(C) 2013 Juha Laiho. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Laiho J, Ståhls G (2013) DNA barcodes identify Central Asian Colias butterflies (Lepidoptera, Pieridae). In: Nagy ZT, Backeljau T, De Meyer M, Jordaens K (Eds) DNA barcoding: a practical tool for fundamental and applied biodiversity research. ZooKeys 365: 175–196. doi: 10.3897/zookeys.365.5879
A majority of the known Colias species (Lepidoptera: Pieridae, Coliadinae) occur in the mountainous regions of Central-Asia, vast areas that are hard to access, rendering the knowledge of many species limited due to the lack of extensive sampling. Two gene regions, the mitochondrial COI ‘barcode’ region and the nuclear ribosomal protein RpS2 gene region were used for exploring the utility of these DNA markers for species identification. A comprehensive sampling of COI barcodes for Central Asian Colias butterflies showed that the barcodes facilitated identification of most of the included species. Phylogenetic reconstruction based on parsimony and Neighbour-Joining recovered most species as monophyletic entities. For the RpS2 gene region species-specific sequences were registered for some of the included Colias spp. Nevertheless, this gene region was not deemed useful as additional molecular ‘barcode’. A parsimony analysis of the combined COI and RpS2 data did not support the current subgeneric classification based on morphological characteristics.
Barcoding, COI, Colias, Central-Asia, RpS2
The use of a standardized gene region, i.e. a 650 bp fragment of the 5’-region of the mitochondrial cytochrome c oxidase subunit I (hereafter COI), as a DNA barcode (
The butterfly genus Colias Fabricius, 1807 is a genus of the family Pieridae (subfamily Coliadinae), comprising about 85 species. Most of its species have a limited distribution in the Arctic and Alpine regions of the Holarctic realm, but two species occur in the Afrotropical and seven are known from the Neotropical regions (
The Central Asian mountainous regions harbour nearly half of all Colias species. The distribution, ecology and taxonomy are still incompletely documented for most of these species, mainly due to their remote occurrences (
The first contribution to the species classification of Colias was given by
The aim of the present study was to test the usefulness of COI barcodes for species identification of a broad representation of Central Asian Colias species, including nine Colias species overlapping with
This study includes material from the mountain regions of Kirgizistan, Tadzhikistan, northern Afghanistan, northern Pakistan and India (e.g. mountain ranges Tian Shan, Hindu Kush, Karakorum, Himalaya) and the mountain regions in the Chinese provinces Qinghai, Gansu, Sichuan, Yunnan and the autonomous regions Tibet and Xinjiang Uygur. The Colias fauna of these Central Asian regions comprises about 34 species (
The taxon sampling aimed to cover as many of the Colias species from this area as possible. Additionally, a few Colias species occurring in adjacent territories (e.g. Buryatia) were also available for molecular study. Whenever possible, several individuals of each species were analysed to assess intraspecific variation. The available specimens used for molecular study consisted of a total of 56 adult specimens covering 27 species of Central Asian Colias and two Colias species from adjacent territories (Table 1). The specimens are preserved as DNA voucher specimens and labelled accordingly, to be deposited in the collections of the Zoological Museum of Finnish Museum of Natural History, Helsinki, Finland (MZH) (DNA voucher specimens MZH_JL1-JL71). Species identifications were verified by JL based on easily recognizable diagnostic characters using the monograph by
List of specimens used for molecular analyses including GenBank accession numbers.
| Species | Sex | Locality and date | Lab code | COI accession number | RpS2 accession number |
|---|---|---|---|---|---|
| subgenus Colias Fabricius, 1807 | |||||
| Colias hyale (Linnaeus, 1758) irkutskana Stauder, 1923 | male | Russia, SW Transbaikalia, Buryatia, Selenga river district, Gusinoye Ozero village env., steppe rivulet valley, 7.6.2003 | MZH_JL35 | HE775142 | HE775198 |
| Colias hyale (Linnaeus, 1758) irkutskana Stauder, 1923 | male | Russia, SW Transbaikalia, Buryatia, Selenga river district, Gusinoye Ozero village env., steppe rivulet valley, 7.6.2003 | MZH_JL44 | HE775143 | HE775199 |
| subgenus Eriocolias Berger, 1986 | |||||
| Colias adelaidae adelaidae Verhulst, 1991 | male | China, Gansu, Xia-He, 3400 m, 35°11'N, 102°31'E, 25.6.2004 | MZH_JL61 | HE775187 | HE775243 |
| Colias alpherakii alpherakii Staudinger, 1882 | female | Kyrgyzstan, Alai mts., 4 km SE Tengizbai pass, 3400 m, 3.7.2001 | MZH_JL37 | HE775169 | HE775225 |
| Colias alpherakii alpherakii Staudinger, 1882 | female | Kyrgyzstan, Alai mts., 4 km SE Tengizbai pass, 3400 m, 3.7.2001 | MZH_JL51 | HE775180 | HE775236 |
| Colias berylla berylla Fawcett, 1904 | male | China, S Tibet, Himalaya Mts., Lablungla pass, 4800 m, 18–22.7.2001 | MZH_JL48 | HE775178 | HE775234 |
| Colias berylla berylla Fawcett, 1904 | male | China, Tibet, Lhodak, 4600 m, 15.7.2002 | MZH_JL55 | HE775182 | HE775238 |
| Colias christophi christophi Grum Grshimailo, 1885 | female | Tadjikistan, Turkestanskyi Mts., Kumbel pass, 3000 m, July 2002 | MZH_JL45 | HE775175 | HE775231 |
| Colias christophi helialaica Schulte, 1988 | male | Kyrgyzstan, Alai Mts., W end of Tengizbai pass, 3700 m, 5–6.7.2001 | MZH_JL67 | HE775192 | HE775246 |
| Colias cocandica cocandica Erschoff, 1874 | male | Kyrgyzstan, Suusamyr Mt. r., Alabel pass, 3200 m, 10.7.2002 | MZH_JL43 | HE775174 | HE775230 |
| Colias cocandica hinducucica Verity, 1911 | male | Tajikistan, E Pamir, Ak-Buura Mts., 4250 m, 14–15.7.2003 | MZH_JL34 | HE775168 | HE775224 |
| Colias cocandica pljushtchi Verhulst, 2000 | male | Kyrgyzstan, Sary Dzhaz riv. bas., Kaindy-Ketta mts., Tashkoro village, 3000 m 10.7.2003 | MZH_JL19 | HE775160 | HE775216 |
| Colias eogene C. et R. Felder, [1865] elissa Grum Grshimailo, 1890 | male | Kyrgyzstan, W end of Tengizbai pass, 3700 m, 5–6.7.2001 | MZH_JL1 | HE775144 | HE775200 |
| Colias eogene C. et R. Felder, [1865] elissa Grum Grshimailo, 1890 | male | Kyrgyzstan, W end of Tengizbai pass, 3700 m, 5–6.7.2001 | MZH_JL40 | HE775171 | HE775227 |
| Colias fieldii Ménétriés, 1855 chinensis Verity, 1909 | male | China, Sichuan, Zhangia, 3000 m, 32°47'N, 103°36'E, 6.6.2002 | MZH_JL50 | HE775179 | HE775235 |
| Colias fieldii Ménétriés, 1855 chinensis Verity, 1909 | female | China, Gansu, Shin-Long-Shan, 2800 m, 35°48'N, 103°59'E, 29.6.2004 | MZH_JL60 | HE775186 | HE775242 |
| Colias grumi grumi Alphéraky, 1897 | female | China, Gansu, Altun Shan, road from Aksay to Danjing pass, 2500–2800 m, 22–23.7.2002 | MZH_JL54 | HE775197 | - |
| Colias heos heos (Herbst, 1792) | male | Russia, SW Transbaikalia, Buryatia, Selenga river district, Gusinoye Ozero village env., steppe rivulet valley, 1.7.2003 | MZH_JL39 | HE775170 | HE775226 |
| Colias heos heos (Herbst, 1792) | male | Russia, SW Transbaikalia, Buryatia, Selenga river district, Gusinoye Ozero village env., steppe rivulet valley, 1.7.2003 | MZH_JL46 | HE775176 | HE775232 |
| Colias lada lada Grum Grshimailo, 1891 | male | China, Sichuan, Maningano surr., 31°56'N, 99°12'E, 4500 m, 15.6.2002 | MZH_JL7 | HE775150 | HE775206 |
| Colias lada lada Grum Grshimailo, 1891 | male | China, Sichuan, Maningano surr., 31°56'N, 99°12'E, 4500 m, 15.6.2002 | MZH_JL27 | HE775165 | HE775221 |
| Colias ladakensis Felder, 1865 seitzi Bollow, 1939 | male | China, SW Tibet, Himalaya Mts., 100km W Paryang, 4650–5000 m, 13.6.2004 | MZH_JL4 | HE775147 | HE775203 |
| Colias ladakensis Felder, 1865 seitzi Bollow, 1939 | male | China, SW Tibet, Himalaya Mts., 100km W Paryang, 4650–5000 m, 13.6.2004 | MZH_JL57 | HE775183 | HE775239 |
| Colias marcopolo marcopolo Grum Grshimailo, 1888 | male | Tadjikistan, E Pamir, Dunkeldyk Lake, 4400 m, 25.7.2003 | MZH_JL30 | HE775166 | HE775222 |
| Colias marcopolo marcopolo Grum Grshimailo, 1888 | male | Tadjikistan, E Pamir, Dunkeldyk Lake, 4400 m, 25.7.2003 | MZH_JL33 | HE775167 | HE775223 |
| Colias marcopolo marcopolo Grum Grshimailo, 1888 | male | Tadjikistan, E Pamir, Dunkeldyk Lake, 4400 m, 25.7.2003 | MZH_JL41 | HE775172 | HE775228 |
| Colias montium montium Oberthür, 1886 | male | China, Sichuan, Maningano surr., 31°55'N, 99°12'E, 4000 m, 9–18.6.2004 | MZH_JL59 | HE775185 | HE775241 |
| Colias nebulosa Oberthür, 1894 sungpani Bang-Haas, 1927 | male | China, Sichuan, Maningano surr., 31°56'N, 99°12'E, 4500 m, 15.6.2002 | MZH_JL9 | HE775152 | HE775208 |
| Colias nebulosa Oberthür, 1894 sungpani Bang-Haas, 1927 | male | China, Sichuan, Maningano surr., 31°56'N, 99°12'E, 4500 m, 15.6.2002 | MZH_JL24 | HE775162 | HE775218 |
| Colias nebulosa Oberthür, 1894 sungpani Bang-Haas, 1927 | male | China, Sichuan, Maningano surr., 31°56'N, 99°12'E, 4500 m, 15.6.2002 | MZH_JL26 | HE775164 | HE775220 |
| Colias nina Fawcett, 1904 hingstoni Riley, 1923 | male | China, SW Tibet, Himalaya Mts., 60 km S Saga, 4600–5000 m, 7–8.6.2004 | MZH_JL53 | HE775181 | HE775237 |
| Colias nina Fawcett, 1904 hingstoni Riley, 1923 | male | China, SW Tibet, Himalaya Mts., Lablongla pass, 4800 m, 5.6.2004 | MZH_JL58 | HE775184 | HE775240 |
| Colias regia regia Grum Grshimailo, 1887 | male | Kyrgyzstan, Kaindy-Ketta Mt. r., Kumar pass, 3200 m, 12.7.2003 | MZH_JL8 | HE775151 | HE775207 |
| Colias regia regia Grum Grshimailo, 1887 | male | Kyrgyzstan, Kaindy-Ketta Mt. r., Kumar pass, 3200 m, 12.7.2003 | MZH_JL42 | HE775173 | HE775229 |
| Colias romanovi romanovi Grum Grshimailo, 1885 | male | Kyrgyzstan, Alai mts., 4 km SE Tengizbai pass, 3400 m, 7–8.7.2001 | MZH_JL3 | HE775146 | HE775202 |
| Colias romanovi romanovi Grum Grshimailo, 1885 | male | Kyrgyzstan, Alai mts., 4 km SE Tengizbai pass, 3400 m, 7–8.7.2001 | MZH_JL47 | HE775177 | HE775233 |
| Colias sieversi sieversi Grum Grshimailo, 1887 | male | Tadjikistan, Peter I Mts., Ganishob, 2400 m, 17.6.2004 | MZH_JL70 | HE775195 | - |
| Colias sifanica sifanica Grum Grshimailo, 1891 | male | China, Gansu, Xia-He, 3400 m, 35°11'N, 102°31'E, 25.6.2004 | MZH_JL11 | HE775154 | HE775210 |
| Colias sifanica sifanica Grum Grshimailo, 1891 | male | China, Gansu, Xia-He, 3400 m, 35°11'N, 102°31'E, 25.6.2004 | MZH_JL64 | HE775189 | HE775245 |
| Colias staudingeri Alphéraky, 1881 pamira Grum Grshimailo, 1890 | male | Kyrgyzstan, Zaalaisky (Transalai) Mts., Altyn Dara river, 3000 m, 25.7.2000 | MZH_JL2 | HE775145 | HE775201 |
| Colias staudingeri Alphéraky, 1881 pamira Grum Grshimailo, 1890 | male | Kyrgyzstan, Zaalaisky (Transalai) Mts., Altyn Dara river, 3000 m, 25.7.2000 | MZH_JL13 | HE775156 | HE775212 |
| Colias staudingeri Alphéraky, 1881 pamira Grum Grshimailo, 1890 | male | Kyrgyzstan, Zaalaisky (Transalai) Mts., Altyn Dara river, 3000 m, 25.7.2000 | MZH_JL23 | HE775161 | HE775217 |
| Colias stoliczkana stoliczkana Moore, 1882 | male | India, Jammu Kashmir, Ladakh Range, Markha Valley, Ganda Pass, 4600 m, 12.7.2001 | MZH_JL15 | HE775158 | HE775214 |
| Colias thisoa Ménétriés, 1832 aeolides Grum Grshimailo, 1890 | male | Kyrgyzstan, Sary Dzhaz riv. bas., Kaindy-Ketta mts., Tashkoro village, 3000 m, 10.7.2003 | MZH_JL10 | HE775153 | HE775209 |
| Colias thisoa Ménétriés, 1832 aeolides Grum Grshimailo, 1890 | female | Kyrgyzstan, Sary Dzhaz riv. bas., Kaindy-Ketta mts., Tashkoro village, 3000 m, 10.7.2003 | MZH_JL17 | HE775159 | HE775215 |
| Colias thisoa Ménétriés, 1832 aeolides Grum Grshimailo, 1890 | female | Kyrgyzstan, Sary Dzhaz riv. bas., Kaindy-Ketta mts., Tashkoro village, 3000 m, 10.7.2003 | MZH_JL25 | HE775163 | HE775219 |
| Colias thrasibulus thrasibulus Fruhstorfer, 1910 | male | China, W Tibet, Mandhata Mt., 4900 m, 15–16.7.2003 | MZH_JL14 | HE775157 | HE775213 |
| Colias tibetana tibetana Riley, 1922 | male | China, Tibet, Himalaya Mts., Nyalam, 4200 m, 8.7.2003 | MZH_JL6 | HE775149 | HE775205 |
| Colias tibetana tibetana Riley, 1922 | male | China, SW Tibet, Himalaya Mts., Nyalam, 3700–4200 m, 28–30.6.2004 | MZH_JL63 | HE775188 | HE775244 |
| Colias wanda wanda Grum Grshimailo, 1907 | male | China, Qinghai, 20km NW of Zhidoi City, 4700–5000 m, 16.7.2000 | MZH_JL66 | HE775191 | - |
| Colias wanda wanda Grum Grshimailo, 1907 | male | China, S. Tibet, Cona, 4500–4700 m, 24–25.6.2004 | MZH_JL69 | HE775194 | - |
| Colias wiskotti Staudinger, 1882 draconis Grum Grshimailo, 1891 | male | Uzbekistan, Chandalas Mts., Chakmksh village, 2600 m, 27.6.2004 | MZH_JL71 | HE775196 | - |
| Colias wiskotti Staudinger, 1882 hofmannorum Eckweiler, 2000 | male | Iran, Khorasan, 75km SE of Birjand, 2200 m, 18–20.5.2002 | MZH_JL68 | HE775193 | - |
| Colias wiskotti Staudinger, 1882 separata Grum Grshimailo, 1888 | male | Kyrgyzstan, Alai mts., 4km SE Tengizbai pass, 3400 m, 3.7.2001 | MZH_JL65 | HE775190 | - |
| subgenus Eucolias Berger, 1986 | |||||
| Colias tyche tyche (de Boeber, 1812) | male | Russia, East Siberia, Lake Baikal, Khamar-Daban Mts., Slyudyanka river, taiga, 800 m, 14.6.2003 | MZH_JL5 | HE775148 | HE775204 |
| Colias tyche tyche (de Boeber, 1812) | male | Russia, East Sayan, Buryatia, Mondy env., Huruma river, 1500 m, 6.6.2002 | MZH_JL12 | HE775155 | HE775211 |
List of Colias GenBank samples of the COI barcode used in this study.
| Species | GenBank accession number |
|---|---|
| Colias alpherakii | FJ663407 |
| Colias christophi | FJ663409 |
| Colias chrysotheme elena | FJ663410 |
| Colias chrysotheme elena | FJ663411 |
| Colias croceus | EF457737 |
| Colias croceus | FJ663412 |
| Colias croceus | GU688507 |
| Colias croceus | HQ004279 |
| Colias croceus | HQ004282 |
| Colias eogene | FJ663415 |
| Colias eogene | FJ663416 |
| Colias erate amdensis | EF457736 |
| Colias erate poliographus | EF457735 |
| Colias erate poliographus | EU583852 |
| Colias erate poliographus | GU372561 |
| Colias fieldii | EF584859 |
| Colias hyale | FJ663418 |
| Colias hyale | FJ663421 |
| Colias hyale | HQ004297 |
| Colias hyperborea | EF457739 |
| Colias marcopolo | FJ663422 |
| Colias marcopolo | FJ663423 |
| Colias myrmidone | HQ004303 |
| Colias phicomone | HM393178 |
| Colias regia | FJ663427 |
| Colias tamerlana mongola | FJ663424 |
| Colias tamerlana mongola | FJ663425 |
| Colias tamerlana mongola | FJ663426 |
| Colias thisoa thisoa | FJ663429 |
| Colias tyche | FJ663430 |
| Colias wiskotti chrysoptera | FJ663431 |
| Colias wiskotti chrysoptera | FJ663432 |
| Colias wiskotti chrysoptera | FJ663433 |
| Colias wiskotti wiskotti | FJ663435 |
| Colias wiskotti wiskotti | FJ663436 |
Total genomic DNA was extracted form 2-5 legs of dried, pinned butterfly specimens using NucleoSpin® Tissue Kit (Machery-Nagel), according to manufacturer’s protocols, and resuspended in 50 µl ultrapure water.
The primer pair LCO-1490 (5’-GGTCAACAAATCATAAAGATATTGG-3’) and HCO-2198 (5’-TAAACTTCAGGGTGACCAAAAAATCA-3’) (Folmer et al. 1994) was used to amplify a ca. 650 bp fragment of the mitochondrial COI gene. The polymerase chain reactions (PCR) were done under the following parameters: initial heating 95 °C for 2 min, following 30 cycles of 94 °C for 30 s, 49 °C for 30 s and 72 °C for 2 min, followed by a final extension of 72 °C for 7 min. The primer pair RpS2 nF (5’-ATCWCGYGGTGGYGATAGAG-3’) and RpS2 nR (5’-ATGRGGCTTKCCRATCTTGT-3’) (
We analysed and clustered our sequence data using parsimony and Neighbour-Joining (NJ) of K2P-distances. We used parsimony and NJ for our newly generated COI sequence dataset, NJ for RpS2 sequences, parsimony for the concatenated COI and RpS2 sequences, and, finally, NJ for the combined COI sequences generated in this study and those in GB. All trees were rooted using Papilio glaucus (family Papilionidae) and Aporia crategi (Pieridae, subfamily Pierinae) as outgroup taxa.
Parsimony analysis was performed using NONA (
We obtained a 643 bp COI barcode for 56 Colias specimens, and a 409 bp fragment of RpS2 was obtained for 49 specimens (Table 1). A+T content of the COI sequences was 69.22%, and of the RpS2 45.0%. There were 115 parsimony informative sites for COI and 39 for RpS2.
Uncorrected pairwise divergences between ingroup taxa ranged between 1.09 and 4.09% (mean 2.77%) for COI and 0.0–1.7% (mean 1.0%) for RpS2. GenBank accession numbers are given in Table 1. Intraspecific uncorrected distances were up to 1.09% (in Colias thisoa) for COI, with specimens of most species differing by less than 4 nucleotide changes.
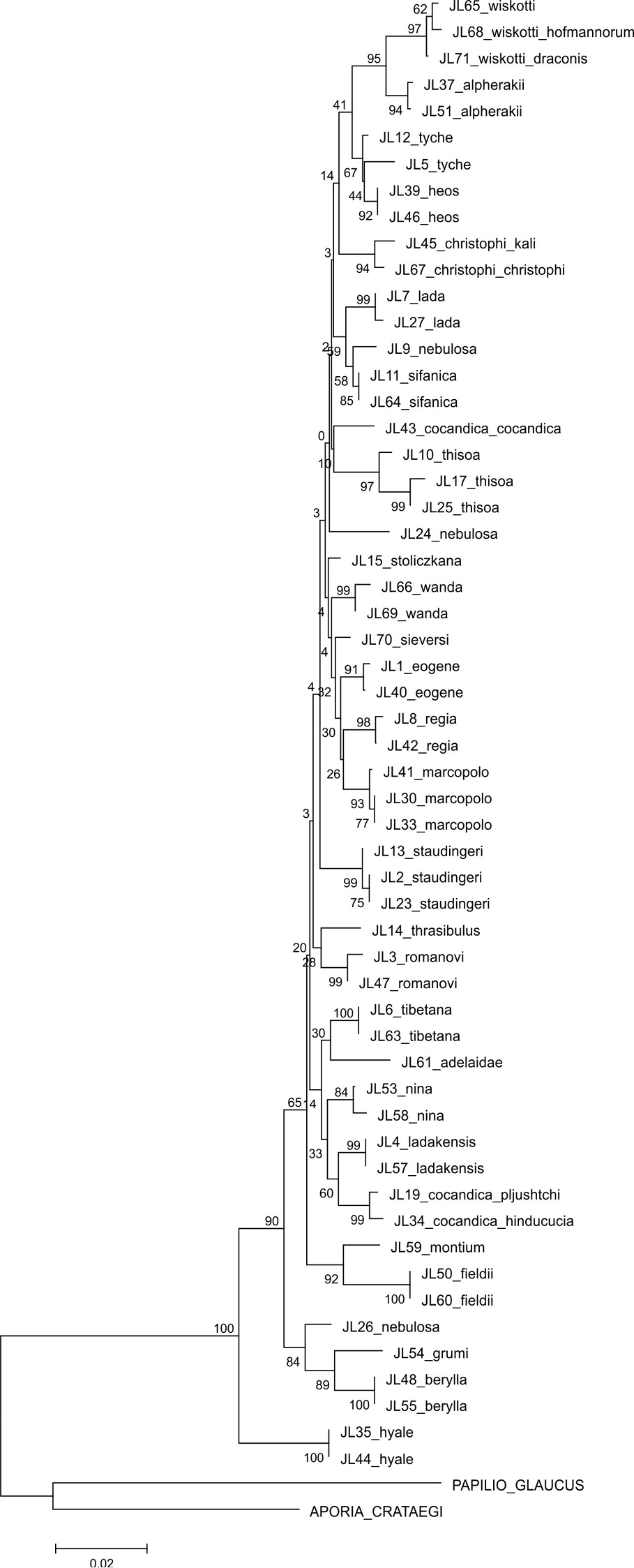
The parsimony analysis of the new COI sequences yielded four equally parsimonious trees (CI = 0.59, RI = 0.75) the strict consensus tree of which is presented in Figure 1. The NJ tree is presented in Figure 2.
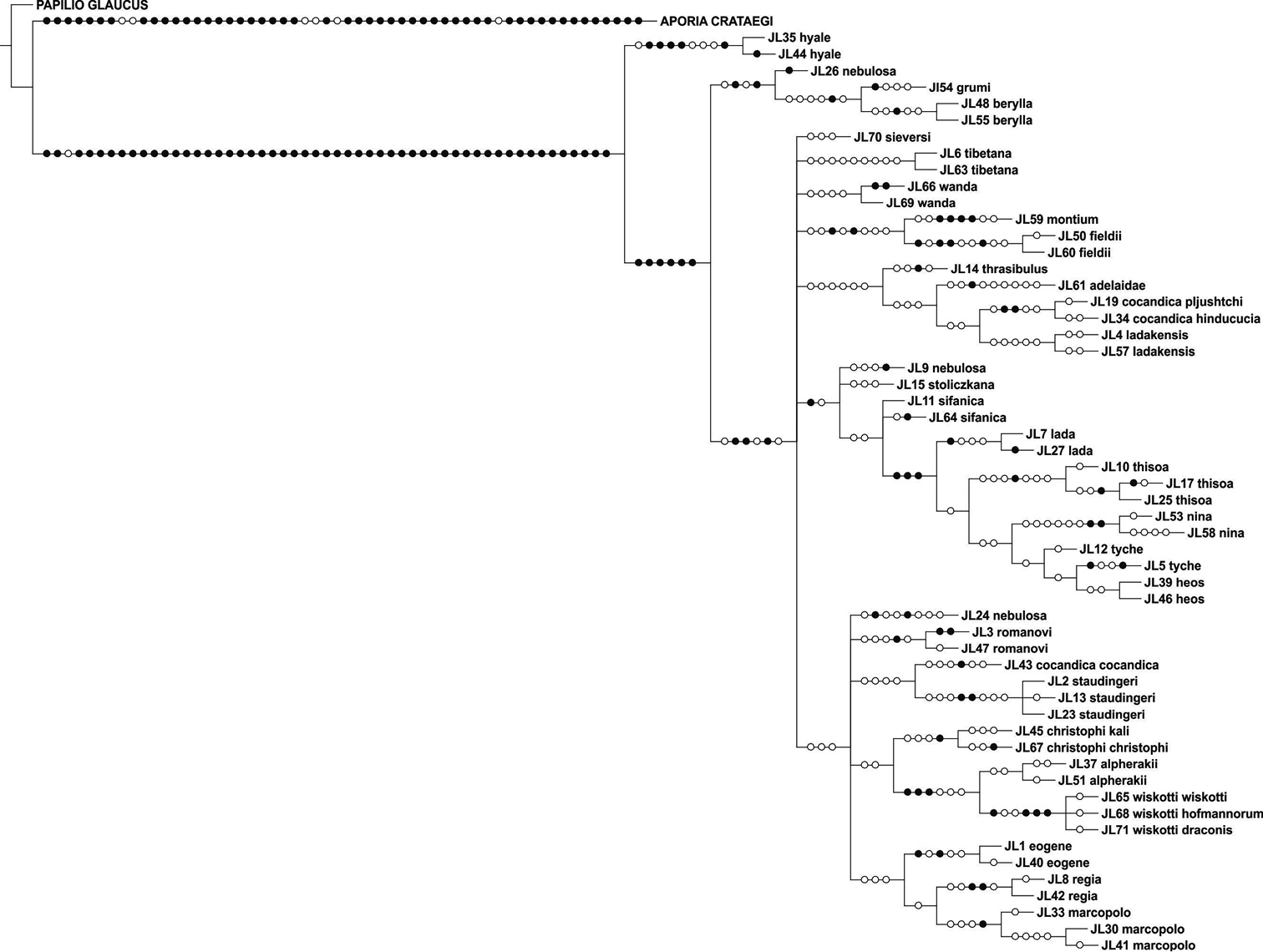
The majority of the species could be identified with COI alone, as no COI haplotypes were shared between species. Both parsimony and NJ trees recovered 25 (out of 28) species as monophyletic groups (Figures 1–2). Neither Colias cocandica, nor Colias nebulosa formed monophyletic entities, as their sequences were scattered over various parts of the trees. The two samples of Colias tyche were not recovered as sister taxa, for sample MZH_JL5 appeared as sister taxon of Colias heos. The overall topologies of the parsimony and NJ trees were identical, except for the placement of Colias thrasibulus. Parsimony placed the taxon as sister to a clade of five taxa (Figure 1), while NJ placed it as sister to Colias romanovi (Figure 2). The external morphology of Colias thrasibulus is rather different from that of Colias romanovi, while some similarities can be found between Colias thrasibulus and Colias nina, Colias ladakensis, Colias tibetana and Colias cocandica (Figure 1). Only 17 of the 39 parsimony informative sites of RpS2 were variable among the 49 ingroup members. NJ only recovered few species as separate lineages due to the shallow divergences (Figure 3). The information content of this gene region is best interpreted as a character-based diagnostic table, as suggested by
Strict consensus cladogram of Colias COI sequences obtained in this study.
Neighbour-Joining tree using the K2P-model for the COI sequences obtained in this study.
Neighbour-Joining tree using the Tamura-Nei model with gamma distributed rates for the RpS2 sequences.
Species haplotypes for 17 variable positions of RpS2 for Central Asian Colias species (RpS2 data matrix positions no 14, 152, 170, 176, 189, 191, 194, 195, 218, 284, 287, 302, 341, 353, 356, 365, 380).
| Haplotype | positions of RpS2 |
|---|---|
| MZH_JL35_hyale | TCCCCGGGTCCATTTTC |
| MZH_JL44_hyale | TCCCCGGGTCCATTTTC |
| MZH_JL02_staudingeri | TCCTCGAGTTCAAATCC |
| MZH_JL13_staudingeri | TCCTCGAGTTCAAATCC |
| MZH_JL23_staudingeri | TCCTCGAGTTCAAATCC |
| MZH_JL43_cocandica_cocandica | TCCCCGAGTTCAAATCC |
| MZH_JL41_marcopolo | TACCCGAGTTCAAAACC |
| MZH_JL30_marcopolo | TACCCGAGTTCAAAACC |
| MZH_JL07_lada | TCCCAAAAGTCGATTCC |
| MZH_JL27_lada | TCCCAAAAGTCGATTCC |
| MZH_JL25_thisoa | TCCCAAAAGTCGATTCC |
| MZH_JL10_thisoa | TCCCAAAAGTCGATTCC |
| MZH_JL17_thisoa | TCCCAAAAGTCGATTCC |
| MZH_JL05_tyche | TCCCAAAAGTCGATTCC |
| MZH_JL12_tyche | TCCCAAAAGTCGTTTCC |
| MZH_JL39_heos | TCCCAAAAGTCGATTCC |
| MZH_JL46_heos | TCCCAAAAGTCGATTCC |
| MZH_JL53_nina | TCCCAAAAGTCGATTCC |
| MZH_JL58_nina | CCCCCGAAGTCGATTCC |
| MZH_JL11_sifanica | TCCCCGAGGTCGWTTCC |
| MZH_JL64_sifanica | TCTCCGAGGTCGATTCC |
| MZH_JL57_ladakensis | TCCCCGAGGTCGATTCC |
| MZH_JL06_tibetana | TCCTCGAGGTTATTTCC |
| MZH_JL09_nebulosa | TCCTCGAGGTTATTTCC |
| MZH_JL26_nebulosa | TCCTCGAGGTTATTTCC |
| MZH_JL14_thrasibulus | TCCTCGAGGTTATTTCC |
| MZH_JL01_eogene | TCCTCGAGGTTATTTCT |
| MZH_JL04_ladakensis | TCTCCGAGGTTATTTCC |
| MZH_JL15_stoliczkana | TCTCCGAGGTTGTTTCT |
| MZH_JL19_cocandica_pljushtchi | TCCTCGAGTTCATTTCC |
| MZH_JL34_cocandica_hinducucia | TCCTCGAGTTCATTTCC |
| MZH_JL03_romanovi | TCCTCGAGTTCATTTCC |
| MZH_JL08_regia | TCCCCGAGTTCATTTCT |
| MZH_JL42_regia | TCCCCGAGTTCATTTCT |
| MZH_JL47_romanovi | CCCTCGAGTTCATTTCC |
| MZH_JL51_alpherakii | TCCCCGAGTTCATTTCC |
| MZH_JL37_alpherakii | CACCCGAGTTCATTTCC |
| MZH_JL67_christophi_christophi | TCCTCGAGTTCATTTCC |
| MZH_JL45_christophi_kali | TCCTCGAGTTCGTTTCC |
| MZH_JL40_eogene | TCCTCGAGGTTGTTTCT |
| MZH_JL24_nebulosa | TCCTCGAGGTCGTTTCC |
| MZH_JL59_montium | CCCTCGAGGTTGTTTCC |
| MZH_JL61_adelaidae | TCCTCGAGGTCGTTTCC |
| MZH_JL60_fieldii | TCCTCGAGGTTATTTCC |
| MZH_JL50_fieldii | TCCTCGAGGTTATTTCT |
| MZH_JL33_marcopolo | TCCCCGAGGTCATTACT |
| MZH_JL63_tibetana | TCCTCGAGGTTATWTCC |
| MZH_JL48_berylla | TCCCCGAGGTCGAATCC |
| MZH_JL55_berylla | TCCCCGAGGTCGAATCC |
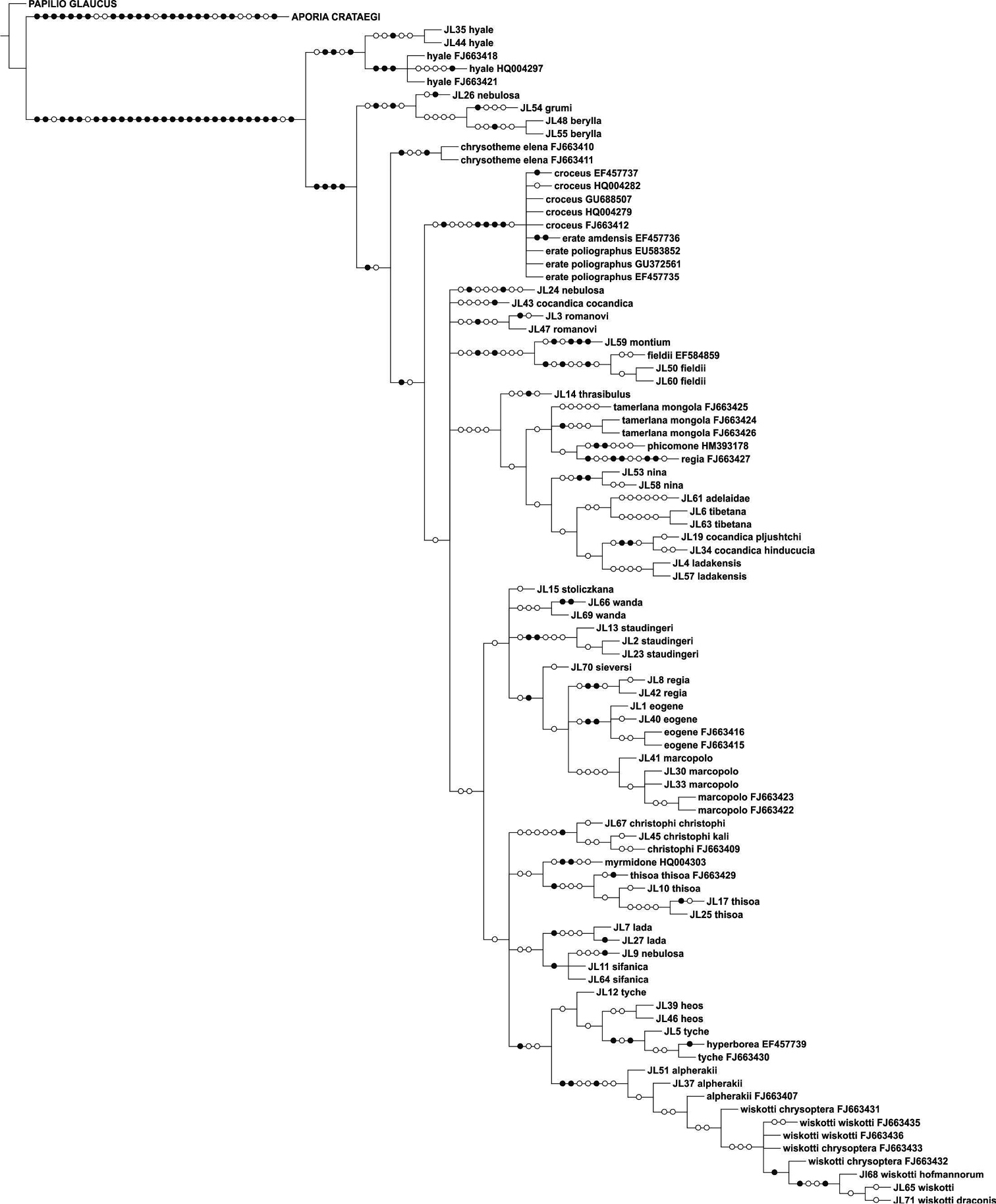
The parsimony analysis of COI + RpS2 yielded nine trees of length 560 steps (CI = 0.63, RI = 0.72), the strict consensus tree of which is shown in Figure 4. Colias cocandica, Colias nebulosa and Colias tyche were not monophyletic and Colias thrasibulus had the same position as in the COI cladogram (Figure 1).
Strict consensus cladogram of the concatenated data set of COI + RpS2.
The strict consensus cladogram for all the available COI data resolved the taxa in the same positions as in the tree of the new COI sequences only. For ten species of the present study sequences were also available from GB. Sequences of most species clustered together as monophyletic entities, except for Colias nebulosa, Colias cocandica, Colias tyche and Colias regia. For Colias regia the GB sequence (GB accession no FJ663427) did not cluster together with our sequences. The GB barcodes of Colias erate and Colias croceus were shared by these two taxa.
Neither the Himalayan and south Tibetan adjacent mountain Colias fauna (Colias berylla, Colias ladakensis, Colias nina, Colias stoliczkana, Colias thrasibulus, Colias tibetana), nor the east Tibetan, Qinghai, Gansu and Sichuan species aggregates (Colias adelaidae, Colias grumi, Colias lada, Colias montium, Colias nebulosa, Colias sifanica, Colias wanda) were resolved as species clusters similar to the Tian Shan, Pamir and Hindukush species.
Several COI haplotypes were noted for a few species, even among specimens obtained from the same locality (e.g. Colias staudingeri and Colias thisoa). Taxa not resolved as monophyletic clusters were the species Colias cocandica and Colias nebulosa. All the included subspecies of Colias cocandica (Colias cocandica cocandica, Colias cocandica pljutshtshi and Colias cocandica hinducucia) showed distinct COI sequences, with Colias cocandica cocandica as most different.
The fact that the three Colias nebulosa samples were scattered over different parts of the COI tree might be the result of a laboratory contamination due to carry over between samples. The Colias nebulosa samples were collected on the same day and in the same place. Colias nebulosa is morphologically distinct from other Colias species, excluding possible misidentification. The RpS2 data, however, could point to two morphologically cryptic species in sympatry (samples MZH_JL24 vs. MZH_JL9 and MZH_JL26), so that the different COI barcodes might represent numts, despite no apparent ‘signs’ (no indels). This discrepancy between morphology and DNA sequence data emphasises the necessity to use multiple samples to detect this sort of challenging issues.
Even though Colias cocandica and Colias nebulosa did not form monophyletic groups our results show that COI barcodes are useful for (1) identifying Palaearctic and Central Asian Colias, (2) pointing to a possible cryptic species, and (3) highlighting the necessity to further investigate the question on the subspecific rank of Colias cocandica cocandica.
The utility of RpS2 as a species barcode for Colias spp. is clearly more limited, since e.g. Colias heos, Colias lada, Colias nina, Colias thisoa of the subgenus Eriocolias and Colias tyche (subgenus Eucolias) have identical sequences (Table 3, Figure 3). Still, RpS2 yielded species specific (diagnostic) haplotypes for 11 species of the subgenus Eriocolias and for Colias hyale (subgenus Colias s.str.).
The strict consensus tree was more resolved than either of the trees resulting from separate analyses of the gene regions (Figure 4).
Although the concatenated data did not resolve the phylogenetic relationships among all Colias species, some observations can be made. The majority of the species confined to the adjacent Tian Shan, Pamir and Hindukush mountain ranges form a well supported clade. This includes Colias eogene, Colias regia, Colias romanovi, Colias marcopolo, Colias staudingeri, Colias christophi, Colias alpherakii and Colias wiskotti. Yet, Colias sieversi, which also occurs in these mountain ranges (Peter I and Khozratishoh mountains), was not included in this clade. Colias sieversi is morphologically most similar to Colias alpherakii, thus showing another case of disagreement between morphological and DNA sequence data. Colias thisoa, too, lives in the aforementioned mountain ranges, but it has a wider distribution, stretching from Turkey to the Altai Mountains. A third taxon, Colias cocandica cocandica, is considered closely related to Colias tamerlana (e.g.
The analyses did not support the monophyly of the subgenera Eucolias and Eriocolias sensu
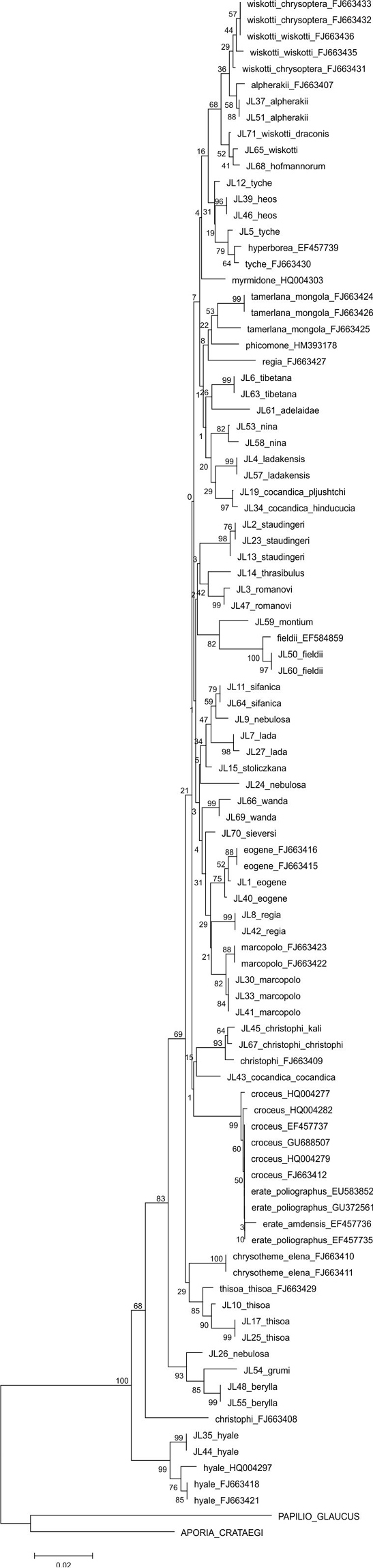
The parsimony (Figure 5) and NJ analyses (Figure 6) of the larger matrix of Palaearctic COI barcodes (total COI) recovered the same species clusters, but some of the species show different placements (e.g. Colias thisoa, Colias christophi). This is not surprising as all internal nodes are very shallow. The samples of Colias tyche and Colias hyperborea show very low sequence difference, morphologically these taxa are different, and they largely share the same distribution area. An example of species that share the same distribution and that exhibit clear morphological similarities, and which as such were resolved as sister species in both analyses, includes Colias wiskotti and Colias alpherakii. Identification of Palaearctic Colias based on COI barcodes is in most cases possible, since shared haplotypes were recorded only for Colias erate and Colias croceus.
Strict consensus cladogram of COI sequences for Palaearctic Colias taxa.
Neighbour-Joining tree using the K2P-model of COI sequences for Palaearctc Colias taxa.
Intraspecific variation is notable between some of the recognized subspecies, both among our own samples and those downloaded from GB. The intraspecific variation can partly be explained by morphologically clearly distinct subspecies, such as those of Colias wiskotti, or by specimens from widely different localities, such the different specimens of Colias hyale (sample FJ663418 from Russia, FJ663421 from Kazakhstan, HQ004297 from Romania and MZH_JL35 and MZH_JL44 from SW Transbaikalia). However, notable intraspecific variation also occurs within populations, such as Colias thisoa aeolides with all samples originating from the same locality and date, but the limited sampling prevents conclusions on the reasons for this. It is apparent that the understanding of intraspecific variability of the COI barcode for Colias is presently very limited.
The combined COI data of our sequences and sequences downloaded from GB include species belonging to one additional subgenus, Neocolias, represented by Colias myrmidone and Colias erate. Only the subgenus Colias, represented by Colias hyale, is well supported as distinct lineage. Yet, one specimen of Colias hyale (FJ663419) clustered together with Colias erate (Neocolias) and Colias croceus (Eriocolias). The other subgenera were not resolved as clades according to present classification, in agreement with our results for the combined analysis.
Our findings generally support COI as a species specific barcode for Colias, but we also highlight the necessity of including multiple individuals of species in molecular barcoding studies. Problematic ‘cases’ of widely divergent barcodes or conflicting morphological and molecular ‘signals’ are found in most if not all barcoding studies, and this study makes no exception.
JL thanks the Societas Entomologica Helsingforsiensis for support for the DNA work.