| Taxon names | Citations | Turn highlighting On/Off |







(C) 2012 Randall T. Schuh. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Arguments are presented for the merit of integrating specimen databases into the practice of revisionary systematics. Work flows, data connections, data outputs, and data standardization are enumerated as critical aspects of such integration. Background information is provided on the use of “barcodes” as unique specimen identifiers and on methods for efficient data capture. Examples are provided on how to achieve efficient workflows and data standardization, as well as data outputs and data integration.
specimen databases, workflows, revisionary systematics
The creation of specimen databases–a subset of a field that has frequently been referred to as biodiversity informatics (
One manifestation of the maturity of biodiversity informatics can be seen in the United States National Science Foundation (NSF) program Advancing Digitization of Biological Collections (
Most of the tools applied in specimen data capture—such as databases and barcodes–were initially developed for use in industry. Their application in the realm of biological collections was originally in collection management, rather than as an adjunct to the preparation of scientific publications such as taxonomic revisions. Even though the technology is available, the full integration of biodiversity databases into revisionary studies is far from a fully realized objective. The reasons may include the foreign nature of the technology to older investigators, the lack of direct access to the tools, the lack of technical expertise for implementation of the technology, and simple reluctance to alter traditional approaches to the preparation of revisions.
In the following pages I will argue for the adoption of database tools as an integral part of the revisionary process. This is not just an argument for the adoption of modern technology. Experience suggests that the benefits accrued will more than justify the costs incurred, both in terms of money spent to acquire the necessary equipment and software as well as time spent learning to incorporate “databasing” into one’s day-to-day taxonomic labors.
I have already written about aspects of this subject in two prior papers which focused on the methods for the solution of large-scale taxonomic problems (
The arguments to be made in this paper assume that one has access to a specimen database with certain “basic” features. These include the capability to efficiently capture all relevant and necessary data in a highly structured format, the capability to organize those data in ways useful to the reviser, and the capacity to output data for direct use in revisions as well as for the production of maps and other visual aids. A number of such database products exist, some free of charge, and most capable of performing the necessary functions. They exist as stand-alone products, as institutional tools functioning on a local area network, or as Internet-based tools. Because information on these databases is not the primary intent of this article, and because the logic of choice is beyond the scope of this article, I will not dwell further on the issue database choice. As sources of further information the reader might wish to consult
The use of barcodes to uniquely identify individual specimens goes back at least to the work of Daniel Janzen and the InBio collections in Costa Rica (
Linear barcode label (left), matrix code label (right).
Machine readability, although not an essential component of a USI, is a valuable aspect of barcode and matrix code labels. At $250 or less, the cost of code readers is now about one-tenth what it was in 1994 (
Production of barcode labels can be contracted out to specialized suppliers or can be done in house. Because of the widespread use of the technology, appropriate tools for their preparation and printing are readily available. Nonetheless, a distinct difference between the commercial application of these technologies and their use in biological collections is that the latter group of users expects the labels to be permanent, suitable for alcohol and dry storage, and for the printed matter to be of high resolution, whereas none of those criteria is important in industrial applications such as package delivery and airline baggage identification. Although most any printer can be used to print barcodes, specialized software is required to produce individual labels with sequential numbering (e.g.,
Curators of biological collections have long applied catalog numbers to specimens, although such practice has been much less common with insect collections than with those of recent vertebrates, fossils, and plants, for example. Although these “catalog” numbers were often not unique within institutions, let alone across institutions, they did offer a way to uniquely associate specimens with log-books of data, accession information, field notebooks, and other written resources. Most barcode implementations come much closer to globally-unique identification than was the case with traditional catalog numbers, through the use of codes that combine an institution code + a collection code + plus a catalog number. This approach complies the with Darwin Core standard promoted by the
The use of barcodes has resulted in the frequent attachment of multiple codes to individual specimens, often in addition to traditional catalog numbers. Several factors are at play, including the use of barcodes as the modern equivalent of catalog numbers as well as to identify specimens used in independent research projects. Sometimes these two uses are included in a single label, sometimes on separate labels. Recent Internet-based discussions suggest that prevailing opinion regards the attachment of multiple labels as acceptable, often unavoidable, and that the all of the codes should remain on the specimens in perpetuity. Some or all of these codes may be globally unique.
Verbatim vs Transformed Data: A choice mediated by the use of USIsA recent symposium organized for the 2011 meeting of the Entomological Collections Network (Reno, Nevada; http://www.ecnweb.org/dev/AnnualMeeting/Program ), included a more or less equal number of presentations arguing for 1) the verbatim capture of all label data in a single text field with subsequent transformation into a more highly structured format, or for 2) transformation of label data into a publication-ready format as an integral part of the data-capture process.
Verbatim data capture allows for data acquisition with minimum training of the data-entry personnel. The only real requirement would seem to be the ability to read the labels and convert them into a text string. Those data must then be transformed into a structured format and written to the database tables by the use of some software algorithm or other automated data-parsing approach. Finally, the accuracy of the transcription must to be checked, an additional step, and one that will require greater expertise in interpretation of label data than did the initial data entry.
Transforming data as part of the data-capture process, so that the data are in the exact form used by the database requires additional training of personnel over what is needed for verbatim data capture. Nonetheless, because the data are structured during the process of data capture, these data are ready for straightforward review for accuracy, at which point they can be considered “publication ready” and the additional training effort will be available for all subsequent data capture.
Even though errors may be made under either approach, the use of USIs allows for subsequent investigators to return to individual specimens with substantial confidence concerning the correspondence of original and transcribed data. It is my view, and that of many of my colleagues, that the capture of transformed data is more efficient because it is a one step process that allows for immediate use of the data. Data captured en masse from collections will not be available until they have undergone algorithmic transformation and been approved for upload, thus potentially presenting a time lag that will hinder the progress of the reviser or other data user.
Data-capture Work Flow in Revisionary Studies Label generation: Capture field data to the database and generate all labels from itMany specimens used in revisionary studies, possibly most particularly in entomology, come from the dedicated fieldwork of the reviser. Thus, the opportunity to use appropriate technology in conjunction with fieldwork would seem to be a straightforward choice. This would include the capture of latitude/longitude and altitude data in the field through the use of a GPS (global positioning system) device in the form used by geographical information systems software and the recording of field data in exactly the format to be used in the specimen database. Thus, the choice should be degrees and decimal parts thereof for lat/long data and meters for altitude. Locality and collection-event data can be directly captured in digital form in the field, or recorded to an archival field notebook and captured in digital form at the earliest subsequent opportunity. GPS data can be downloaded directly, an approach that precludes mistakes during transcription of numbers, one of the most common errors made in the capture of field data.
The argument for using a database to capture/store field data and to produce specimen labels is bolstered by the many examples of specimens in collections where multiple collectors on the same field trip produced their own labels. Although such labels contain similar information, they are frequently not identical and thus may end up in a database as representing distinct localities. The drawbacks are one or more of the following: 1) what was actually a single locality will likely end up being georeferenced multiple times, or if lat/long data were captured in the field, those data may still not be identical on the labels; 2) one or more renderings of the collection locality may contain errors; 3) the locality may be easily interpreted in one rendering but difficult to interpret in another; and 4) some of the labels may be substandard from a curatorial point of view. Using the database from the outset, including for the generation of labels, facilitates data standardization and the uniform presentation of data in all of its subsequent uses. It also greatly facilitates the retrospective capture of data for specimens whose localities are already in the database. This last point has economic implications, because even though the personnel time available to enter all specimens collected at a given locality may not be available at the time the specimens are mounted and labeled, the cost of entering just the locality/collection event data at the time of the fieldwork will never be an issue.
Specimen data: Enter specimen data early in the revisionary processAlthough it has been said many times, and therefore may seem trite, the use of a database can save many key strokes. Once the data have been entered and checked for accuracy for a given locality, they can be re-used in the generation of labels, for preparation of reports of “specimens examined”, and for many other purposes. If for any reason an error is found, it can be corrected and all subsequent and varied uses of those data will be accurate and uniform. The capture early on in the revisionary process of as much specimen data as possible allows for the structuring and examination of those data in ways that are otherwise difficult and cumbersome. What is paramount is that the data are captured once but useable in many ways without the need for re-keyboarding. Nonetheless, it is probably fair to say that in the traditional preparation of a revision, the last thing to be done was to capture specimen data, whether using a word-processing file, spreadsheet, or relational database. The use of a specimen database facilitates the capture of specimen data much closer to the beginning of the revisionary process, so that all relevant observations on specimens can be managed through the medium of the database and available over the entire course of the revisionary process. In addition to locality data, such observations might include host data, habitat descriptions, museum depository information, dissections, images, measurement data, and DNA sequence files, to name just some of the possibilities.
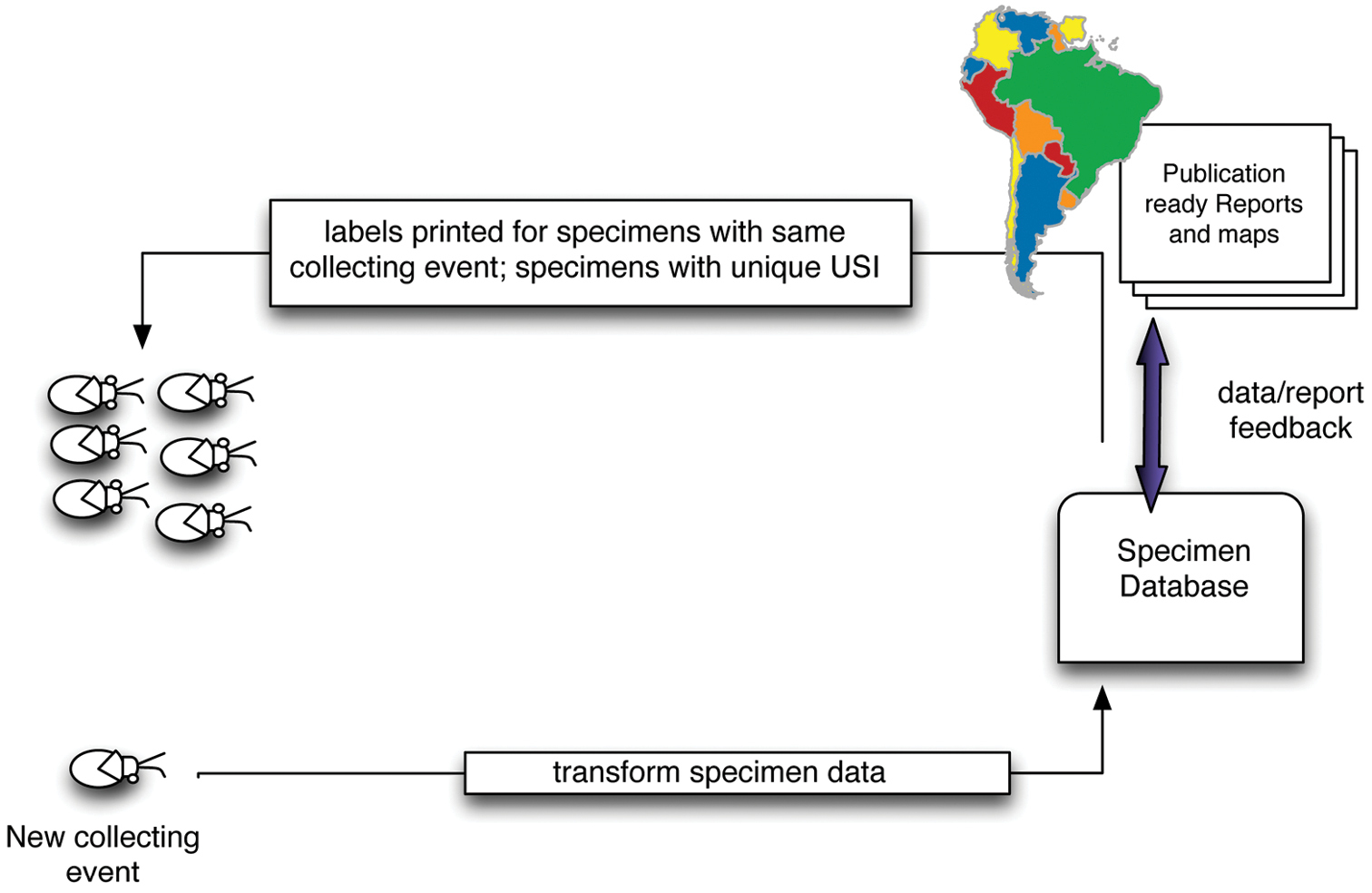
Capturing specimen data: Organize specimens before capturing dataWith some forethought and advance preparation the process of retrospective specimen data capture can be made more efficient and also facilitate other aspects of the revisionary process. Collective experience of participants on the Planetary Biodiversity Inventory project, and other colleagues, recommends the following sequence of events for dealing with specimens from any given institution (Fig. 2):
1. Sort specimens by provisional species criteria (morpho species, etc.)
2. Sort specimens by locality
3. Sort specimens by sex
4. Affix sequential unique specimen identifiers (barcodes, matrix codes)
5. Enter data in database
Diagram of specimen data connections and work flows.
This workflow is efficient because it allows for series of specimens of the same species, sex, and locality to bear USI codes in sequential order and for data for all of those specimens to be captured as a single action. Of course, this approach is most important in those cases where there are multiple examples of a species from a single collecting event.
Although sexing specimens may not be necessary or possible for all taxa, in many groups the standard description is based on one sex, or the other. Sorting by sex before specimen data are entered facilitates comparisons, adds a logical aspect to the organization of the material in collections, and helps to produce sequential USIs, which saves space in presenting data on specimens examined. If during the course of preparing a revision specimens are found to have been initially misidentified, the records for those specimens can be readily retrieved via the barcode and the identifications in the database can be corrected.
Data Connections Georeferencing and mapping: Using the database as an analytic toolGeoreferencing–the addition of latitude/longitude data to individual specimen records–permits the mapping of specimen distributions in space. Such mapping should be part of the revisionary process, rather than taking place near the end, as has traditionally been the case. As a matter of standard practice, lat/long data should be available on all specimen labels being produced as a result of fieldwork in this day and time. And, as mentioned above, data from modern fieldwork should desirably be captured to a database for the preparation of all labels, such that no manual georeferencing will be required. Under this approach, georeferencing is intimately related to the issue of workflow, because the earlier in the revisionary process the specimen data can be mapped, the more useful they will be. Nonetheless, lat/long data will have to be determined for legacy material.
Georeferencing was at one time a time-consuming and tedious process. It is now much easier, due to the ready availability of automated tools such as
Even if your database application does not have integrated mapping tools, the simple ability to export lat-long data will permit the easy visualization of those data and the creation of maps (fig. 3). Some of the tools freely available are the Simple Mapper (
Map of species distributions in western North America created using the Simple Mapper.
As is the case with georeferencing early in the study of specimens, the use of USIs as labels for images, measurement data, and DNA sequences allows these data sources to become an integral part of the data record for the specimens under study, and for tracking those data in an unequivocal manner.
Data outputs: Organizing data through the power of report writing Reports of specimens examinedOnce specimen data have been captured, checked for accuracy, and georeferenced, the real power of the database for revisionary studies comes from the ability to generate reports. Possibly most valuable is the preparation of reports of specimens examined, a core component of traditional revisions (Fig. 4). The reports can be written, revised, and rewritten in a matter of seconds or minutes, and preclude retyping and reformatting of data; the same can be said for the preparation of maps. Other types of reports, such as species by locality, hosts by species, and range of collection dates–among many other possibilities–are also easily produced and complement the contents of many revisions.
Report of specimens examined, including unique specimen identifiers.
The power of database query languages facilitates the preparation of counts of total specimens examined, specimens examined by museum, specimens dissected, and other summary information that helps to clarify the sources and uses of data.
Species pages: Integrating all data sources in electronic formSpecies pages have become the Internet equivalent of species treatments in traditional print publications. The Encyclopedia of Life (
More has probably been written on the use of descriptive databases in revisionary systematics than has been the case for specimen databases. These products allow for the creation of character descriptions, natural language descriptions, interactive keys, and phylogenetic matrices. The most longstanding version of such a database is DELTA (
In my own work, I have created matrices in the program Winclada (
In summary, the affordable technology for capture, manipulation, and sharing of specimen data awaits revisers to avail themselves of the opportunity to harness the power of these tools (see
The approaches described in this paper were derived from experience gained during participation in, and administration of, a USA National Science Foundation-funded Planetary Biodiversity Inventory award for the study of the true bug family Miridae. Many PBI project colleagues helped shape my views on specimen databases and their place in revisionary studies and collection management. Among others, these include co-principal investigator Gerry Cassis, Michael Schwartz, Sheridan Hewson-Smith, Christiane Weirauch, Denise Wyniger, Fedor Konstantinov, and Dimitri Forero. I am also indebted to the late James S. Asche and his colleagues at the University of Kansas, Lawrence, for generous discussion of their own experiences with specimen databasing and to John S. Ascher (AMNH) for his contributions to the logic of specimen data capture. I thank: Katja Seltmann (AMNH) and Michael Schwartz for discussion and encouragement and for their critical comments on earlier versions of the MS which helped to form the final product; Dimitri Forero, Ruth Salas, and two anonymous reviewers for comments on the manuscript; and Katja for the conception and creation of figure 2. Finally, I thank Nina Gregorev (AMNH) for her expert programming contributions and many suggestions on how to produce the truly user-friendly database implementation used in the PBI and TCN projects. This work was funded by NSF awards DEB 0316495 (PBI) and EF1115080 (ADBC-TCN) to the American Museum of Natural History.