(C) 2011 David R. Maddison. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The phylogeny of ground beetles of supertribe Trechitae is inferred using DNA sequences of genes that code for 28S ribosomal RNA, 18S ribosomal RNA, and wingless. Within the outgroups, austral psydrines are inferred to be monophyletic, and separate from the three genera of true Psydrina (Psydrus, Nomius, Laccocenus); the austral psydrines are formally removed from Psydrini and are treated herein as their own tribe, Moriomorphini Sloane. All three genes place Gehringia with Psydrina. Trechitae is inferred to be monophyletic, and sister to Patrobini.
Within trechites, evidence is presented that Tasmanitachoides is not a tachyine, but is instead a member of Trechini. Perileptus is a member of subtribe Trechodina. Against Erwin’s hypothesis of anillines as a polyphyletic lineage derived from the tachyine genus Paratachys, the anillines sampled are monophyletic, and not related to Paratachys. Zolini, Pogonini, Tachyina, and Xystosomina are all monophyletic, with the latter two being sister groups. The relationships of the subtribe Bembidiina were studied in greater detail. Phrypeus is only distantly related to Bembidion, and there is no evidence from sequence data that it belongs within Bembidiina. Three groups that have been recently considered to be outside of the large genus Bembidion are shown to be derived members of Bembidion, related to subgroups: Cillenus is related to the Ocydromus complex of Bembidion, Zecillenus is related to the New Zealand subgenus Zeplataphus, and Hydrium is close to subgenus Metallina. The relationships among major lineages of Trechitae are not, however, resolved with these data.
ground beetles, DNA, molecular phylogeny, Bembidiini, Trechinae, Carabidae, Trechitae,
The supertribe Trechitae comprises over 5, 300 described species (
The basic structure of the phylogeny of trechites is not well known. The only explicit, modern analyses have been based upon limited characters of adult and larval structure (
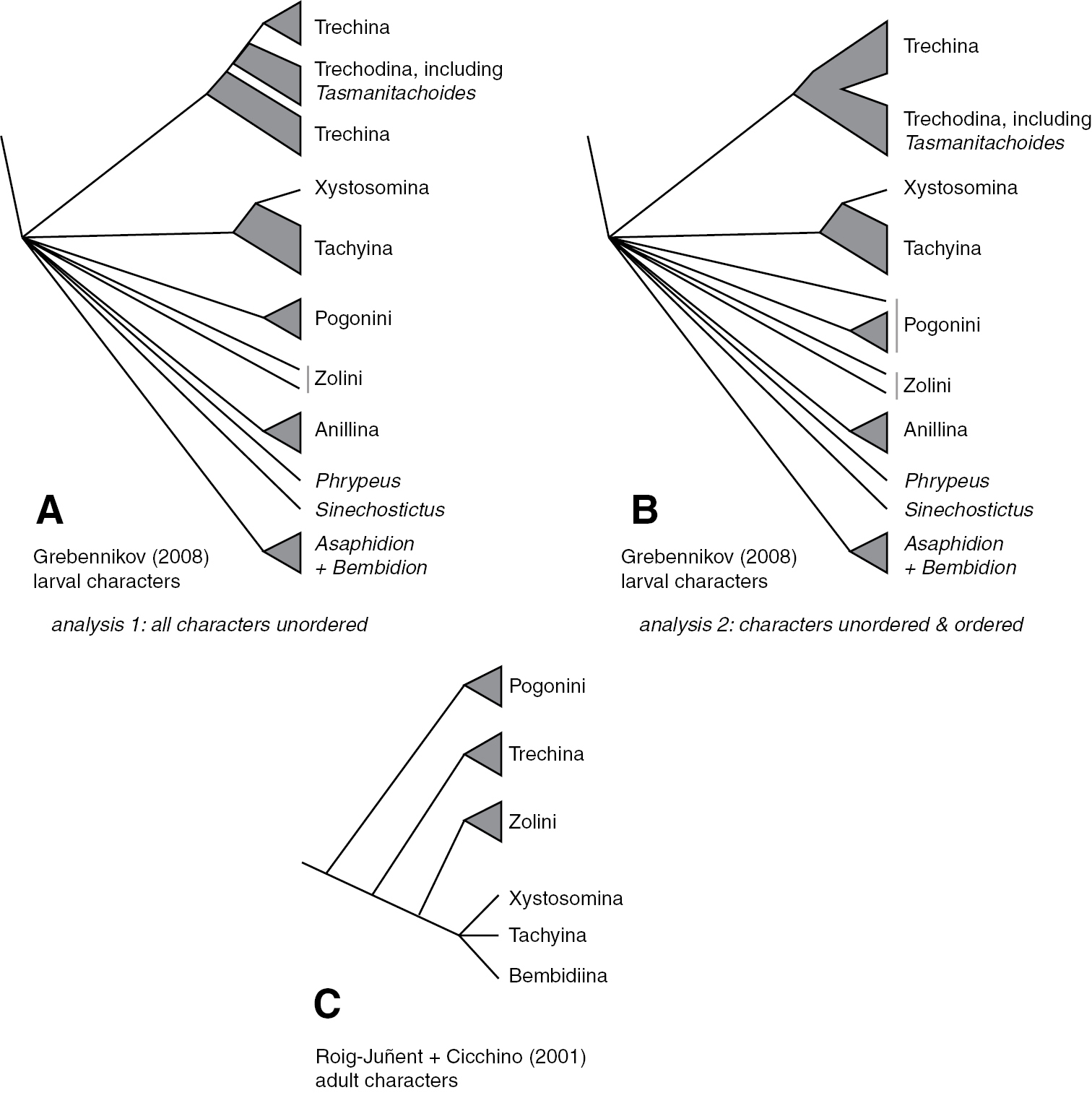
Phylogenies of Trechitae from morphological studies A Strict consensus tree of most parsimonious trees from larval data, with all characters treated as unordered, from
We present here the first detailed examination of relationships within Trechitae based upon DNA sequences, using portions of genes for small (18S) and large (28S) subunits of ribosomal RNA, as well as the nuclear protein-coding gene wingless.
This paper has been over a decade in gestation, and some results have already been reported in other publications. For example, the discovery from the DNA sequence data reported herein of Tasmanitachoides place in Trechini rather than in Tachyina was the inspiration for Grebennikov’s search for Tasmanitachoides larvae, which as he recently reported (
Taxa examined. The 14 outgroup species included are listed in Table 1. Morphological data and previously collected 18S rDNA data suggests that the sister group of Trechitae is likely Patrobini (
Outgroup taxon sampling. Four-digit numbers in entries are D.R. Maddison voucher numbers; further information on the specimens is given in the Appendix; where two numbers are listed, the sequence was formed by combining data from both specimens. Other entries are GenBank numbers of previously published sequences from
| 18S | 28S | wingless | |
|---|---|---|---|
| Pterostichini | |||
| Pterostichus melanarius Illiger | AF002779 | AF398707 | AF398623 |
| Moriomorphini | |||
| Amblytelus curtus (Fabricius) | AF012484 | AF398683 | AF398566 |
| Mecyclothorax vulcanus (Blackburn) | AF012482 | AF398648 | AF398601 |
| Melisodera picipennis Westwood | AF012481 | AF398640 | AF398602 |
| Meonis sp. | AF398722 | AF398692 | AF398603 |
| Sitaphe parallelipennis Baehr | 0669 | 2247 | 0669 |
| Tropopterus sp. | AF012483 | 2200 | 2200 |
| Psydrini | |||
| Laccocenus ambiguus Sloane | AF012486 | AF398675 | AF398596 |
| Psydrus piceus LeConte | AF002784 | AF398684 | 1627 |
| Nomius pygmaeus (Dejean) | 0893 | AF438100 | AF437971 |
| Gehringiini | |||
| Gehringia olympica Darlington | AF012512 | AF398702 | AF398591 |
| Patrobini | |||
| Diplous californicus (Motschulsky) | AF002785 | AF398699 | AF398587 |
| Patrobus longicornis (Say) | AF002786 | AF398700 | AF398613 |
| Penetretus temporalis Bedel | 0631, 1710 | 0631 | 0631 |
Within Trechitae, 64 species in 40 genera are sampled, with all tribes represented, and an emphasis on subtribe Bembidiina (Table 2). The classification used here is modified version of
Taxon sampling of trechites. Four-digit numbers in entries are D.R. Maddison voucher numbers; further information on the specimens is given in the Appendix; where two numbers are listed, the sequence was formed by combining data from both specimens. Other entries are GenBank numbers of previously published sequences from
| 18S | 28S | wingless | |
|---|---|---|---|
| Trechini: Trechodina | |||
| Cnides sp. | 1808 | 0691 | |
| Pachydesus sp. | 0678 | AF438112 | AF437978 |
| Perileptus areolatus (Creutzer) | 1707 | 0824 | 1707 |
| Perileptus sloanei Moore | 0767 | ||
| Thalassophilus longicornis (Sturm) | 0823 | 0823 | |
| Trechodes bipartitus (MacLeay) | 0705 | 0705 | 0705 |
| Trechodes jeanneli jeanneli Mateu | 0606 | ||
| Trechosiella sp. | 1709 | 0723 | 1709 |
| Tasmanitachoides fitzroyi (Darlington) | 1575 | 0762 | |
| Trechini: Trechina | |||
| Trechus chalybeus species group | AF002793 | ||
| Trechus oregonensis Hatch | AF398673 | 0587 | |
| Omalodera limbata Blanchard | 0571 | 0571 | 0571 |
| Homaloderodes germaini Jeannel | 1066 | ||
| Kenodactylus audouini (Guérin-Méneville) | 0670 | 0670 | |
| Paratrechus sp. | 1076 | ||
| Trechinotus flavocinctus Jeannel | 0575 | 0575 | 0575 |
| Zolini | |||
| Merizodus angusticollis Solier | AF012487 | 0453 | 0453 |
| Oopterus sp. | AF012488 | 0387 | 0387 |
| Oopterus helmsi (Sharp) | AF002787 | 0354 | 0354 |
| Sloaneana tasmaniae (Sloane) | AF002788 | 0339 | 0339 |
| Pogonini | |||
| Diplochaetus planatus (G.H. Horn) | AF002789 | AF438060 | AF437938 |
| Pogonus (Pogonus) chalceus (Marsham) | 1711 | 0679 | 0679 |
| Thalassotrechus barbarae (G.H.Horn) | 0703 | 0530 | |
| Bembidiini: Tachyina | |||
| Lymnastis sp. | 0988 | 0988 | 0988 |
| Micratopus sp. | 0605 | 0605 | |
| Paratachys vorax (LeConte) | 0410 | 0410 | |
| Elaphropus obesulus LeConte | 0411 | 0411 | 0411 |
| Elaphropus cf. nigrolimbatus Peringuey | 0761 | ||
| Elaphropus sp. 3 | 0713 | 0713 | |
| Pericompsus laetulus LeConte | AF002790 | 0429 | 0429 |
| Polyderis rufotestacea (Hayward) | 0717, 0718 | 0718 | |
| Tachys vittiger LeConte | 0760 | ||
| Tachys corax LeConte | 0604 | 0604 | |
| Tachyta nana inornata (Say) | 0573 | AF438141 | AF438002 |
| Bembidiini: Xystosomina | |||
| Erwiniana hilaris (Bates) | AF012489 | 0409 | 0409 |
| Erwiniana crassa (Erwin) | 0989 | 0989 | |
| Mioptachys flavicauda Say | 0684 | 0684 | 0684 |
| Philipis bicolor Baehr | 0592 | 0592 | |
| Bembidiini: Anillina | |||
| Anillinus (langdoni group) sp. | 0690 | 0690 | |
| Serranillus sp. | 1084 | 1084 | |
| Typhlocharis armata Coiffait | 0572, 1718 | 0572 | 0572 |
| Nesamblyops sp. | 0696 | ||
| Bembidiini: Bembidiina | |||
| Asaphidion alaskanum Wickham | 0585 | 0585 | |
| Asaphidion championi Andrewes | 0574 | ||
| Asaphidion curtum (Heyden) | AF002792 | 0267 | 0267 |
| Amerizus (Amerizus) sp. | 0576 | 0576 | |
| Ocys harpaloides (Audinet-Serville) | 0569 | 0569 | |
| Phrypeus rickseckeri Hayward | 0776 | 0692 | |
| Sinechostichus solarii (G. Müller) | 0603 | 0603 | |
| Bembidion (Antiperyphanes)sp. nr. chilense Solier | 0714 | 0714 | |
| Bembidion (Hoquedela) cf. csikii Jedlicka | 0916 | 0916 | |
| Bembidion (Cillenus) laterale (Samouelle) | 0602 | 0602 | |
| Bembidion (Notaphus) insulatum (LeConte) | 0444 | 0444 | |
| Bembidion (Bracteon) balli Lindroth | EF648613 | EF648838 | EF649474 |
| Bembidion (Odontium) coxendix Say | EF648618 | EF648837 | EF649481 |
| Bembidion (Metallina) dyschirinum LeConte | 0896 | 0896 | |
| Bembidion (Pseudoperyphus) integrum Casey | EF648659 | EF649056 | EF649609 |
| Bembidion (Bracteon) levettei carrianum Casey | EF648620 | EF648842 | EF649480 |
| Bembidion (Hydrium) levigatum Say | 0763 | 0763 | |
| Bembidion (Ocydromus) mexicanum Dejean | AF012490 | 0260 | 0262 |
| Bembidion (Phyla) obtusum Audinet-Serville | 0895 | 0895 | |
| Bembidion (Melomalus) planatum (LeConte) | 0601 | 0601 | |
| Bembidion (Bembidion) quadrimaculatum dubitans (LeConte) | 0676 | 0676 | 0676 |
| Bembidion (Zeplataphus) tairuense Bates | 0607 | 0607 | 0607 |
| Bembidion (Zecillenus) sp. | 0595 | 0595 | 0595 |
Locality information for the specimens newly sequenced in this paper is given in the Appendix. Voucher specimens are deposited in the David Maddison voucher collection in the Oregon State Arthropod Collection at Oregon State University.
DNA sequencing. Methods for obtaining DNA sequences, including extraction methods and cycling reactions, are described in
Primers used for DNA amplification and sequencing. Dir: direction of primer, either forward (F) or reverse (R). Syn: primer synonym. Kind: primer used for original PCR amplification and sequencing (A) or primer used only for sequencing (S). Original references for primer sequences are given in
| Gene | Primer | Syn | Dir | Kind | Sequence |
|---|---|---|---|---|---|
| 28S | LS58F | D1 | F | A | GGGAGGAAAAGAAACTAAC |
| LS998R | D3 | R | A | GCATAGTTCACCATCTTTC | |
| 18S | SS27F | 518S | F | A | TATGCTTGTCTCAAAGATTAA |
| S1893R | 18L | R | A | CACCYACGGAAACCTTGTTACGACTT | |
| SS398F | 18Sai | F | S | CCTGAGAAACGGCTACCACATC | |
| SS1054F | 760F | F | S | ATCAAGAACGAAAGT | |
| SS1090R | 18Sbi | R | S | GAGTCTCGTTCGTTATCGGA | |
| SS1554R | 909R | R | S | GTCCTGTTCCATTATTCCAT | |
| wg | wg550F | F | A | ATGCGTCAGGARTGYAARTGYCAYGGYATGTC | |
| wgAbRZ | R | A | CACTTNACYTCRCARCACCARTG | ||
| wg578F | F | A | TGCACNGTGAARACYTGCTGGATG | ||
| wgAbR | R | A | YTCGCAGCACCARTGGAA | ||
| B5wg1 | F | A | GARTGYAAGTGTCAYGGYATGTCTGG | ||
| 5wg | F | A | GARTGYAARTCYCAYGGYATGTCTGG | ||
| 5wgB | F | A | ACBTGYTGGATGCGNCTKCC | ||
| 3wg2 | R | A | CTCGCARCACCARTGGAATGTRCA | ||
| B3wg2 | R | A | ACTCGCARCACCAGTGGAATGTRCA | ||
| 3wg | R | A | ACTCGCARCACCARTGGAATGTRCA |
Assembly of multiple chromatograms for each gene fragment and initial base calls were made with Sequencher (Gene Codes Corporation) or using Phred (
Newly obtained sequences have been deposited in GenBank with accession numbers GU556024 through GU556153.
Alignment and resulting matrices. The two ribosomal genes, 18S and 28S, were aligned using ClustalW 1.8.3, with a gap opening cost of 10, gap extension of 0.1, then adjusted by eye; areas of uncertain alignment were excluded.
The amino acid translation of the wingless gene was aligned using Clustal W version 1.83 (
Phylogenetic inference. Each of the four matrices (28S, 18S, and the two wingless matrices) were subjected to parsimony, Bayesian, and maximum likelihood analyses.
Most-parsimonious trees were sought using PAUP* (
For parsimony bootstrap analyses in PAUP*, 1000 bootstrap replicates were conducted, each of which used a heuristic search with five replicates, each beginning with a starting tree formed by the random addition sequence option, with TBR branch rearrangement, with each replicate saving no more than 25 trees.
Models of nucleotide evolution chosen with the aid of ModelTest (
Bayesian analyses were conducted using MrBayes (
Likelihood analyses of nucleotide data were conducted using RAxML version 7.0.4 (
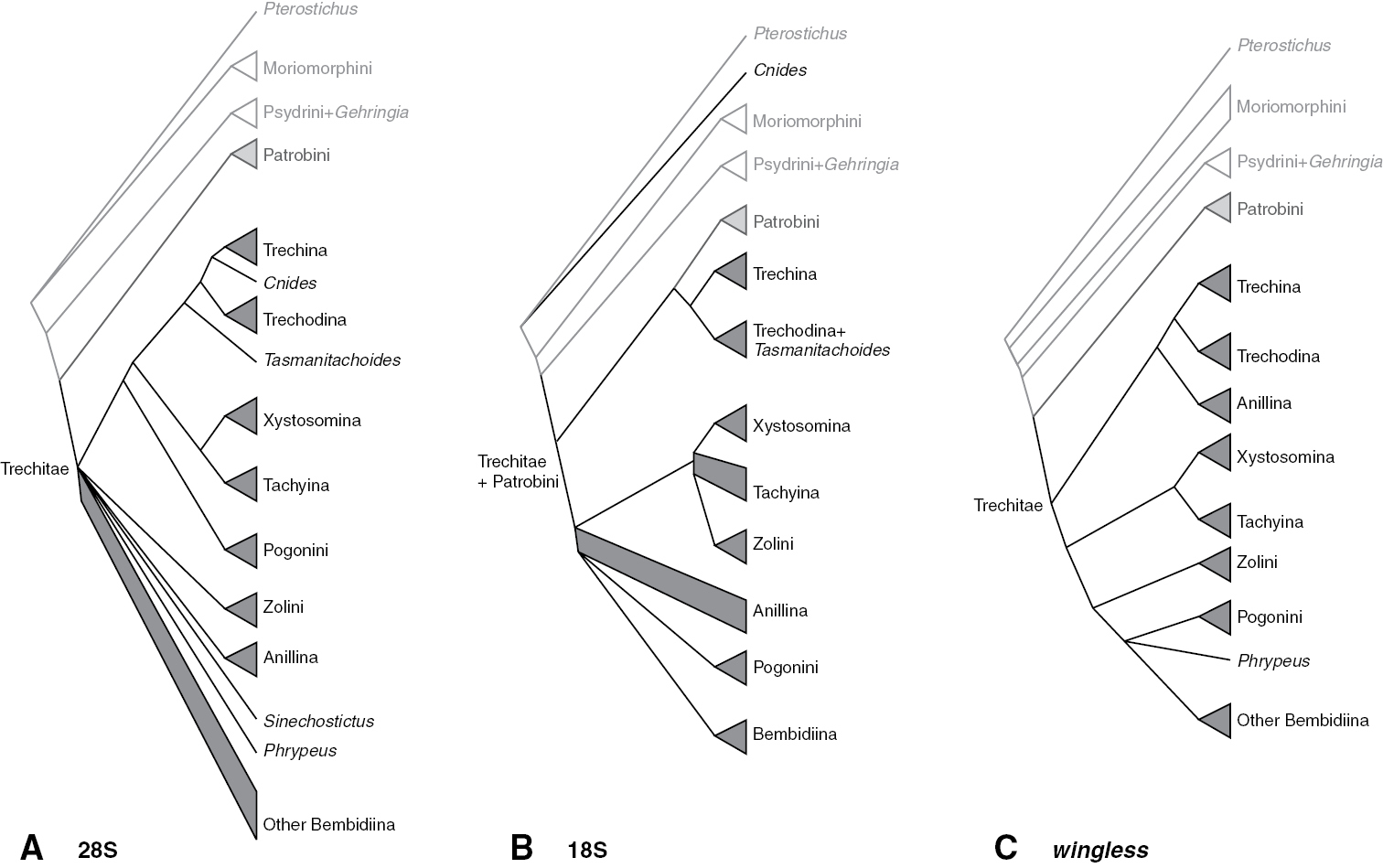
Several of the analyses of 18S rDNA yielded trees with the trechine Cnides, whose terminal branch was extremely long, well outside of Trechitae. As morphological data indicates definitively that Cnides is a trechite, some analyses were performed that forced it to reside within Trechitae. Two full suites of constrained analyses were conducted, one with Trechitae constrained to be monophyletic, and other with Trechini constrained to be monophyletic. For the latter, the position of Tasmanitachoides was not constrained in likelihood and parsimony analyses, allowing it to move anywhere on the tree.
All trees are shown rooted within the outgroup, arbitrarily next to Pterostichus.
Grebennikov’s (2008) larval morphological data were reanalyzed using TNT version 1.1 (
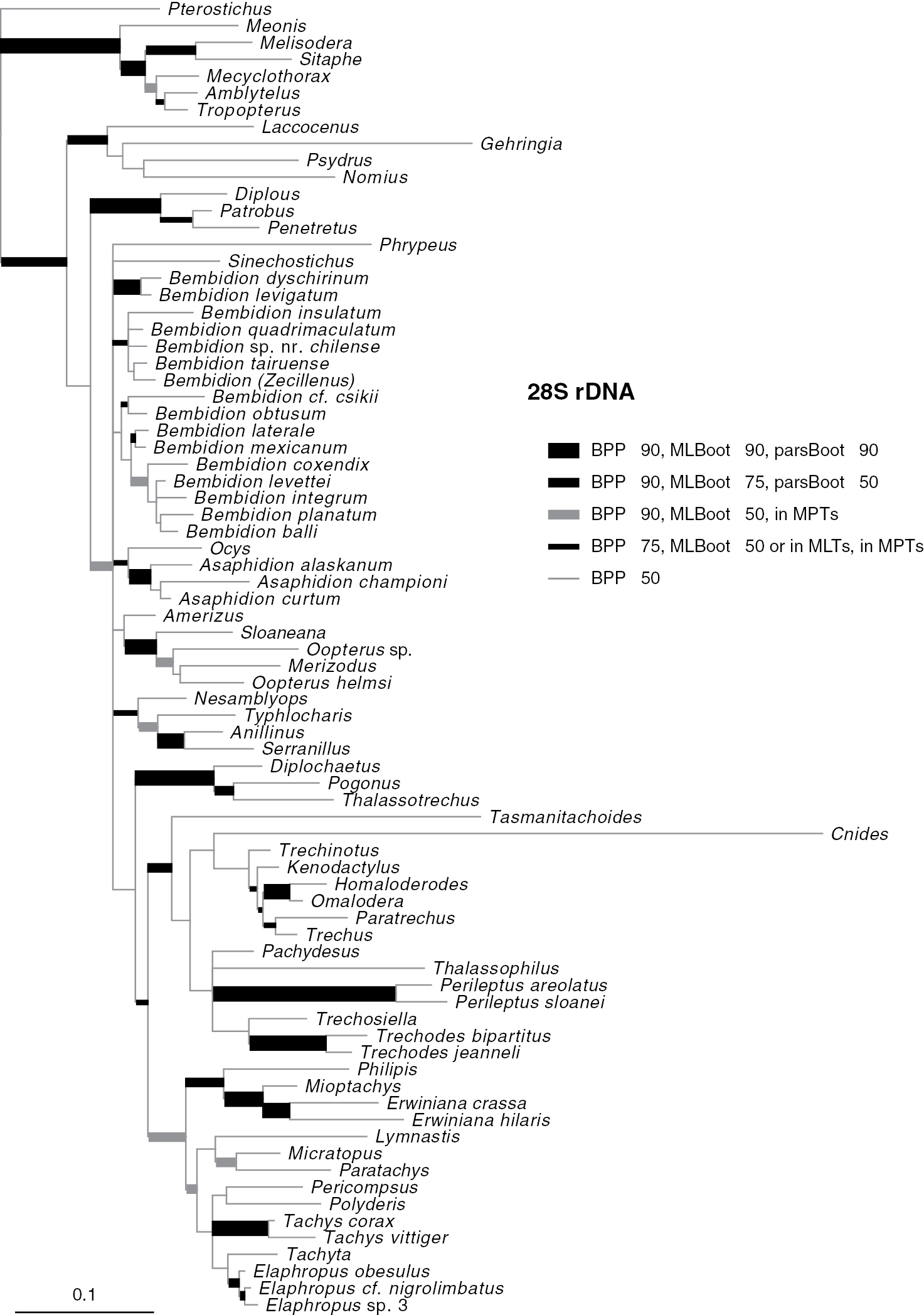
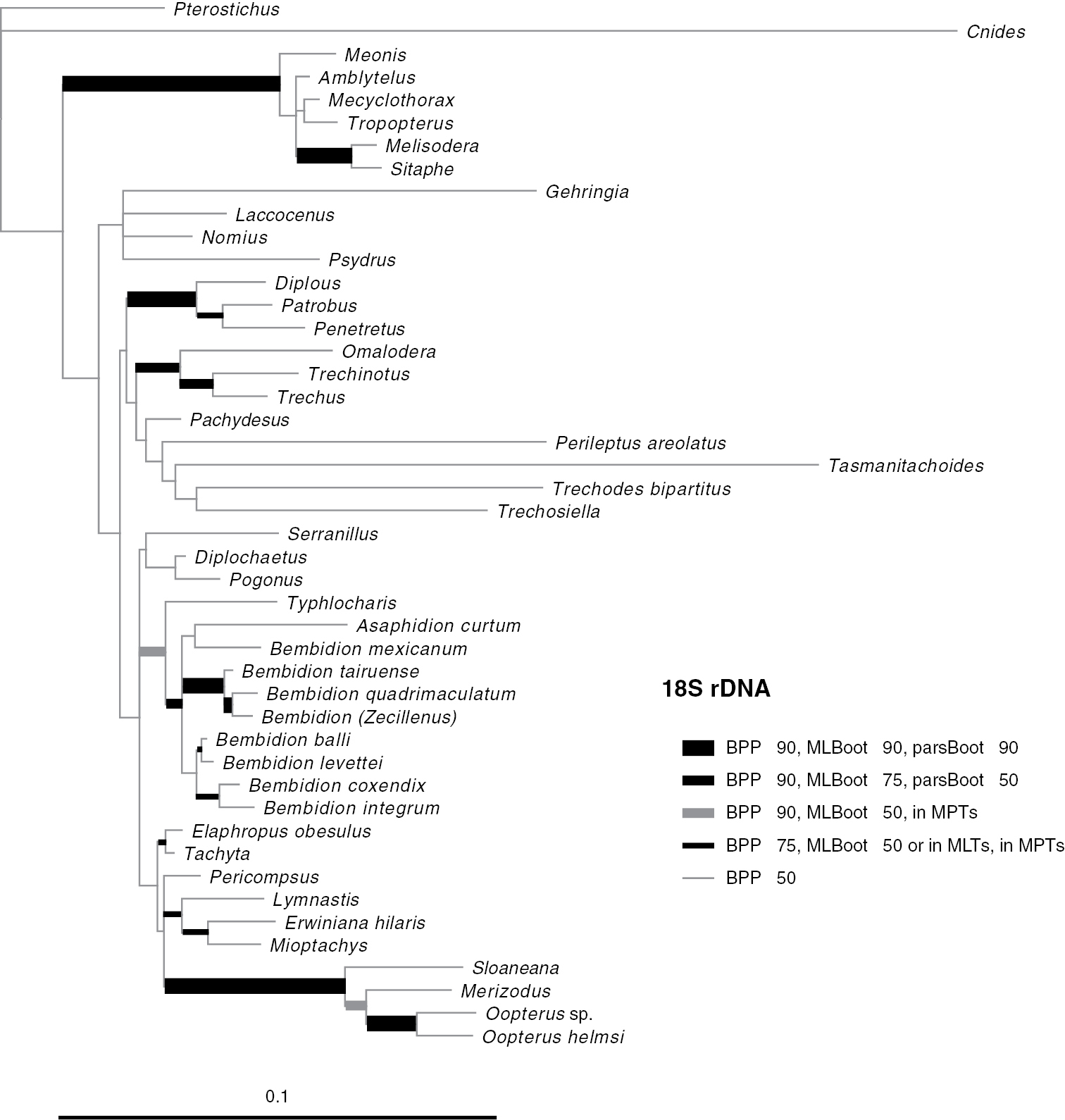
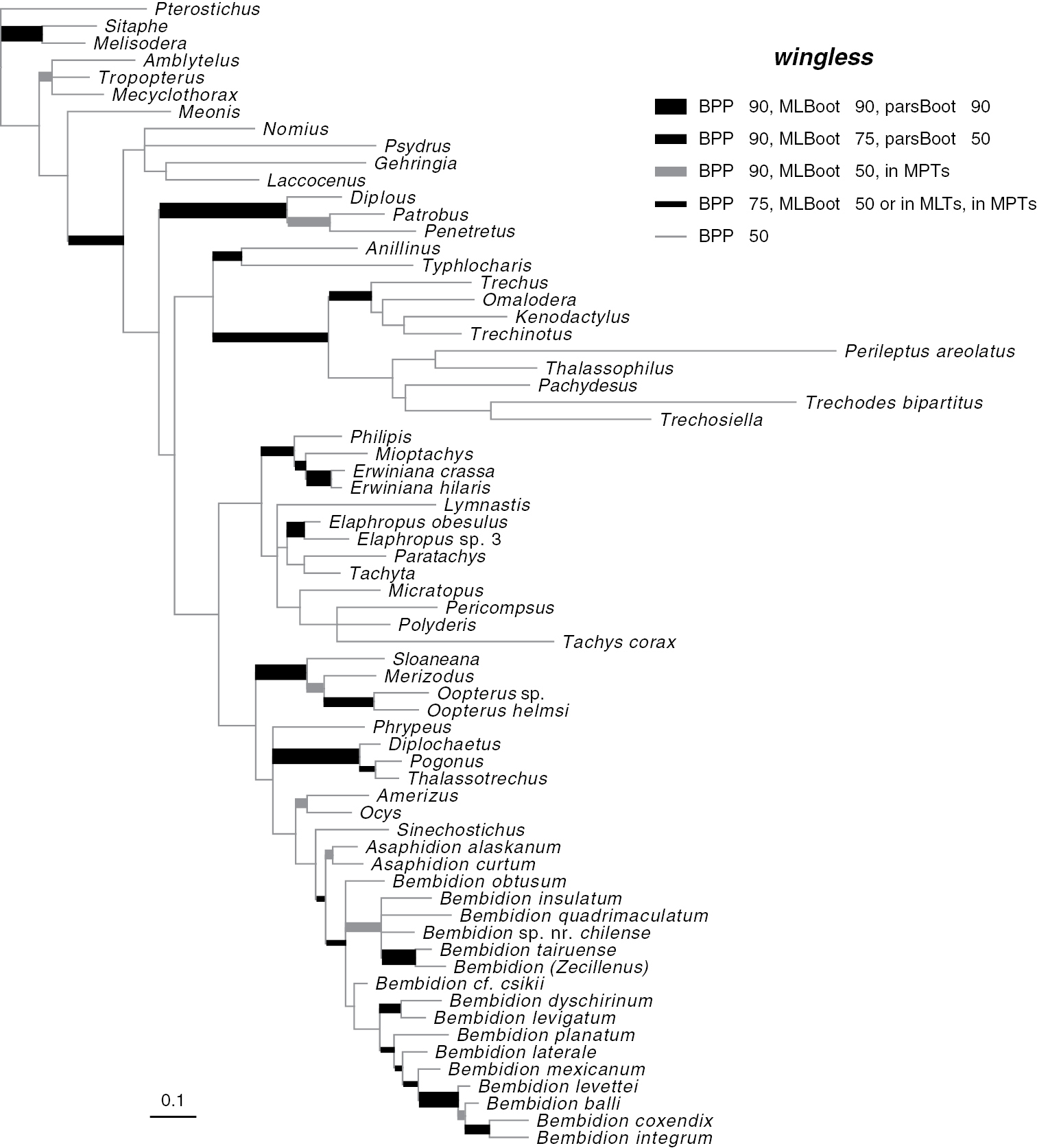
Phylogenetic trees inferred from individual genes are shown in Figs 2–4, and from the merged matrix in Fig. 5. Support values for various hypotheses are shown in Tables 4 and 5. Summaries of supported phylogenetic hypotheses are presented in Figs 6, 7.
Majority-rule consensus tree of trees sampled in Bayesian analysis, with branch lengths proportional to average branch lengths across trees that contain that branch, for 28S rDNA data. Branch lengths were reconstructed by MrBayes; scale bar units are substitutions per site. Thickness and shade of branches indicate support for that clade, based upon estimated Bayesian Posterior Probability percentages (BPP), Maximum Likelihood bootstrap values (MLBoot), and parsimony bootstrap values (parsBoot).
Majority-rule consensus tree of trees sampled in Bayesian analysis, with branch lengths proportional to average branch lengths across trees that contain that branch, for 18S rDNA data. See caption of Fig. 2 for additional details.
Majority-rule consensus tree of trees sampled in Bayesian analysis, with branch lengths proportional to average branch lengths across trees that contain that branch, for the complete wingless data. See caption of Fig. 2 for additional details.
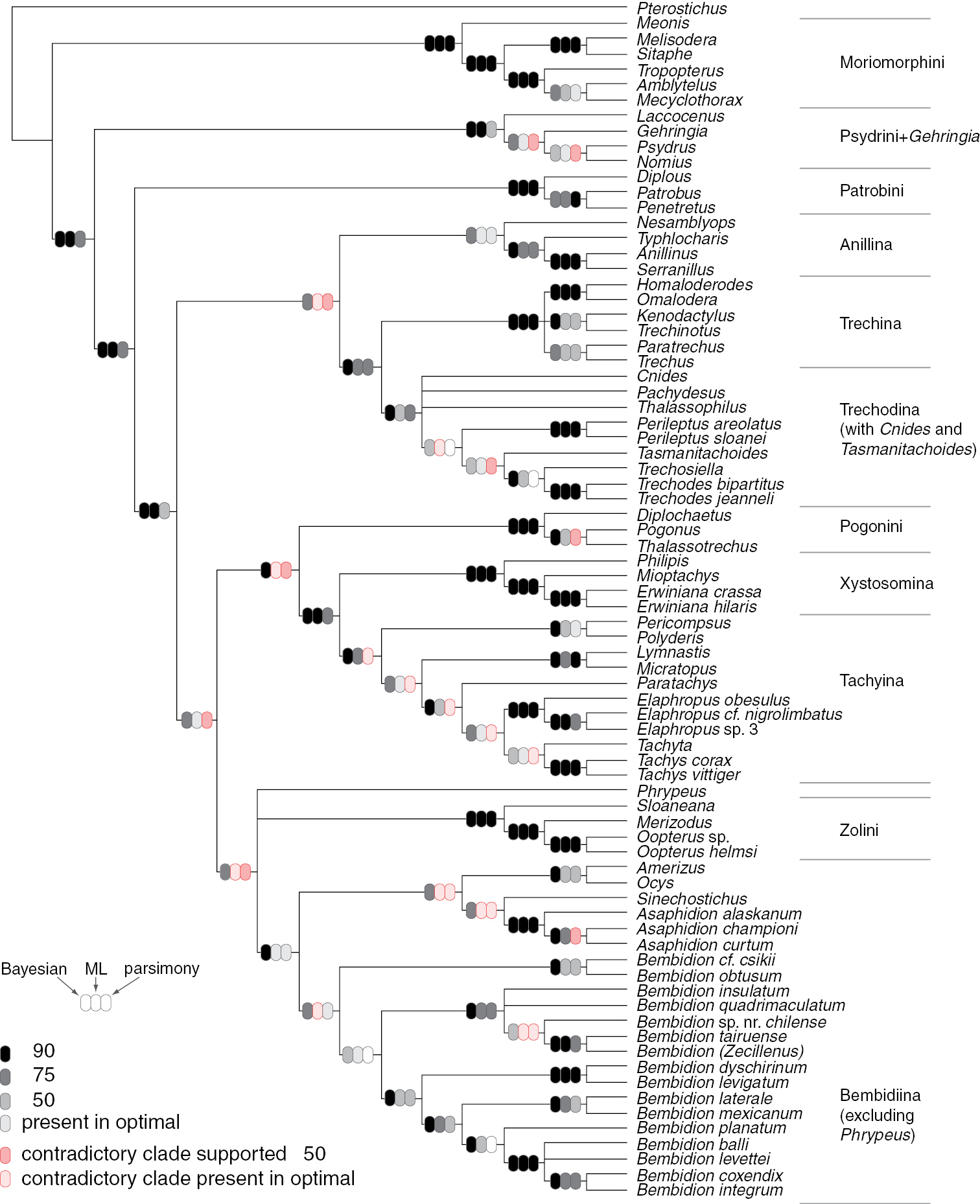
Majority-rule consensus tree of trees sampled in Bayesian analysis for all three genes analyzed together. Ovals on branches indicate support for the clade based upon Bayesian (left), maximum likelihood (center), and parsimony (right) analyses. Darkest tones indicate strongest support for (grays and black) or against (pinks) the clade, with values indicating posterior probability expressed as a percentage (Bayesian), or bootstrap percentage (likelihood and parsimony).
Support values for various groups outside of Bembidiini (sensu lat.): B: Bayesian posterior probability, expressed as a percentage; ML: Maximum likelihood analysis; P: parsimony analysis. For maximum likelihood and parsimony analyses, numbers indicate the bootstrap support expressed as a percentage; check marks indicate that the clade is present in the optimal (maximum likelihood or post parsimonious) trees but with bootstrap value below 50; x indicates that a contradictory clade was present in the optimal (maximum likelihood or post parsimonious) trees but with bootstrap value below 50; negative values indicate Bayesian posterior probability or bootstrap support for a contradictory clade. Boxes in gray to black indicate support for the clade; boxes in pink to red indicate support against that clade, with darker colors indicating stronger support. Dashes indicate no support for or against the clade because of insufficient taxon sampling for that gene; blank boxes indicate no support for or against the clade because of lack of resolution in the inferred trees. Abbreviations: “inc.” = “including”, “exc.” = “excluding”.
| merged | 28S rDNA | 18S rDNA | wg, all nucleotides | wg, well-aligned nucleotides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | ML | P | B | ML | P | B | ML | P | B | ML | P | B | ML | P | |
| Moriomorphini | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | -100 | -78 | -75 | x | ✓ | |
| Psydrini+ Gehringia | 100 | 99 | 70 | 100 | 99 | 70 | 56 | x | x | 88 | ✓ | 88 | ✓ | ✓ | |
| Patrobini+ Trechitae | 100 | 92 | 80 | 54 | 58 | 66 | -66 | x | x | 100 | 58 | 100 | 59 | ✓ | |
| Trechitae | 100 | 93 | 66 | 100 | 70 | 53 | -66 | x | x | 100 | 77 | 100 | 75 | ✓ | |
| Trechini | 99 | 79 | 86 | 100 | 83 | 64 | -66 | x | x | 100 | 98 | 78 | 100 | 98 | 75 |
| Trechina | 100 | 95 | 98 | 62 | 70 | 96 | 100 | 80 | 70 | 100 | 99 | 80 | 100 | 100 | 79 |
| Trechodina | 94 | 64 | 78 | -66 | x | ✓ | -89 | x | x | 100 | 82 | 100 | 79 | ||
| Tasman. with Trechini | 99 | 79 | 86 | 100 | 83 | 64 | 95 | ✓ | 60 | - | - | - | - | - | - |
| Pogonini | 100 | 100 | 100 | 100 | 98 | 97 | 100 | 72 | 72 | 100 | 100 | 100 | 100 | 100 | 100 |
| Zolini | 100 | 100 | 100 | 100 | 96 | 96 | 100 | 100 | 100 | 100 | 99 | 97 | 100 | 97 | 92 |
Support values for various groups of Bembidiini (sens. lat.). See legend of Table 4 for more explanation.
| merged | 28S rDNA | 18S rDNA | wg, all nucleotides | wg, well-aligned nucleotides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | ML | P | B | ML | P | B | ML | P | B | ML | P | B | ML | P | |
| Bembidiini (sens. lat.) | -88 | x | x | -85 | x | x | -98 | x | x | -100 | -63 | x | -100 | -56 | x |
| Tachyina+ Xystosomina | 100 | 92 | 85 | 100 | 72 | 66 | -62 | x | x | 68 | x | x | 80 | ✓ | ✓ |
| Tachyina | 100 | 88 | x | 99 | 67 | x | -62 | x | x | 99 | ✓ | x | 87 | ✓ | x |
| Xystosomina | 100 | 99 | 100 | 100 | 80 | 75 | 89 | 71 | 69 | 99 | 88 | 85 | 100 | 88 | 89 |
| Anillina (inc. Nesamblyops) | 87 | ✓ | ✓ | 93 | ✓ | ✓ | - | - | - | - | - | - | - | - | - |
| Bembidiina inc. Phrypeus | x | x | x | x | - | - | - | - | x | 51 | x | x | |||
| Bembidiina exc. Phrypeus | 91 | ✓ | ✓ | x | x | 100 | 76 | 78 | 100 | ✓ | ✓ | 89 | x | x | |
| Bembidion (sens. lat.) | 75 | x | ✓ | x | -54 | x | -57 | 89 | ✓ | ✓ | 83 | x | |||
| Zecillenus in Bembidion | 100 | 91 | 89 | 91 | ✓ | ✓ | 100 | 98 | 98 | 100 | 98 | 96 | 100 | 95 | 93 |
| Bembidion taiurense + Zecillenus | 100 | 91 | 89 | 52 | ✓ | ✓ | -100 | -91 | -91 | 100 | 98 | 96 | 100 | 95 | 93 |
| Bembidion series | 100 | 87 | 88 | 91 | ✓ | ✓ | 100 | 98 | 98 | 98 | 60 | 67 | 97 | 57 | 66 |
| Cillenus in Bembidion | 100 | 84 | 64 | 99 | 79 | 78 | - | - | - | 99 | ✓ | ✓ | 93 | ✓ | ✓ |
Summary of subtribal and tribal relationships supported by individual genes. Triangles indicate monophyletic groups; quadrangles represent paraphyletic groups A 28S rDNA B 18S rDNA C wingless.
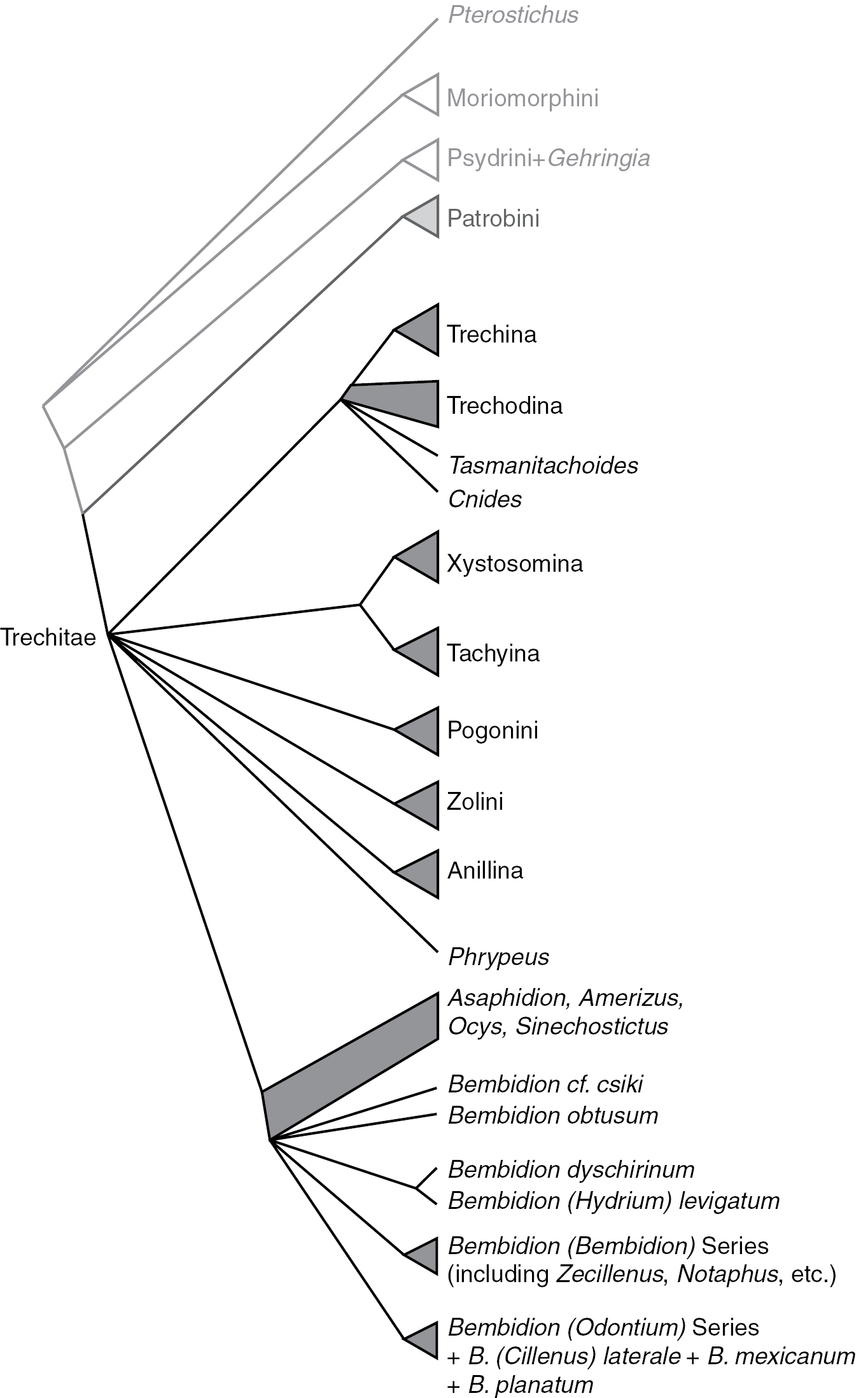
Summary of relationships in Trechitae and related taxa. Branches (including those subtended by triangles) indicate monophyletic groups supported by the combined analyses and at least two of the genes; quadrangles indicate groups whose status is unresolved.
Support values for various groups in analyses of 18S constrained to keep Trechitae or Trechini monophyletic are shown in Table 6.
Results for the reanalysis of Grebennikov’s (2008) larval data are presented in Fig. 1B, and described in more detail in the text, below.
Support values for various groups outside of Bembidiini (sensu lat.): comparison of results for 18S rDNA between unconstrained and constrained analyses. For the “Trechitae constrained” analyses, all of Trechitae was constrained to be monophyletic. For the “Trechini constrained analyses”, all of Trechini was constrained to be monophyletic for the Bayesian analysis; for the remaining analyses, Trechini was constrained to be monophyletic, except that Tasmanitachoides was unconstrained, and could move anywhere in the tree if that were optimal. “n/a” indicates that support values for that group are irrelevant as the group was forced to be monophyletic; for meaning of other symbols and colors, see legend of Table 4.
| No Constraints | Trechitae constrained | Trechini constrained | |||||||
|---|---|---|---|---|---|---|---|---|---|
| B | ML | P | B | ML | P | B | ML | P | |
| Psydrini+ Gehringia | 56 | x | x | 59 | x | -63 | 50 | ✓ | x |
| Patrobini+ Trechitae | -66 | x | x | 99 | 95 | 94 | 94 | ✓ | 53 |
| Trechitae | -66 | x | x | n/a | n/a | n/a | x | ✓ | |
| Trechini | -66 | x | x | -85 | x | ✓ | n/a | n/a | n/a |
| Trechina | 100 | 80 | 70 | 99 | 82 | 69 | 99 | 82 | 70 |
| Trechodina | -89 | x | x | -85 | x | 64 | 3 | x | |
| Tasman. with Trechini | 95 | ✓ | 60 | 89 | ✓ | 67 | 100 | 76 | 84 |
We here discuss in turn the evidence available for various relationships within trechites. As significant results have been found within the outgroups sampled, we will discuss these first.
Outgroup structure: Monophyly of MoriomorphiniAmong the carabids currently considered to belong to Psydrini (sens. lat.), only three genera belong to Psydrina in the strict sense: Psydrus (North America), Nomius (Holarctic and Africa), and Laccocenus (Australia). The remaining genera are arrayed in multiple subtribes (
Our data indicate that Psydrina are not closely related to austral psydrines. While a strong test of this hypothesis with 28S and wingless would require more extensive sampling of non-trechites than we have done, all three genes we studied suggest that Psydrini in the classical sense, containing Psydrus, Nomius, Laccocenus, and the austral psydrines, is not monophyletic (Figs 2–5). Combined with evidence provided by the more extensive 18S rDNA taxon sampling of
Our results indicate strong support for monophyly of Moriomorphini from both 18S rDNA and 28S rDNA (Table 4). Most analyses of the wingless gene suggest instead that the moriomorphines form a grade, although parsimony analysis of the well-aligned nucleotides does support monophyly of the group. More extensive sampling of wingless sequences of non-trechites is needed to examine this further.
Outgroup structure: relationship of GehringiiniGehringiini are a small group (five known species;
Trechitae + Patrobini share a number of synapomorphies in adult structure (
Consistent with most morphological data, our 28S and wingless data support monophyly of Patrobini plus Trechitae (Table 5), as does 18S if Trechini is constrained to be monophyletic (Table 6).
Monophyly of TrechitaeShared derived characters that provide evidence for monophyly of Trechitae are found in multiple character systems. Protarsomeres of adult males are uniquely dentate and dilated on the mesal side (
Trechitae is strongly supported as monophyletic in 28S, wingless, and the merged matrix (Table 5). In contrast, because of the placement of Cnides outside of Trechitae (Fig. 3), as discussed in the next section, 18S provides evidence against the monophyly of Trechitae. If Cnides is forced to stay within Trechini, however, 18S provides no clear signal for or against monophyly (Table 6).
Monophyly and phylogeny of TrechiniThe primary synapomorphy suggesting the monophyly of Trechini is the presence of deep furrows on the dorsal surface of the head (
Our results from 28S, wingless, and the merged matrices indicate strong support for monophyly of Trechini (Table 4), if Tasmanitachoides is included within the tribe (as discussed in the next section). In contrast, 18S provides moderate evidence against the monophyly of trechines, because of the placement of Cnides outside of Trechitae (Fig. 3). However, the exceedingly divergent Cnides 18S sequence (note the length of its branch in Fig. 3) makes artificial attraction of the long branch (
In Jeannel’s great work
Trechini is a diverse group, with complex patterns of morphological variation, and has been subject to many different classification schemes. For example, some classifications view homaloderines as members of Trechina proper (e.g.,
In recent years larval structure and DNA sequences have been used in a few explicit phylogenetic studies within trechines. The most complete available larval data (
Our results (Figs 2–5, Table 4) confirm or refute several previous proposals. Trechina (including Jeannel’s homaloderines and the aepiine Kenodactylus) is monophyletic, supported by all three genes individually, and by analyses of the combined data. Perileptus is a trechodine, as supported by all three genes, and by the combined data, as predicted by
In the original description of Tasmanitachoides,
Although our study cannot be a strong test of the monophyly of zolines, as we do not have members of two subtribes (Sinozolina and Chalteniina), the three genera we have examined (Oopterus, Merizodus, and Sloaneana) are strongly supported as a clade, in all three genes and in all analyses (Table 4).
Monophyly of PogoniniPogonini is a tribe of about 70 species, most of which live in saline habitats (
As delimited in this paper, Bembidiini includes four subtribes: Bembidiina, a large group of over 1, 200 species primarily found in temperate regions, and which includes the larger bembidiines; Tachyina, the second large group, mostly ground-dwelling, centered in warmer regions; Xystosomina, a primarily arboreal group most abundant in the Neotropics; and Anillina, containing very small, often blind, mostly soil-dwelling carabids. In some classifications, these groups are treated as separate tribes, in part as there is only one derived character that suggests that the group is monophyletic (the small terminal article of the maxillary and labial palps of adults, a character that occurs in some trechines, e.g., Perileptus), and in part as the subtribes are relatively uniform within themselves, but with several characters that distinguish them one from another. An analysis of larval characters (
Our results are consistent with the view that Bembidiini is a heterogeneous group, with all three genes and the combined analysis indicating non-monophyly (Table 5), although with no consistent pattern of particular subgroups of Bembidiini being related to non-bembidiines. More details are provided under the discussions of each subtribe, below.
Monophyly of XystosominaThis subtribe was established by
Monophyly of Tachyina exclusive of Xystosomina is supported by the obliquely notched front tibia of adults (
Bayesian analysis of 28S, wingless, and the merged matrix supports the monophyly of Tachyina (Table 5). However, this is result is not supported by Bayesian analysis of 18S rDNA, and parsimony analyses of all genes speak against monophyly of tachyines. A denser taxon sampling of both tachyines and xystosomines is needed to resolve this conflict.
Monophyly and origin of AnillinaMost anillines are minute, blind, wingless, pale inhabitants of soil and deep leaf litter; members of a few genera have small eyes, e.g., Nesamblyops from New Zealand (
If
The exclusion of anillines from the Tachyina + Xystosomina clade (Fig. 7) suggests that the obliquely notched anterior tibia in anillines and Tachyina (
Bembidiina comprises all Bembidiini that do not belong to the other tribes; the group is defined by the lack of derived characters of its members. As such, evidence for monophyly is not evident in morphological data (
The majority of species within subtribe Bembidiina belong to the genus Bembidion. Bembidion was regarded by Carl
More recently, other groups have been excluded from Bembidion in most classifications, including Ocys, Cillenus, Amerizus, Sinechostictus, Orzolina Machado, and Caecidium Uéno. Thus, current classifications have a very large, poorly-defined genus Bembidion surrounded by a number of small “satellite” genera that are each defined in good part based upon autapomorphies. Bembidion in either the traditional or modern senses has no known synapomorphies of its members, and thus it would not be surprising if some of these satellite genera were found to be derived lineages within Bembidion. However, as there have been no comprehensive phylogenetic analyses at this level within Bembidiina, the lack of known derived states does not indicate that Bembidion as currently defined is non-monophyletic. Only some recent papers on larval characters (
Our data suggest that some of these smaller genera are indeed outside of Bembidion. The distant relationship of Phrypeus to Bembidion is discussed above. Both Amerizus and Ocys fall outside of Bembidion in the broad sense in analyses of 28S rDNA, wingless, and the merged matrix (Figs 2, 4, 5). The placement of Sinechostictus has varied through time. These beetles very closely resemble in general habitus members of the Ocydromus complex of Bembidion, with which they have been placed by several authors (e.g.,
On the other hand, some of the groups that have been recently considered outside of Bembidion are evidently derived members of that genus.
Hydrium, considered by most as a subgenus of Bembidion (e.g.,
Cillenus was considered by
When described,
The only well-supported result we obtained about the relationships between the tribes or subtribes of trechites was the sister-group relationship between Tachyina and Xystosomina. This relationship is supported by the common presence of a recurrent groove on the elytra (
While our results have clarified the position of a number of enigmatic lineages within Trechitae, including Tasmanitachoides, Cillenus, and Zecillenus, our data have surprisingly little to say about deep relationships within Trechitae (Fig. 7). This is perhaps a result of the shortness of the deep branches in ribosomal gene trees (Figs 2, 3). Whether these might indicate a rapid radiation or limited ribosomal evolution during that period, they make inference of the relationships difficult.
Future work should increase taxon sampling, to split long branches in the tree, and add missing lineages. Additional genes, especially those with relatively longer lengths for the deeper branches (such as those seen in wingless, Fig. 4), are also needed. These efforts should increase our understanding of the phylogeny of this diverse group of small beetles.
Our heartfelt thanks go to Ross and Joyce Bell, who have provided inspiration over the years to both of us, through their excellent work and generous natures.
Many people aided this project by providing valuable specimens, and we thank them all: David Kavanaugh, Alfred Newton, Margaret Thayer, Wendy Moore, Geoff Monteith, Wayne Maddison, Konjev Desender, Betsy Arnold, Julia Amerongen Maddison, A.S. Gerber, P.M. Johns, Allan Ashworth, Matthias Hartmann, J. M. Pérez Zaballos, J.I. Townsend, Wolfgang Dormann, Dietrich Mossakowski, D.J. Cook, S. Endrödy-Younga, O. Junco, W. Reeves, Rich Leschen, Chuck Bellamy, Karl Kjer, Roger Blahnik, Vasily Grebennikov, and Kipling Will.
For help in identifying specimens, we thank Terry Erwin, Konjev Desender, J. M. Pérez Zaballos, Matthias Hartmann, Paolo Bonavita, Geoff Monteith, Igor Sokolov, and Vasily Grebennikov.
I thank Vasily Grebennikov for making the original data matrix available from his 2008 study. Thanks as well to the Willi Hennig Society for making TNT available through its sponsorship.
This project was funded primarily by NSF grants DEB-9420219 and DEB-9981935 to DRM, as well as funds generously provided by the University of Arizona, and the Harold E. and Leona M. Rice Endowment Fund at Oregon State University.
Locality data for specimens sequenced in this study. # is the D.R. Maddison voucher number.
| Taxon | # | Locality |
|---|---|---|
| Moriomorphini | ||
| Sitaphe parallelipennis | 0669 | Australia: North Queensland: Devil’s Thumb via Mossman |
| Sitaphe parallelipennis | 2247 | Australia: North Queensland: Upper Whitehall Gully |
| Tropopterus sp. | 2200 | Chile: Reg. IX: Parque Nacional Nahuelbuta |
| Psydrini | ||
| Psydrus piceus | 1627 | USA: New Mexico: Gila National Forest, Pine Flats Campground |
| Nomius pygmaeus | 0893 | USA: Arizona: Pima Co., Rincon Peak |
| Trechini: Trechodina | ||
| Cnides sp. | 1808, 0691 | México: Sonora: Alamos, Rio Cuchujaqui |
| Pachydesus sp. | 0678 | Republic of South Africa: Kwazulu-Natal, Ngome Forest Reserve |
| Perileptus areolatus | 1707 | Spain: Madrid: Rio Cofio |
| Perileptus areolatus | 0824 | Russia: NW Caucasus: Krasnodar Dist., r. Belaya, Nikel |
| Perileptus sloanei | 0767 | Australia: Queensland: Gray’s Waterhole, Gayndah. |
| Thalassophilus longicornis | 0823 | Russia: NW Caucasus: Krasnodar Dist., r. Belaya, Nikel |
| Trechodes bipartitus | 0705 | Australia: Queensland: Cow Bay |
| Trechodes jeanneli jeanneli | 0606 | Madagascar: Fianarantsoa Province: Ranomafana National Park |
| Trechosiella sp. | 1709 | Republic of South Africa: North Cape Prov.: Farm Klipdam |
| Tasmanitachoides fitzroyi | 1575, 0762 | Australia: Queensland: Gayndah, Gray’s Waterhole |
| Trechini: Trechina | ||
| Trechus oregonensis | 0587 | USA: Montana: Glacier Co., Divide Creek |
| Omalodera limbata | 0571 | Chile: Malleco Pr.: Coimallin area, 8.2 km NW Los Portones |
| Homaloderodes germaini | 1066 | Chile: Malleco Pr.: Coimallin area, 8.2 km NW Los Portones |
| Kenodactylus audouini | 0670 | Argentina: Tierra Del Fuego: 78 km E. of Ushuaia. |
| Paratrechus sp. | 1076 | Costa Rica: Cerro de la Muerte |
| Trechinotus flavocinctus | 0575 | Chile: Palena Pr.: Austral Highway km 67.9 (11 km S. Contao turnoff) |
| Zolini | ||
| Merizodus angusticollis | 0453 | Chile: Valdivia Province: Rincón de la Piedra, 14.8 km SE Valdivia |
| Oopterus sp. | 0387 | New Zealand: South Island, Canterbury Prov, Arthur’s Pass Nat. Park |
| Oopterus helmsi | 0354 | New Zealand: South Island, Canterbury Prov, Arthur’s Pass Nat. Park |
| Sloaneana tasmaniae | 0339 | Australia: Tasmania: Mount Field N.P., Lake Dobson Rd., E end Wombat Moor |
| Pogonini | ||
| Pogonus (Pogonus) chalceus | 1711, 0679 | Spain: Cádiz: El Puerto de Sta. Maria |
| Thalassotrechus barbarae | 0703, 0530 | USA: California: Marin Co., Tiburon Peninsula |
| Bembidiini: Tachyina | ||
| Lymnastis sp. | 0988 | Malaysia: Sabah: Kinabatangan River |
| Micratopus sp. | 0605 | USA: Arizona: Pima Co., Tucson |
| Paratachys vorax | 0410 | USA: Arizona: Santa Cruz Co., Santa Cruz River near Tumacacori |
| Elaphropus obesulus | 0411 | USA: Arizona: Santa Cruz Co., Santa Cruz River near Tumacacori |
| Elaphropus cf. nigrolimbatus | 0761 | Republic of South Africa: Kwa-Zulu-Natal: Near Bayala |
| Elaphropus sp. 3 | 0713 | Republic of South Africa: North Cape Prov.: Farm Klipdam |
| Pericompsus laetulus | 0429 | USA: Arizona: Pima Co., Arivaca Creek |
| Polyderis rufotestacea | 0717, 0718 | USA: Arizona: Gila Co., Gila River near Winkelman |
| Tachys vittiger | 0760 | USA: California: San Diego Co., Batiquitos Lagoon |
| Tachys corax | 0604 | USA: Arizona: Gila Co., Winkelman |
| Tachyta nana inornata | 0573 | USA: Mississippi: Noxubee Co., Noxubee Nat. Wildlife Refuge |
| Bembidiini: Xystosomina | ||
| Erwiniana hilaris | 0409 | Ecuador: Sucumbios: Cuyabeno Faunal Reserve |
| Erwiniana crassa | 0989 | Ecuador: Orellana Province: Tiputini |
| Mioptachys flavicauda | 0684 | USA: Mississippi: Noxubee Co., Noxubee Nat. Wildlife Refuge |
| Philipis bicolor | 0592 | Australia: Queensland: Mt. Lewis Rd |
| Bembidiini: Anillina | ||
| Anillinus (langdoni group) sp. | 0690 | USA: Georgia: Habersham Co., Big Panther Creek Trail |
| Serranillus sp. | 1084 | USA: North Carolina: Graham Co., Santeetlah Lake |
| Typhlocharis armata | 0572, 1718 | Spain: Cádiz: Tarifa |
| Nesamblyops sp. | 0696 | New Zealand: 3.5 km N Rapahoe |
| Bembidiini: Bembidiina | ||
| Asaphidion alaskanum | 0585 | USA: Alaska: mile 412.3 Dalton Highway |
| Asaphidion championi | 0574 | Nepal: Prov. Karnali, Dolpo, Tripurakot Flußufer |
| Asaphidion curtum | 0267 | USA: Massachusetts: Norfolk Co., Jamaica Plain |
| Amerizus sp. | 0576 | USA: Utah: Abajo Mountains |
| Ocys harpaloides | 0569 | Belgium: Schorisse |
| Phrypeus rickseckeri | 0776 | USA: California: Del Norte Co., Smith River |
| Phrypeus rickseckeri | 0692 | USA: Montana: Jefferson Co., Boulder River at Galena Gulch |
| Sinechostichus solarii | 0603 | Italy: Tuscany: Vallombrosa |
| Bembidion (Antiperyphanes)sp. nr. chilense | 0714 | Peru: Pisac, tributary of Rio Urubamba, 3020m |
| Bembidion (Hoquedela) cf. csikii | 0916 | China: Yunnan Prov.: Gaoligong Shan, Nujiang Prefecture, 13.5 air km SSW of Gof Gonshan |
| Bembidion (Cillenus) laterale | 0602 | Germany: Wadden Sea, Mellum Island |
| Bembidion (Notaphus) insulatum | 0444 | USA: Arizona: Cochise Co., 2.2 km S of Willcox |
| Bembidion (Metallina) dyschirinum | 0896 | USA: Washington: Columbia Co., Blue Mountains |
| Bembidion (Hydrium) levigatum | 0763 | USA: Nebraska: Lancaster Co., Lincoln |
| Bembidion (Ocydromus) mexicanum | 0260 | USA: Arizona: Cochise Co., Turkey Creek, Chiricahua Mtns |
| Bembidion (Phyla) obtusum | 0895 | Canada: Ontario: Burlington |
| Bembidion (Melomalus) planatum | 0601 | Canada: British Columbia: Alexander Creek on highway 3 |
| Bembidion (Bembidion) quadrimaculatum dubitans | 0676 | Canada: Alberta: Bayette Lake near Flatbush |
| Bembidion (Zeplataphus) tairuense | 0607 | New Zealand: Tekapo River Delta, Lake Benmore |
| Bembidion (Zecillenus)sp. | 0595 | New Zealand: Foxton Beach, Manuwatu |