU1335 | Steinegg, Austria | AY987887 | AJ786726 | AY947392 | RBINS Brussels, I.G. 32471 |
| fus-SU2188 | Kreuzen, Austria | NA | KF356221 | NA | RBINS Brussels, I.G. 32471 |
| Arion gilvus Torres Mínguez, 1925 | |||||
| gil-46 | Serra de Pandóls, Valencia, Spain | NA | NA | KF385450 | RBINS Brussels, I.G. 32471 |
| gil-47 | Serra de Pandóls, Valencia, Spain | KF305199 | KF356222 | KF385451 | RBINS Brussels, I.G. 32471 |
| gil-73 | Serra de Pandóls, Valencia, Spain | KF305200 | KF356223 | KF385452 | RBINS Brussels, I.G. 32471 |
| AGIL 49A* | Serra de Pandóls, Valencia, Spain | NA | NA | AY316282 | |
| Arion hispanicus Simroth, 1886 | |||||
| AHIS 52B* | Cáceres, Spain | NA | NA | AY316285 | |
| Arion iratii Garrido, Castillejo and Iglesias, 1995 | |||||
| ira-559 (paratype) | Navarra, Spain | AY987892 | AY947367 | AJ509042 | MNCN Madrid, 15.05/18705 |
| Arion lizarrustii Garrido, Castillejo and Iglesias, 1995 | |||||
| liz-562 (paratype) | Navarra, Spain | AY987893 | AY947368 | AJ509046 | MNCN Madrid, 15.05/18706 |
| ALIZ 47C* | Lizarrusti, Spain | NA | NA | AY316280 | |
| Arion lusitanicus Mabille, 1868 | |||||
| lus-1613 | Feitos, Portugal | KF305203 | KF356224 | NA | RBINS Brussels, I.G. 32471 |
| lus-1631 | Currais, Portugal | KF305204 | KF356225 | NA | RBINS Brussels, I.G. 32471 |
| lus-1641 | Cacia, Portugal | KF305205 | NA | NA | RBINS Brussels, I.G. 32471 |
| lus-1647 | Cacia, Portugal | KF305206 | NA | NA | RBINS Brussels, I.G. 32471 |
| lus-1652 | Forjães, Portugal | KF305207 | KF356226 | NA | RBINS Brussels, I.G. 32471 |
| lus-1654 | Currais, Portugal | NA | KF356227 | NA | RBINS Brussels, I.G. 32471 |
| lus-1655 | Forjães, Portugal | KF305208 | KF356228 | NA | RBINS Brussels, I.G. 32471 |
| lus-79 | Ursel, Belgium | AY987894 | AY947369 | AJ509062 | RBINS Brussels, I.G. 32471 |
| lus-181 | Terceira, Azores, Portugal | NA | NA | KF385453 | RBINS Brussels, I.G. 32471 |
| lus-186 | Namur, Belgium | AY987895 | AY947370 | AJ509061 | RBINS Brussels, I.G. 32471 |
| lus-465 | Görlitz, Germany | NA | NA | AJ509063 | RBINS Brussels, I.G. 32471 |
| lus-509 | Emptinne, Belgium | KF305209 | KF356229 | NA | RBINS Brussels, I.G. 32471 |
| ALUS 42A* | Serra de Arrábida, Portugal | NA | NA | AY316273 | |
| ALUS 42B* | Serra de Arrábida, Portugal | NA | NA | AY316274 | |
| ALUS 42C* | Serra de Arrábida, Portugal | NA | NA | AY316275 | |
| ALUS 42G* | Alpi Carniche, Rivolato, Italy | NA | NA | AY316276 | |
| ALUS 62E* | Montagne Noire, France | NA | NA | AY316289 | |
| ALUS 70C* | Girona, Spain | NA | NA | AY316290 | |
| Arion molinae Garrido, Castillejo and Iglesias, 1995 | |||||
| mol-565 (paratype) | La Molina, Spain | AY987896 | AY947371 | AJ509043 | MNCN Madrid, 15.05/18707 |
| AMOL 48A* | Serra del Cadí, Barcelona, Spain | NA | NA | AY316281 | |
| Arion nobrei Pollonera, 1889 | |||||
| ANOB 41A* | Luso, Portugal | NA | NA | AY316271 | |
| ANOB 41B* | Luso, Portugal | NA | NA | AY316272 | |
| Arion paularensis Wiktor and Parejo, 1989 | |||||
| pau-121 | Sierra de Guadarrama, Spain | KF305210 | NA | KF385454 | RBINS Brussels, I.G. 32471 |
| pau-224 | Sierra de Guadarrama, Spain | AY987899 | AY947374 | AJ509057 | RBINS Brussels, I.G. 32471 |
| pau-226 | Sierra de Guadarrama, Spain | NA | NA | KF385455 | RBINS Brussels, I.G. 32471 |
| APAU 51A* | Sierra de Guadarrama, Spain | NA | NA | AY316284 | |
| Arion ponsi Quintana Cardona, 2007 | |||||
| pon-1959 | Ciutadella de Menorca, Spain | KF305211 | KF356230 | KF385456 | RBINS Brussels, I.G. 32471 |
| pon-1960 | Ferreries, Menorca, Spain | KF305212 | KF356231 | KF385457 | RBINS Brussels, I.G. 32471 |
| pon-1962 | Ciutadella de Menorca, Spain | KF305213 | KF356232 | KF385458 | RBINS Brussels, I.G. 32471 |
| pon-1965 | Ciutadella de Menorca, Spain | KF305214 | KF356233 | KF385459 | RBINS Brussels, I.G. 32471 |
| Arion subfuscus (Draparnaud, 1805) | |||||
| sub1-2312 | Kortrijk, Belgium | KF305215 | KF356238 | KF385461 | RBINS Brussels, I.G. 32471 |
| sub1-2318 | Ingrandes, France | AY987904 | AY860678 | AY860729 | RBINS Brussels, I.G. 32471 |
| sub1-2317 | Burnopfield, UK | AY987906 | AY860672 | AY860726 | RBINS Brussels, I.G. 32471 |
| sub1-SU87 | Barnstable, MA, USA | NA | KF356235 | NA | RBINS Brussels, I.G. 32471 |
| sub1-1618 | Wilrijk, Belgium | KF305216 | KF356236 | NA | RBINS Brussels, I.G. 32471 |
| sub1-1633 | Wilrijk, Belgium | KF305217 | KF356237 | NA | RBINS Brussels, I.G. 32471 |
| sub2-SU2424 | Heppenbach, Belgium | NA | KF356239 | NA | RBINS Brussels, I.G. 32471 |
| sub2-SU349 | Grootrees, Belgium | NA | KF356240 | NA | RBINS Brussels, I.G. 32471 |
| sub2-2309 | Gomzé, Belgium | KF305218 | KF356241 | KF385462 | RBINS Brussels, I.G. 32471 |
| sub2-2313 | Le Landin, France | AY987908 | AY860687 | KF385463 | RBINS Brussels, I.G. 32471 |
| sub2-2314 | Heppenbach, Belgium | AY987909 | AY860709 | KF385464 | RBINS Brussels, I.G. 32471 |
| sub3-2322 | Bucholz, Germany | AY987910 | AY860716 | KF385466 | RBINS Brussels, I.G. 32471 |
| sub3-2310 | La Salle, France | AY987911 | AY860722 | KF385465 | RBINS Brussels, I.G. 32471 |
| sub3-SU2401 | La Salle, France | NA | KF356242 | NA | RBINS Brussels, I.G. 32471 |
| sub4-123 (topotype) | Montagne Noire, France | AY987913 | AY860682 | AY860733 | RBINS Brussels, I.G. 32471 |
| sub4-568 (neotype) | Montagne Noire, France | AY987914 | KF356244 | AJ509044 | MNCN Madrid, 15.05/18704 |
| sub4-2341 | Oulès, France | AY987912 | AY860685 | AY860731 | RBINS Brussels, I.G. 32471 |
| sub4-SU1058 | Col de Peyresourde, France | NA | KF356243 | NA | RBINS Brussels, I.G. 32471 |
| sub5-2321 | Villemont-Baubiat, France | AY987915 | AY860681 | KF385468 | RBINS Brussels, I.G. 32471 |
| sub5-2311 | Villemont-Baubiat, France | AY987916 | AY860679 | KF385467 | RBINS Brussels, I.G. 32471 |
| ASUB 45A* | Montagne Noire, France | NA | NA | AY316279 | |
| Arion transsylvanus Simroth, 1885 | |||||
| tra-SU1088 | Covasna, Romania | AY943858 | AY860798 | AY947393 | RBINS Brussels, I.G. 30412 |
| tra-SU1203 | Lunca Vişagului, Romania | AY943859 | AY860805 | AY947394 | RBINS Brussels, I.G. 30412 |
| tra-SU1296 | Holda, Romania | AY943860 | AY860799 | AY947395 | RBINS Brussels, I.G. 30412 |
| Arion urbiae De Winter, 1986 | |||||
| urb-SU2755 | Saldaña, Spain | AY987919 | AY947381 | AY947396 | RBINS Brussels, I.G. 32471 |
| urb-99 | Sierra da Urbia, Spain | NA | NA | KF385469 | RBINS Brussels, I.G. 32471 |
| AURB 50A* | NA | NA | AY316283 | ||
| Subgenus Kobeltia Seibert, 1873 | |||||
| Arion distinctus Mabille, 1868 | |||||
| dis-106 | Mortsel, Belgium | AY987875 | AY947354 | AJ509040 | RBINS Brussels, I.G. 32471 |
| dis-14 | AY987874 | AY947353 | AJ509038 | RBINS Brussels, I.G. 32471 | |
| Arion hortensis Férussac, 1819 | |||||
| hor-102 | Mortsel, Belgium | AY987888 | AJ518061 | AJ509037 | RBINS Brussels, I.G. 32471 |
| hor-220 | London, UK | AY987889 | AY947364 | AJ509036 | RBINS Brussels, I.G. 32471 |
| Arion intermedius Normand, 1852 | |||||
| int-104 | Rochefort, Belgium | AY987891 | AY947366 | AJ509031 | RBINS Brussels, I.G. 32471 |
| int-52 | Flores, Azores, Portugal | AY987890 | AY947365 | AJ509029 | RBINS Brussels, I.G. 32471 |
| Arion obesoductus Reischütz, 1973 | |||||
| alp-1610 | Žďárské Vrchy, Czech Republic | DQ904249 | DQ904248 | NA | RBINS Brussels, I.G. 32471 |
| alp-208 | Saxony, Germany | AY987867 | AY947346 | AJ509041 | RBINS Brussels, I.G. 32471 |
| Arion owenii Davies, 1979 | |||||
| owe-310 | Devon, UK | AY987897 | AY947372 | AJ509033 | RBINS Brussels, I.G. 32471 |
| owe-316 | Devon, UK | AY987898 | AY947373 | AJ509034 | RBINS Brussels, I.G. 32471 |
| Arion wiktori Parejo & Martín, 1990 | |||||
| wik-SU2693 | Viniegra de Abajo, Spain | AY987921 | AY947383 | AY947397 | RBINS Brussels, I.G. 32471 |
| wik-44 | Burgos, Spain | AY987920 | AY947382 | AJ509060 | RBINS Brussels, I.G. 32471 |
| wik-94 | Burgos, Spain | NA | KF356245 | AJ509059 | RBINS Brussels, I.G. 32471 |
| AWIK 58A* | Demanda Sierra, Burgos, Spain | NA | NA | AY316287 | |
| AWIK 58C* | Urbión Mountains, Soria, Spain | NA | NA | AY316288 | |
| Subgenus Carinarion Hesse, 1926 | |||||
| Arion circumscriptus Johnston, 1828 | |||||
| cir-151 | Aran Island, Kilmurvey, Ireland | AY987872 | AY947351 | AJ509071 | RBINS Brussels, I.G. 32471 |
| Arion fasciatus (Nilsson, 1823) | |||||
| fas-144 | Görlitz, Germany | AY987877 | AY947356 | AJ509068 | RBINS Brussels, I.G. 32471 |
| Arion silvaticus Lohmander, 1937 | |||||
| sil-142 | Poulseur, Belgium | AY987917 | AY947379 | AJ509070 | RBINS Brussels, I.G. 32471 |
| Subgenus Arion s.s. Férussac, 1819 | |||||
| Arion ater-rufus complex | |||||
| ate-SU517 | Musland, Norway | AY987870 | AY947349 | AY947387 | RBINS Brussels, I.G. 32471 |
| ate/ruf-1602 | Manteigas, Portugal | KF305219 | NA | KF385449 | RBINS Brussels, I.G. 32471 |
| ate/ruf-1619 | Santa Leocádia, Portugal | KF305220 | KF356213 | NA | RBINS Brussels, I.G. 32471 |
| ate/ruf-1620 | Gortmore, Ireland | KF305221 | KF356214 | NA | RBINS Brussels, I.G. 32471 |
| ate/ruf-1624 | Oleirinhos, Portugal | KF305222 | KF356215 | NA | RBINS Brussels, I.G. 32471 |
| ate/ruf-1638 | Portulezo, Portugal | KF305223 | KF356216 | NA | RBINS Brussels, I.G. 32471 |
| ate/ruf-1649 | Manteigas, Portugal | KF305224 | KF356217 | NA | RBINS Brussels, I.G. 32471 |
| ruf-105 | St.-Katelijne Waver, Belgium | KF305225 | KF356234 | NA | RBINS Brussels, I.G. 32471 |
| ruf-15 | Santiago de Compostela, Spain | AY987900 | AY947375 | AJ509066 | RBINS Brussels, I.G. 32471 |
| ruf-155 | Brussels, Belgium | AY987901 | AY947376 | AJ509064 | RBINS Brussels, I.G. 32471 |
| ruf-180 | Hoboken, Belgium | AY987902 | AY947377 | AJ509065 | RBINS Brussels, I.G. 32471 |
| ruf-182 | Brecht, Belgium | AY987903 | AY947378 | AJ509067 | RBINS Brussels, I.G. 32471 |
| ruf-624 | Nazareth, Belgium | NA | NA | KF385460 | RBINS Brussels, I.G. 32471 |
| AATE 39A* | Caldas de Gerês, Portugal | NA | NA | AY316268 | |
| AATE 39E* | Valporquero Cave, Leon, Spain | NA | NA | AY316269 | |
| ARUF 40G* | Montagne Noire, France | NA | NA | AY316270 | |
Sequences were aligned in ClustalW (
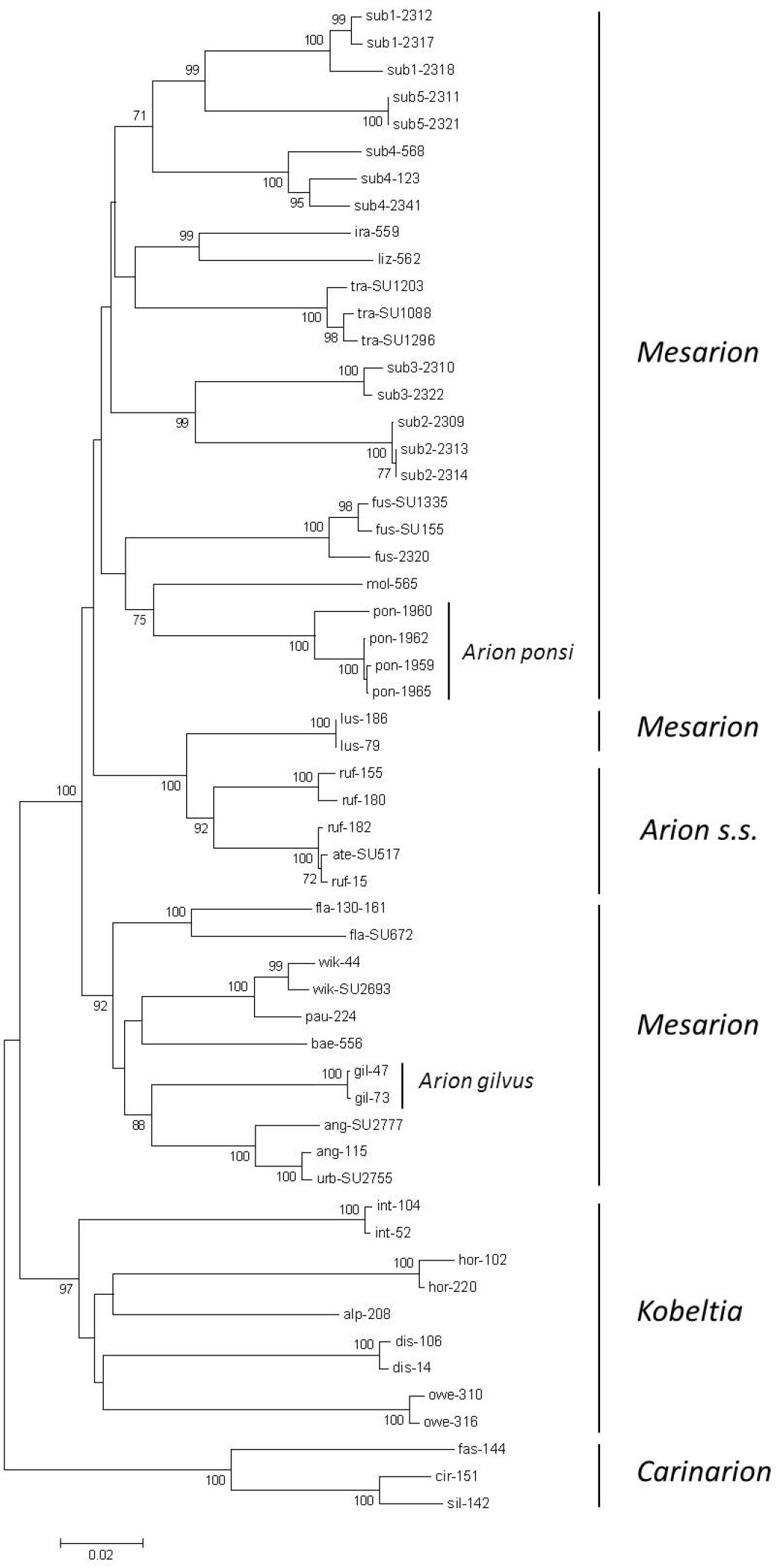
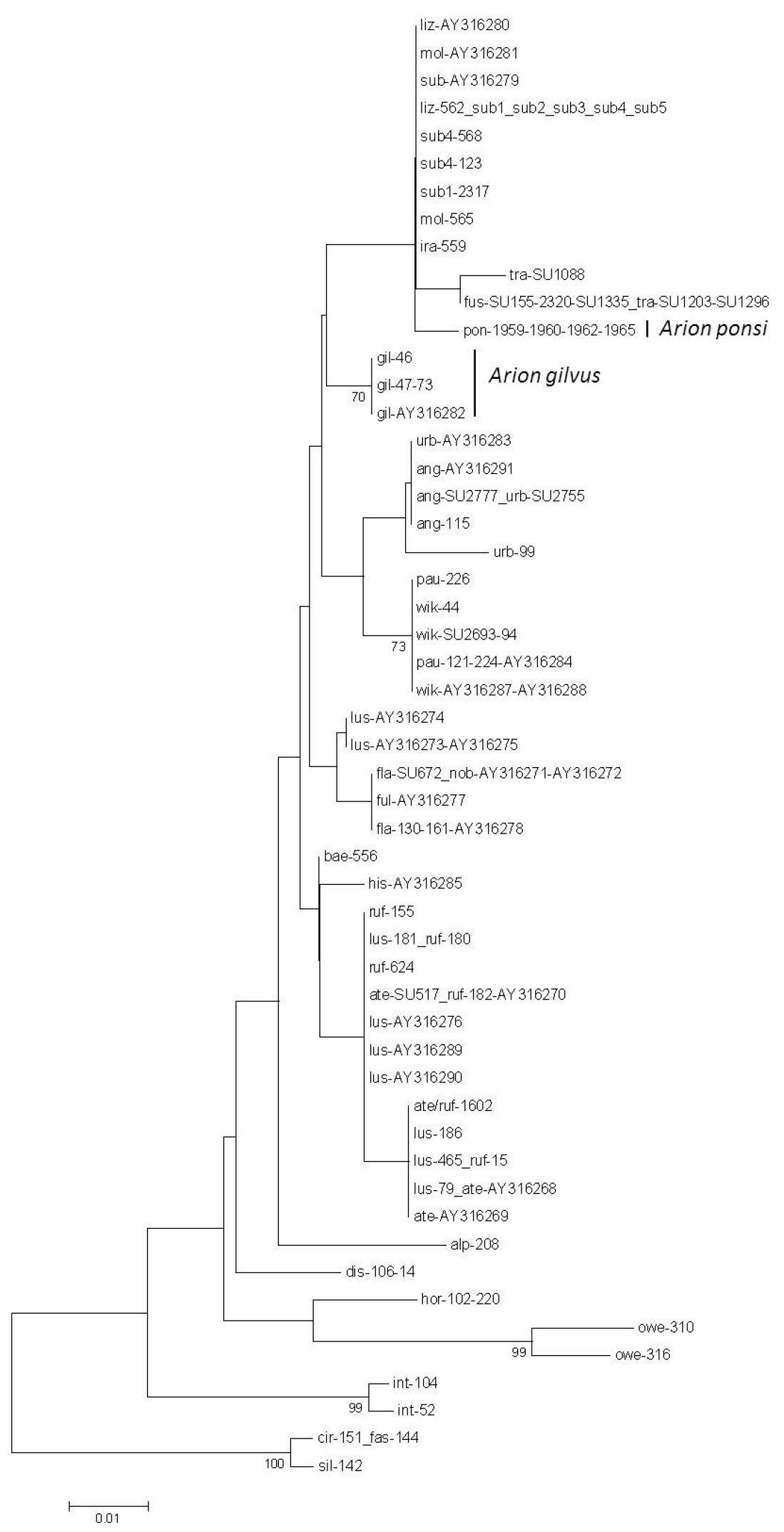
The alignments comprized 504 bp for COI (196 variable sites), 408 bp for 16S (121 sites with alignment gaps, 122 variable sites) and 587 bp for ITS1 (277 sites with alignment gaps, 64 variable sites). For the concatenated dataset, there was strong support for the subgenera Carinarion, Kobeltia (excluding Arion wiktori) and Arion s.s., and for a clade of Arion s.s. + Mesarion (including Arion wiktori) (Figure 3). The subgenus Mesarion was not monophyletic but consisted of (1) a clade of Arion flagellus, Arion wiktori, Arion paularensis, Arion baeticus, Arion urbiae, Arion anguloi, and Arion gilvus, (2) two haplotypes of Arion lusitanicus (lus-79 and lus-186) that formed a sister group of Arion s.s. [insofar Arion lusitanicus is, of course, considered as a member of Mesarion; see e.g.
Neighbour-Joining tree (Kimura 2-parameter model) of a 1499 bp concatenated fragment (504 bp of the mitochondrial cytochrome c oxidase subunit I (COI) gene, 408 bp of the mitochondrial 16S rDNA gene, 587 bp fragment of the nuclear internal transcribed spacer 1 (ITS1) region) for the land slug subgenus Mesarion. Bootstrap values ≥ 70% are shown at the nodes. For sample codes see Table 1.
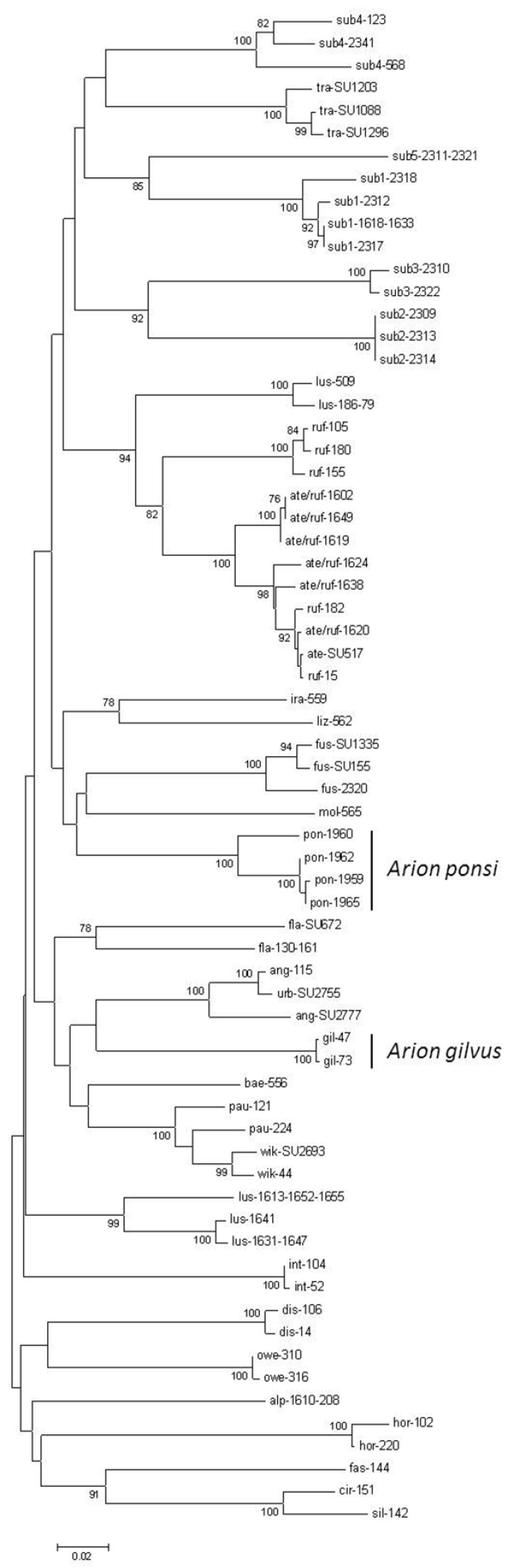
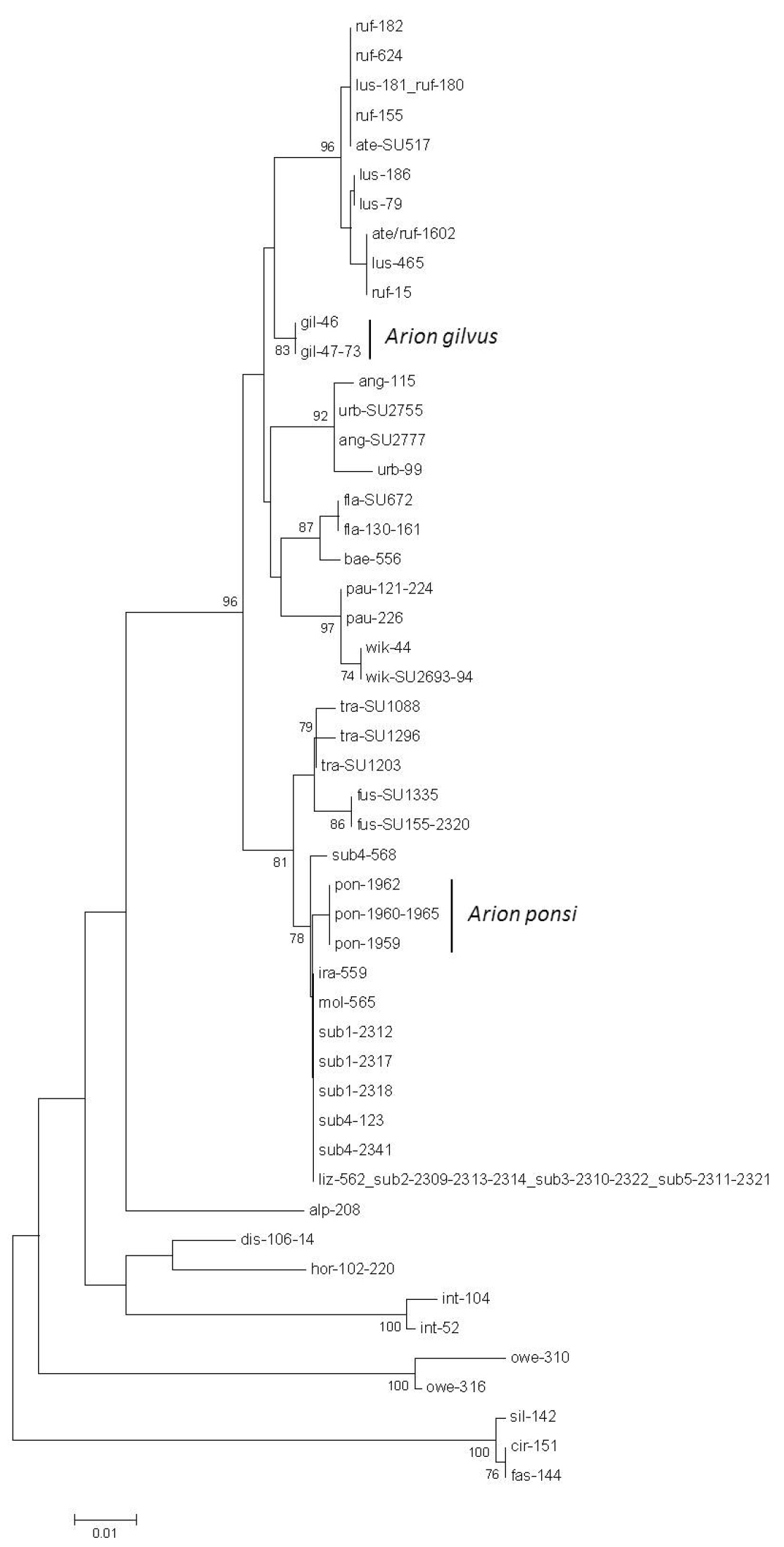
The four individuals of Arion ponsi yielded four COI and three 16S haplotypes (Appendix, Supplementary figures 1–2), yet two 16S haplotypes only differed by an indel of two base pairs at positions 291–292. For both genes Arion molinae showed the smallest p-distance with Arion ponsi (COI: mean p-distance 0.142 ± 0.014; 16S: mean p-distance 0.162 ± 0.019), but a sister species relationship with Arion molinae was only well-supported by 16S. There were three ITS1 haplotypes for Arion ponsi; one of these had a deletion of a poly-T stretch of six base pairs at positions 556–561; the other two differed by a deletion of a G at position 554. These three ITS1 haplotypes of Arion ponsi clustered within a clade of Arion subfuscus S1–5, Arion lizarrustii, Arion molinae, Arion iratii and Arion transsylvanus (Appendix, Supplementary figure 3). The ITS1 analysis with the sequences of
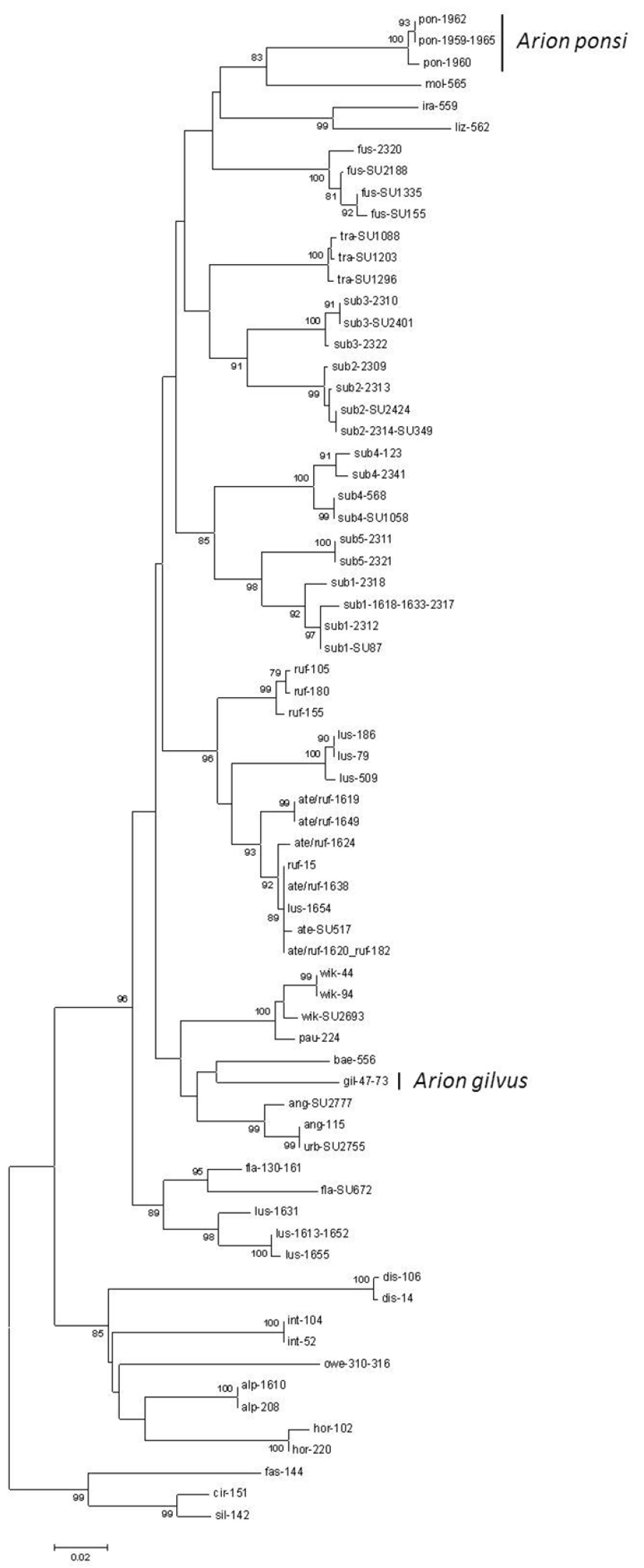
As for 16S, the concatenated tree of the three gene fragments showed a sister species relationship between Arion ponsi and Arion molinae (Figure 3).
The three Arion gilvus specimens yielded two COI (one synonymous A-G substitution at position 366) and one 16S haplotypes. For both genes the smallest mean p-distances were observed relative to Arion urbiae and Arion anguloi (COI: mean p-distance = 0.145 ± 0.013; 16S: mean p-distance = 0.134 ± 0.016). The two Arion gilvus ITS1 haplotypes reduced to one when considering the stretch that overlapped with the
The NJ-tree of the concatenated dataset confirms the major outcomes of previous phylogenetic studies, viz. 1) a strong support for the monophyly of the subgenus Carinarion (
Because Arion gilvus and Arion ponsi were originally described on morphological grounds they are to be interpreted as morphospecies. This phenetic morphological distinction, however, correlates well with a phenetic separation based on mtDNA distances. Indeed, the overall mean p-distance among the Mesarion MOTUs (excluding Arion ponsi and Arion gilvus) dealt with in this study is 16.7% for COI and 13% for 16S. As such, the mean p-distances between Arion ponsi and Arion molinae (COI: 14.2%; 16S: 16.2%) or between Arion gilvus and Arion urbiae (COI: 14.5%; 16S: 13.4%) are perfectly comparable with the mean p-distances among the other MOTUs and morphospecies in Mesarion. Hence, with respect to mtDNA divergence, both Arion ponsi and Arion gilvus, behave as other Mesarion species or putative species-level MOTUs.
Obviously, the strong COI differentiation among Mesarion taxa, and of Arion ponsi and Arion gilvus in particular, suggests that DNA barcoding may be a suitable identification tool for these animals. Yet, this may be a too simplistic conclusion, since stylommatophorans may show extremely high intraspecific mtDNA divergences of sometimes up to 27% (K2P-distances, but note the uncorrected p-distances are almost similar) (
Because DNA barcoding on its own may be unreliable for identifying and detecting species-level taxa in stylommatophorans, it its necessary to backup this sort of data with, amongst others, phylogenetic analyses. As such, our phylogenetic trees of the DNA sequence data show that the morphospecies Arion ponsi and Arion gilvus, also represent phylogenetic species, since both form well-supported clades that are “significantly” associated with well-defined, but morphologically different sister species. For Arion ponsi, the sister species appears to be Arion molinae, the distribution range of which is located in NE continental Spain (
In conclusion, the present work shows that Arion ponsi and Arion gilvus clearly differ from Arion subfuscus or any other currently recognized arionid species. As such, former records of Arion subfuscus from Menorca (e.g.
This work was supported by BELSPO Action 1 project MO/36/017 and FWO Research Network W0.009.11N “Belgian Network for DNA Barcoding”. We are indebted to Dr. Ramón Martín (Bilbao) for providing us with specimens of Arion gilvus, and to the editor of Spira to give us permission to use the pictures of Arion ponsi. We wish to thank Ben Rowson and one anonymous referee for their helpful comments.
Neighbour-Joining tree (Kimura 2-parameter model) of a 504 bp fragment of the mitochondrial cytochrome c oxidase subunit I (COI) gene for the land slug subgenus Mesarion. Bootstrap values ≥ 70% are shown at the nodes. For sample codes see Table 1.
Neighbour-Joining tree (Kimura 2-parameter model) of a 408 bp fragment of the mitochondrial 16S rDNA gene for the land slug subgenus Mesarion. Bootstrap values ≥ 70% are shown at the nodes. For sample codes see Table 1.
Neighbour-Joining tree (Kimura 2-parameter model) of a 587 bp fragment of the nuclear internal transcribed spacer 1 (ITS1) region for the land slug subgenus Mesarion. Bootstrap values ≥ 70% are shown at the nodes. For sample codes see Table 1.
Neighbour-Joining tree (Kimura 2-parameter model) of a 378 bp fragment of the nuclear internal transcribed spacer 1 (ITS1) region for the land slug subgenus Mesarion. This figure also includes the Iberian Mesarion ITS1 sequences of