(C) 2013 Jonathan Marescaux. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Marescaux J, Van Doninck K (2013) Using DNA barcoding to differentiate invasive Dreissena species (Mollusca, Bivalvia). In: Nagy ZT, Backeljau T, De Meyer M, Jordaens K (Eds) DNA barcoding: a practical tool for fundamental and applied biodiversity research. ZooKeys 365: 235–244. doi: 10.3897/zookeys.365.5905
The zebra mussel (Dreissena polymorpha) and the quagga mussel (Dreissena rostriformis bugensis) are considered as the most competitive invaders in freshwaters of Europe and North America. Although shell characteristics exist to differentiate both species, phenotypic plasticity in the genus Dreissena does not always allow a clear identification. Therefore, the need to find an accurate identification method is essential. DNA barcoding has been proven to be an adequate procedure to discriminate species. The cytochrome c oxidase subunit I mitochondrial gene (COI) is considered as the standard barcode for animals. We tested the use of this gene as an efficient DNA barcode and found that it allow rapid and accurate identification of adult Dreissena individuals.
COI, zebra mussel, quagga mussel, barcoding gap, RFLP
Biological invasions are a topical issue in today’s world since they are the biggest threat to biodiversity after habitat destruction. The first, and probably the biggest, problem for scientists is to deal with widely divergent perceptions of the criteria defining “invasive” species (
The zebra mussel (Dreissena polymorpha (Pallas, 1771) and the quagga mussel (Dreissena rostriformis bugensis Andrusov, 1897) are invasive freshwater bivalves in Europe and North America (
Although Dreissena specialists may discriminate adults of the different species based on internal and external shell features (
Dreissena samples were collected in the Meuse River (see
Total genomic DNA was extracted from 241 Dreissena individuals using the «DNeasy Blood and Tissue» kit (Qiagen) according to manufacturer guidelines. To minimize cost, DNA extraction with the CTAB (hexadecyltrimethylamoniumbromide) protocol proposed by
Sequences were collapsed into unique haplotypes using DnaSP (
GenBank accession numbers and localities of Dreissena spp. sequences included in the network analysis.
| GenBank | Taxon | Location |
|---|---|---|
| DQ840122 | Dreissena polymorpha polymorpha | Black and Caspian Seas |
| DQ840125 | Dreissena polymorpha polymorpha | Liman, Caspian Sea |
| DQ840123 | Dreissena polymorpha polymorpha | Caspian Sea |
| DQ840121 | Dreissena polymorpha polymorpha | Black and Caspian Seas |
| EF414493 | Dreissena polymorpha | Turkey |
| U47653 | Dreissena polymorpha | Lake Ontario |
| AF474404 | Dreissena polymorpha | Poland |
| EU484441 | Dreissena polymorpha | Lake Superior |
| EU484437 | Dreissena polymorpha | Lake Superior |
| EU484448 | Dreissena polymorpha | Lake Superior |
| EU484444 | Dreissena polymorpha | Lake Superior |
| AM748997 | Dreissena polymorpha | Italy |
| AM748986 | Dreissena polymorpha | Germany |
| AM748977 | Dreissena polymorpha | Italy |
| U47651 | Dreissena bugensis | Lake Ontario |
| U47650 | Dreissena bugensis var. profunda | Lake Ontario |
| DQ840132 | Dreissena bugensis | Black Sea |
| EF080861 | Dreissena rostriformis bugensis | Hollandsch Diep |
| AF495877 | Dreissena bugensis | Ukraine |
| AF479637 | Dreissena bugensis | Ukraine |
| AM748999 | Dreissena polymorpha | Germany |
Using the restriction map application (http://www.bioinformatics.org/sms2/rest_map.html), we selected two endonucleases to differentially cut the COI gene of Dreissena species: Hinf I and Nla III. We also tested two other enzymes used in previous studies: Nla IV (
Restriction analysis of the amplified 654 bp COI fragment was carried out on each dreissenid haplotype (using individuals from the Meuse River). For each haplotype, the RFLP was performed in 31 μl total volume including 10 µl of PCR reaction mixture, 18 µl of distilled water, 2 µl of buffer (supplied by the manufacturer with the enzyme), and 1 µl of enzyme. Digests were incubated at 37 °C for 3 hours and then loaded on 2% agarose gels.
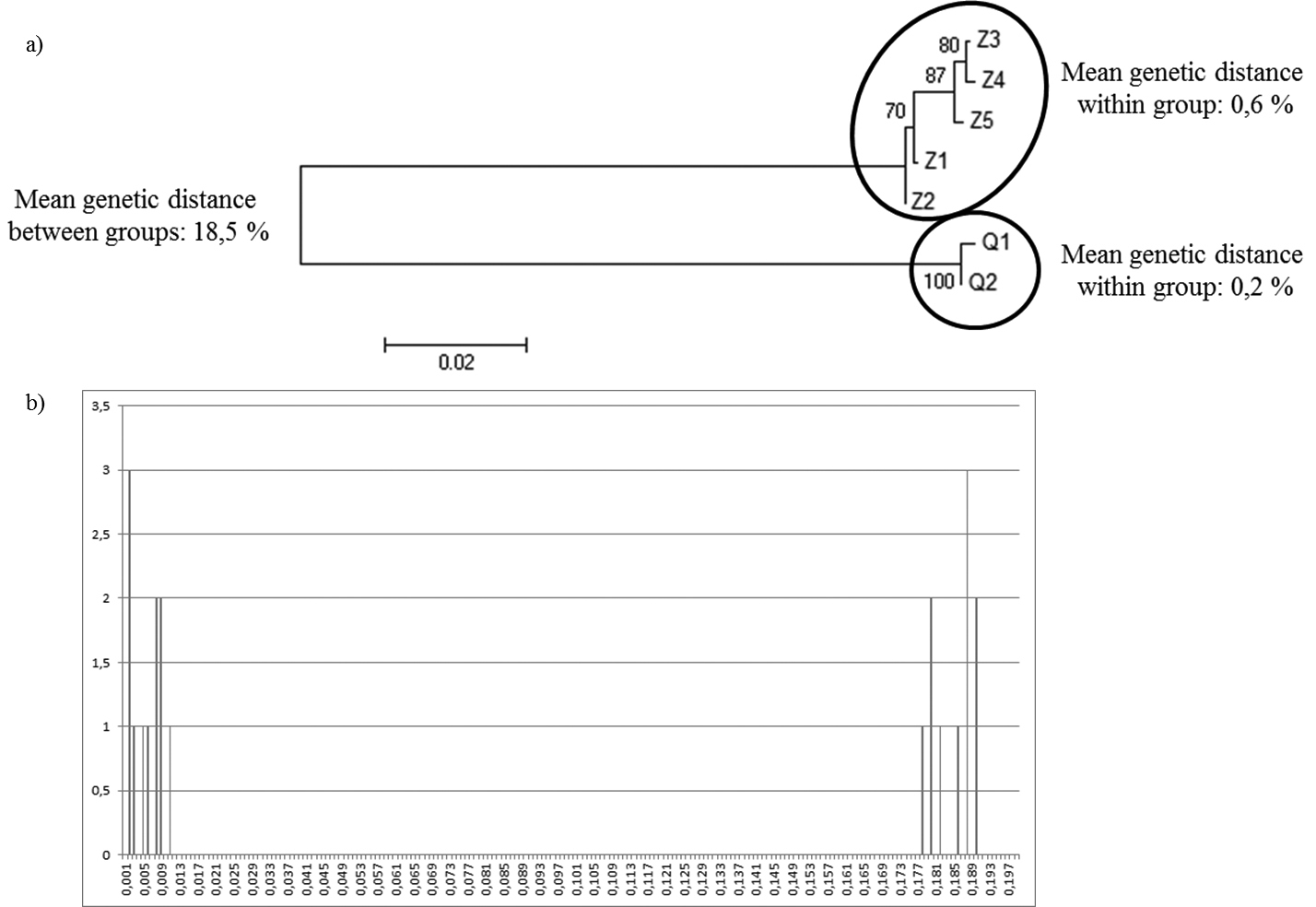
Sequencing of the 654 bp COI fragment revealed seven haplotypes among the 241 Dreissena individuals. The OTU method revealed, by a NJ tree, two clusters separated by a genetic distance of 18.5% (Figure 1a), which is higher than the 3% threshold typically used for species delimitation with COI (
Barcoding analysis based on a fragment of 654 base pairs of the COI gene. a) NJ analysis of K2P-pairwise distances b) “barcoding gap” method based on the K2P-pairwise distance.
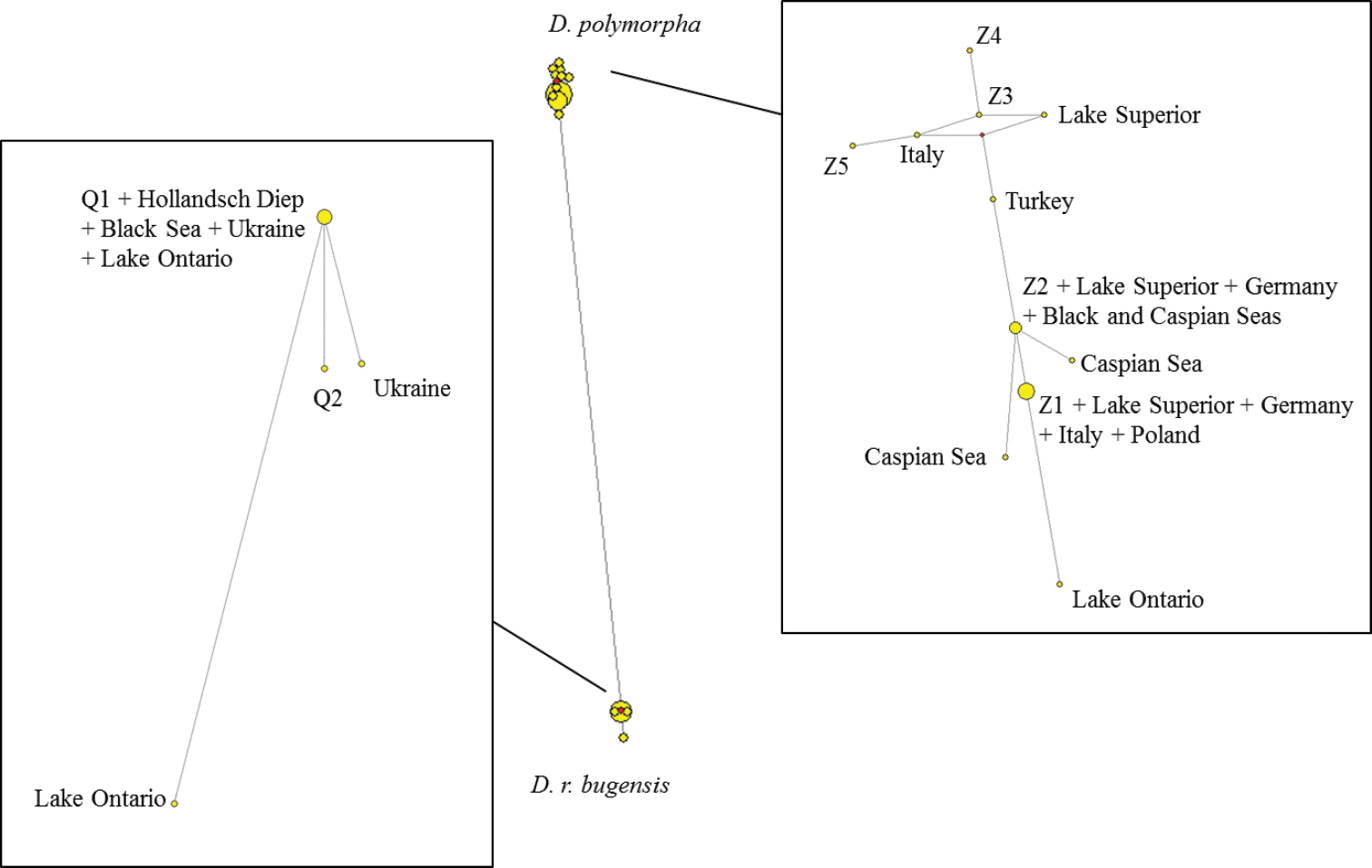
Our network (Figure 2) revealed that haplotypes 1 and 2 (Q1 and Q2) cluster with Dreissena rostriformis bugensis and the five other haplotypes (Z1 to Z5) cluster with Dreissena polymorpha. This, together with the three barcoding methods which each identified two clusters, shows that both Dreissena polymorpha and Dreissena rostriformis bugensis species occur in the Meuse River.
Haplotype networks based on a fragment of 654 base pairs of the COI gene. Our seven haplotypes are labelled: Q1 and Q2 for haplotypes 1 and 2 (belonging to Dreissena rostriformis bugensis) / Z1 to Z5 for the 5 other haplotypes (belonging to Dreissena polymorpha).
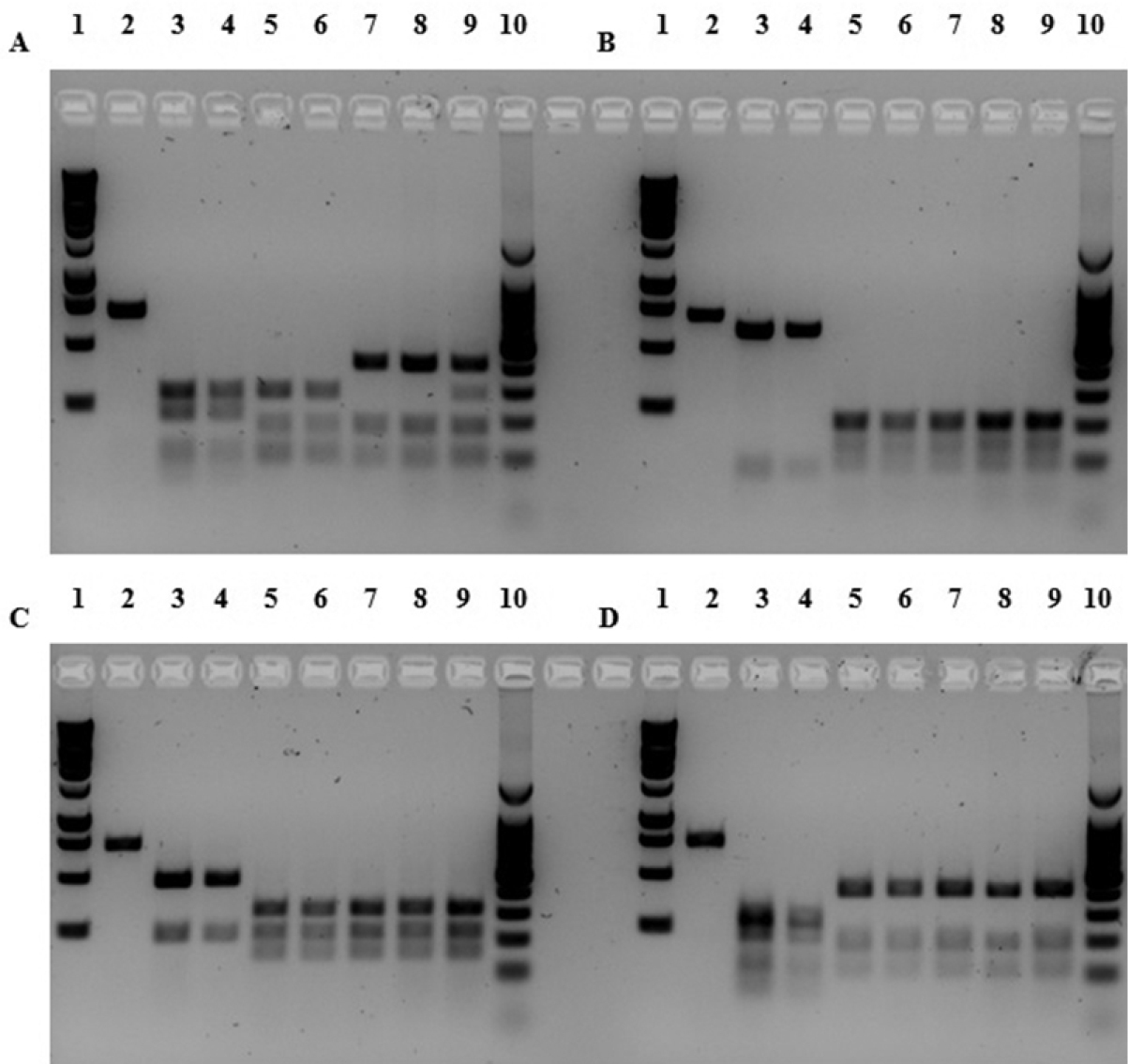
Digestion profiles for each haplotype are illustrated in Figure 3. Each of the four endonucleases tested, yielded distinct restriction patterns between both Dreissena species. Digestion with Nla IV produced four fragments in quagga mussels (Q haplotype) of approximately 70, 79, 211, and 294 bp and three distinct patterns for the zebra mussel (Z haplotype): haplotype Z1 and Z2 (91, 120, 150, and 293 bp), haplotype Z3 and Z4 (91, 150, and 413 bp), and haplotype Z5 (91, 150, 200, and 413 bp). We suggest here that the 200 bp fragment of the haplotype Z5 is an artefact, as confirmed by the restriction map, since the summed fragment lengths do not add up to the expected 654 bp. We infer that haplotype Z5 has the same pattern as haplotype Z3 and Z4. Digestion with Hinf I produced two fragments in quagga mussels of approximately 73 and 581 bp and five fragments in zebra mussels of approximately 31, 101, 114, 195, and 213 bp. The small fragments can not be distinguished on the gel but the difference between quagga and zebra is clear. Digestion with Nla III produced two fragments in quagga mussels of approximately 193 and 461 bp and three fragments in zebra mussels of approximately 193, 319, and 335 bp. Digestion with Scr FI produced five fragments in quagga mussels of approximately 42, 53, 120, 171, and 268 bp and three fragments in zebra mussels of approximately 95, 152, and 407 bp. The digestion pattern for the quagga mussel using the endonuclease Scr FI is not clearly defined (smear) since the five fragments are very short.
RFLP analysis of the COI gene to distinguish Dreissena rostriformis bugensis (Q haplotype) and Dreissena polymorpha (Z haplotype) using the endonucleases (A) Nla IV (B) Hinf I (C) Nla III and (D) Scr FI. Lane 1, 1-kb ladder; lane 2, non-digested fragment of quagga mussel; lane 3, Q1 haplotype; lane 4, Q2 haplotype; lane 5, Z1 haplotype; lane 6, Z2 haplotype; lane 7, Z3 haplotype; lane 8, Z4 haplotype; lane 9, Z5 haplotype; lane 10, 100-bp ladder.
On September 9 2013, the European Commission has published a proposal for a Regulation on the prevention and management of the introduction and spread of invasive alien species. This proposal highlights three types of interventions: prevention, early warning and rapid response, and then management of invasive species (
In order to help managers and national agencies, we propose here the use of the COI mitochondrial gene as a barcode to discriminate Dreissena polymorpha and Dreissena rostriformis bugensis. Moreover, it is possible to conduct a RFLP analysis on this gene to obtain results without sequencing cost. This method could also easily be applied to Dreissena presbensis and Dreissena blanci since the COIgene have already been sequenced by
This study is the first step of an extensive phylogeographical analysis on the invasion of Western Europe by the dreissenids. Further experiments will be needed to assess potential risks of both zebra and quagga mussels on native biodiversity in Western European rivers, e.g. predation on phytoplankton, infestation on native bivalves and alteration of macro-invertebrate communities.
Special thanks to Emilie Etoundi and Doctor Xiang Li for the help with the delimitation metrics. We also thank two reviewers and the editor for their helpful comments and critical reading of this manuscript. This study received financial support from the University of Namur. Jonathan Marescaux is funded by a PhD grant from the Belgian National Fund for Scientific Research (FRS-FNRS).