(C) 2011 Mahir Budak. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Cephinae is traditionally divided into three tribes and about 24 genera based on morphology and host utilization. There has been no study testing the monophyly of taxa under a strict phylogenetic criterion. A molecular phylogeny of Cephinae based on a total of 68 sequences of mtDNA COI gene, representing seven genera of Cephinae, is reconstructed to test the traditional limits and relationships of taxa. Monophyly of the traditional tribes is not supported. Monophyly of the genera are largely supported except for Pachycephus. A few host shift events are suggested based on phylogenetic relationships among taxa. These results indicate that a more robust phylogeny is required for a more plausible conclusion. We also report two species of Cephus for the first time from Turkey.
Hymenoptera, phylogeny, Cephidae, COI, host shift
The Cephidae is a small family of Hymenoptera with a thin integument, usually black or dark colored and commonly with narrow yellow bands on the abdomen. It comprises approximately 160 species in three subfamilies and about 24 genera and is primarily Holarctic (
Cephidae can be easily identified since they are morphologically intermediate between the hymenopteran suborders Symphyta and Aculeata. Because of several apocritan-like characters, such as a weak constriction between the first and second abdominal segments, the lack of cenchri and the rough area on fore wing, and the form of male genitalia, they were once considered as a likely sister group of Apocrita (
Phylogenetic studies of taxa that exhibit adaptive phenotypic variation provide valuable insights into the evolutionary forces driving the origins of diversification (
Here, we selected the mitochondrial cytochrome oxidase subunit I (COI) gene to reconstruct the phylogenetic relationships of the Cephinae and identify systematic position of its tribes and genera by applying phylogenetic inference methods. The selected COI gene region is informative for estimating relationships at both intra- and interspecies level due to possession of both completely conserved and variable regions and having a heterogeneous evolutionary rate across the gene (
Sixty-eight specimens representing three tribes and seven genera of the subfamily Cephinae were collected from localities presented in Table 1. All specimens are deposited in the Entomological Collection of Cumhuriyet University, Sivas (ECCUS). A specimen of Arge sp. (Argidae) was included as an outgroup in the phylogenetic analyses. Several keys were used to identify the specimens (
List of taxa and voucher specimens used for sequencing.
| Genus | Species | Voucher no. | GenBank accession no. | Location | Col. date |
|---|---|---|---|---|---|
| Arge | Arge sp. | ECCUS 201 | JF901916 | ||
| Calameuta | |||||
| Calameuta filiformis (Eversmann, 1847) | ECCUS 210 | JF901849 | İçel | 12.04.2009 | |
| Calameuta filiformis | ECCUS 211 | JF901850 | Sivas | 04.06.2009 | |
| Calameuta haemorrhoidalis (Fabricius, 1781) | ECCUS 212 | JF901852 | Kütahya | 20.05.2009 | |
| Calameuta haemorrhoidalis | ECCUS 213 | JF901853 | Isparta | 17.05.2009 | |
| Calameuta haemorrhoidalis | ECCUS 214 | JF901855 | Kocaeli | 04.05.2010 | |
| Calameuta haemorrhoidalis | ECCUS 215 | JF901856 | Kocaeli | 04.05.2010 | |
| Calameuta haemorrhoidalis | ECCUS 216 | JF901857 | Bayburt | 05.06.2010 | |
| Calameuta haemorrhoidalis | ECCUS 217 | JF901858 | Uşak | 19.05.2009 | |
| Calameuta haemorrhoidalis | ECCUS 218 | JF901859 | Isparta | 17.05.2009 | |
| Calameuta idolon (Rossi, 1794) | ECCUS 219 | JF901851 | Konya | 17.05.2009 | |
| Calameuta pallipes (Klug, 1803) | ECCUS 220 | JF901854 | Sivas | 13.05.2010 | |
| Calameuta pallipes | ECCUS 221 | JF901860 | Hakkari | 11.06.2003 | |
| Calameuta pygmaea (Poda, 1761) | ECCUS 222 | JF901848 | Hatay | 09.04.2009 | |
| Calameuta sp. | ECCUS 223 | JF901861 | Sivas | 17.06.2007 | |
| Cephus | |||||
| Cephus brachycercus Thomson, 1871 | ECCUS 230 | JF901871 | İstanbul | 08.05.2010 | |
| Cephus brachycercus | ECCUS 231 | JF901872 | Sivas | 10.05.2010 | |
| Cephus fumipennis Eversmann, 1847 | ECCUS 232 | JF901873 | Ardahan | 07.06.2010 | |
| Cephus nigrinus Thomson, 1871 | ECCUS 233 | JF901874 | İstanbul | 08.05.2010 | |
| Cephus parvus (Dovnar-Zapolskij, 1931) | ECCUS 234 | JF901875 | Sivas | 17.05.2010 | |
| Cephus parvus | ECCUS 235 | JF901876 | Sivas | 26.05.2010 | |
| Cephus pulcher Tischbein, 1852 | ECCUS 236 | JF901877 | Erzurum | 06.06.2010 | |
| Cephus pygmeus (Linné, 1767) | ECCUS 237 | JF901911 | Denizli | 18.05.2009 | |
| Cephus pygmeus | ECCUS 238 | JF901912 | Hatay | 09.04.2009 | |
| Cephus pygmeus | ECCUS 239 | JF901913 | Hatay | 09.04.2009 | |
| Cephus pygmeus | ECCUS 240 | JF901914 | Bayburt | 07.06.2008 | |
| Cephus pygmeus | ECCUS 241 | JF901915 | Bayburt | 07.06.2008 | |
| Cephus rjabovi Dovnar-Zapolskij, 1926 | ECCUS 242 | JF901878 | Kırıkkale | 20.06.2009 | |
| Cephus rjabovi | ECCUS 243 | JF901879 | Kırıkkale | 20.06.2009 | |
| Cephus runcator Konow, 1896 | ECCUS 244 | JF901880 | Edirne | 07.05.2010 | |
| Cephus runcator | ECCUS 245 | JF901881 | Edirne | 07.05.2010 | |
| Cephus sareptanus Dovnar-Zapolskij, 1928 | ECCUS 246 | JF901882 | Erzurum | 06.06.2010 | |
| Cephus sareptanus | ECCUS 247 | JF901883 | Erzurum | 06.06.2010 | |
| Cephus sp. | ECCUS 248 | JF901884 | Bilecik | 05.05.2010 | |
| Cephus sp. | ECCUS 249 | JF901885 | Bilecik | 05.05.2010 | |
| Cephus sp. | ECCUS 250 | JF901886 | Çanakkale | 06.05.2010 | |
| Cephus sp. | ECCUS 251 | JF901887 | Amasya | 02.05.2010 | |
| Cephus sp. | ECCUS 252 | JF901888 | Amasya | 02.05.2010 | |
| Cephus sp. | ECCUS 253 | JF901889 | Tekirdağ | 08.05.2010 | |
| Cephus sp. | ECCUS 254 | JF901890 | Sivas | 18.05.2010 | |
| Cephus sp. | ECCUS 255 | JF901891 | Erzurum | 06.06.2010 | |
| Cephus sp. | ECCUS 256 | JF901892 | Kars | 07.06.2010 | |
| Cephus sp. | ECCUS 257 | JF901893 | Kars | 07.06.2010 | |
| Cephus sp. | ECCUS 258 | JF901894 | Bolu | 04.05.2010 | |
| Trachelus | |||||
| Trachelus iudaicus (Konow, 1907) | ECCUS 260 | JF901865 | Bayburt | 05.06.2010 | |
| Trachelus iudaicus | ECCUS 261 | JF901866 | Bayburt | 05.06.2010 | |
| Trachelus libanensis (André, 1881) | ECCUS 262 | JF901867 | İçel | 13.04.2009 | |
| Trachelus libanensis | ECCUS 263 | JF901868 | İçel | 13.04.2009 | |
| Trachelus sp. | ECCUS 264 | JF901862 | Sivas | 12.06.2010 | |
| Trachelus sp. | ECCUS 265 | JF901863 | Sivas | 30.05.2010 | |
| Trachelus tabidus (Fabricius, 1775) | ECCUS 266 | JF901869 | İçel | 12.04.2009 | |
| Trachelus tabidus | ECCUS 267 | JF901870 | Çanakkale | 06.05.2010 | |
| Trachelus troglodyta (Fabricius, 1787) | ECCUS 268 | JF901864 | Zonguldak | 03.05.2010 | |
| Hartigia | |||||
| Hartigia linearis (Schrank, 1781) | ECCUS 270 | JF901896 | Ardahan | 07.06.2010 | |
| Hartigia linearis | ECCUS 271 | JF901897 | Kırşehir | 03.06.2003 | |
| Hartigia linearis | ECCUS 272 | JF901898 | Kırşehir | 03.06.2003 | |
| Hartigia nigra (M. Harris, 1779) | ECCUS 273 | JF901899 | Konya | 17.05.2009 | |
| Hartigia sp. | ECCUS 274 | JF901900 | Sivas | 17.05.2010 | |
| Hartigia sp. | ECCUS 275 | JF901901 | Sivas | 13.05.2010 | |
| Hartigia xanthostoma (Eversmann, 1847) | ECCUS 276 | JF901902 | Zonguldak | 03.05.2010 | |
| Hartigia xanthostoma | ECCUS 277 | JF901903 | Zonguldak | 03.05.2010 | |
| Syrista | |||||
| Syrista parreyssii (Spinola, 1843) | ECUUS 280 | JF901906 | Sivas | 26.05.2007 | |
| Syrista parreyssii | ECUUS 281 | JF901907 | Adana | 05.06.2003 | |
| Characopygus | |||||
| Characopygus sp. | ECCUS 290 | JF901895 | İçel | 13.04.2009 | |
| Pachycephus | |||||
| Pachycephus cruentatus (Eversmann, 1847) | ECCUS 300 | JF901904 | Sivas | 06.06.2009 | |
| Pachycephus smyrnensis J.P.E.F. Stein, 1876 | ECCUS 301 | JF901908 | Edirne | 07.05.2010 | |
| Pachycephus smyrnensis | ECCUS 302 | JF901909 | Edirne | 07.05.2010 | |
| Pachycephus smyrnensis | ECCUS 303 | JF901910 | Sivas | 11.06.2010 | |
| Pachycephus sp. | ECCUS 304 | JF901905 | Sivas | 12.06.2010 | |
Alcohol-preserved specimens were allowed to dry on filter paper, and DNA was extracted from left legs of the specimens using the High Pure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany) following the protocol for DNA isolation from mammalian tissue. Each DNA sample was dissolved in 200 µl elution buffer and stored at -20oC. The partial mitochondrial COI gene (750 bp) was amplified by using the conserved COI primers with the following sequence: COI–s1859, 5’ – GGAACIGGATGAACWGTTTAYCCICC – 3’ and COI–a2590 5’ – GCTCCTATTGATARWACATARTGRAAATG – 3’ (
Estimates of evolutionary divergence analyses were conducted in MEGA5 (
In order to investigate the phylogenetic relationship of Cephinae, phylogenetic trees were constructed using maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) methods. Nucleotides were used as discrete and unordered characters. The best-fit model of DNA substitution and the parameter estimates used for tree constructions were chosen according to the Akaike Information Criterion (AIC) as implemented in Modeltest version 3.7 (
Evaluation of the material collected after publication of
Turkey: Sivas [39°42.71'N, 37°01.30'E] 1300 m, 26.05.2010, 1♀, 17.05.2010, 1♂.
Palearctic region.
Turkey: Edirne [40°39.32'N, 26°17.82'E] 50 m, 07.05.2010, 6♀, 1♂.
Turkey, S. E. Europe.
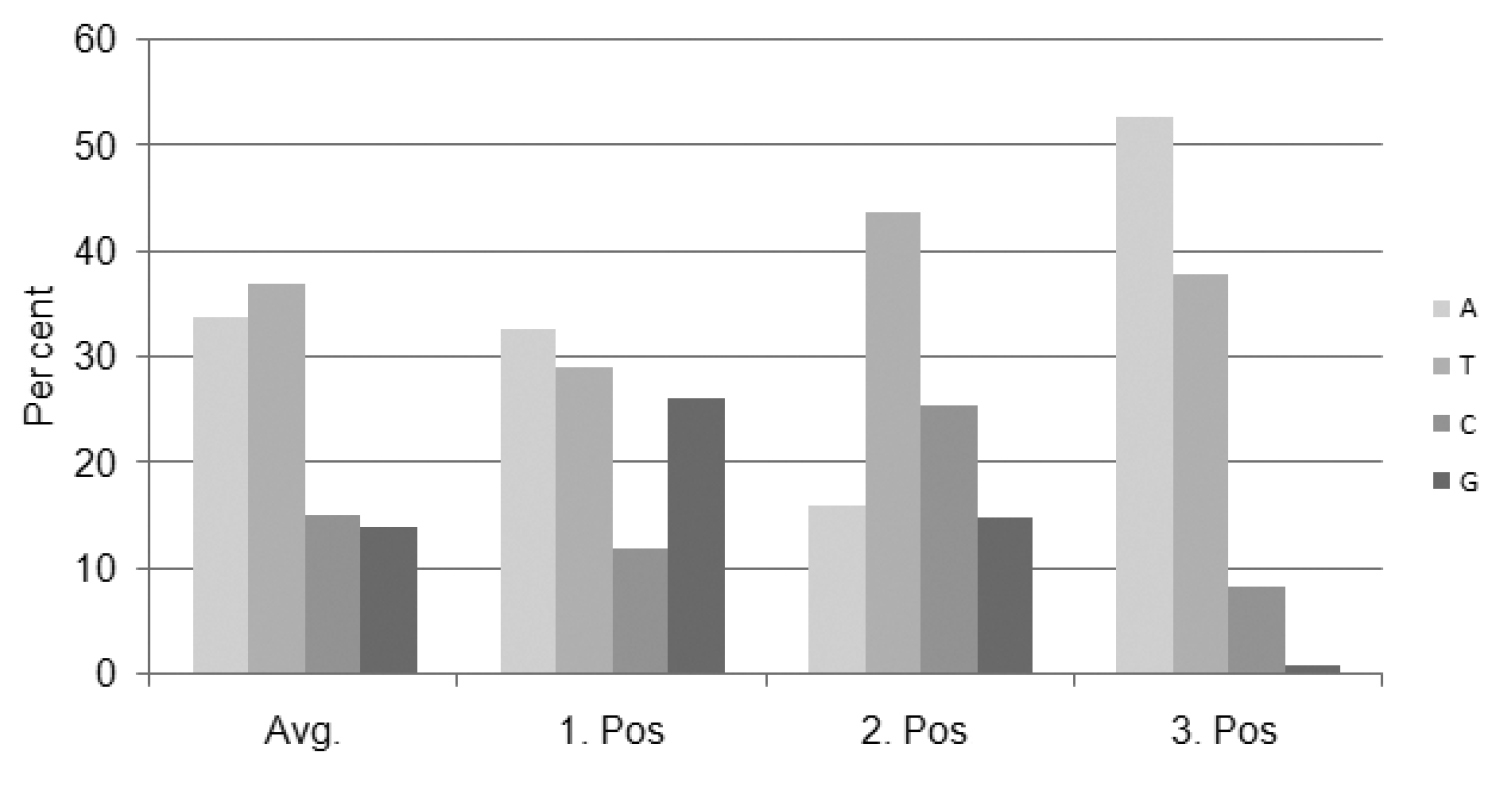
The complete alignment of the partial mitochondrial COI gene sequences from 68 cephid specimens, including representatives of these two new records, resulted in a fragment containing 658 base pairs, among which 287 nucleotide positions were variable and 223 sites of which were parsimony-informative. The analyzed sequences correspond to a functional mitochondrial gene region because of the presence of singular peaks in each chromatograph and absence of in–del and premature stop codons, and presence of the highest nucleotide substitutions at the third codon position (
Percentage of nucleotide composition at each codon position.
The numbers of base substitutions per site from averaging over all sequence pairs between genera are shown in Table 2. The least diverged genera appears to be Characopygus and Pachycephus (p= 0.062) and, the most are Hartigia and Syristra (p= 0.161) also with highest standard error value of 0.017.
Estimates of evolutionary divergence over sequence pairs between genera. The number of base substitutions per site from averaging over all sequence pairs between groups are shown. Standard error estimates are shown above the diagonal.
| Genera | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| 1. Calameuta | 0.010 | 0.011 | 0.010 | 0.013 | 0.012 | 0.015 | 0.025 | |
| 2. Trachelus | 0.110 | 0.010 | 0.009 | 0.013 | 0.012 | 0.014 | 0.025 | |
| 3. Cephus | 0.108 | 0.119 | 0.007 | 0.013 | 0.010 | 0.016 | 0.026 | |
| 4. Characopygus | 0.078 | 0.094 | 0.069 | 0.012 | 0.007 | 0.012 | 0.028 | |
| 5. Hartigia | 0.136 | 0.146 | 0.143 | 0.114 | 0.013 | 0.017 | 0.029 | |
| 6. Pachycephus | 0.116 | 0.125 | 0.113 | 0.062 | 0.137 | 0.013 | 0.029 | |
| 7. Syrista | 0.124 | 0.145 | 0.156 | 0.102 | 0.161 | 0.125 | 0.030 | |
| 8. Outgrup | 0.248 | 0.263 | 0.256 | 0.249 | 0.292 | 0.278 | 0.279 |
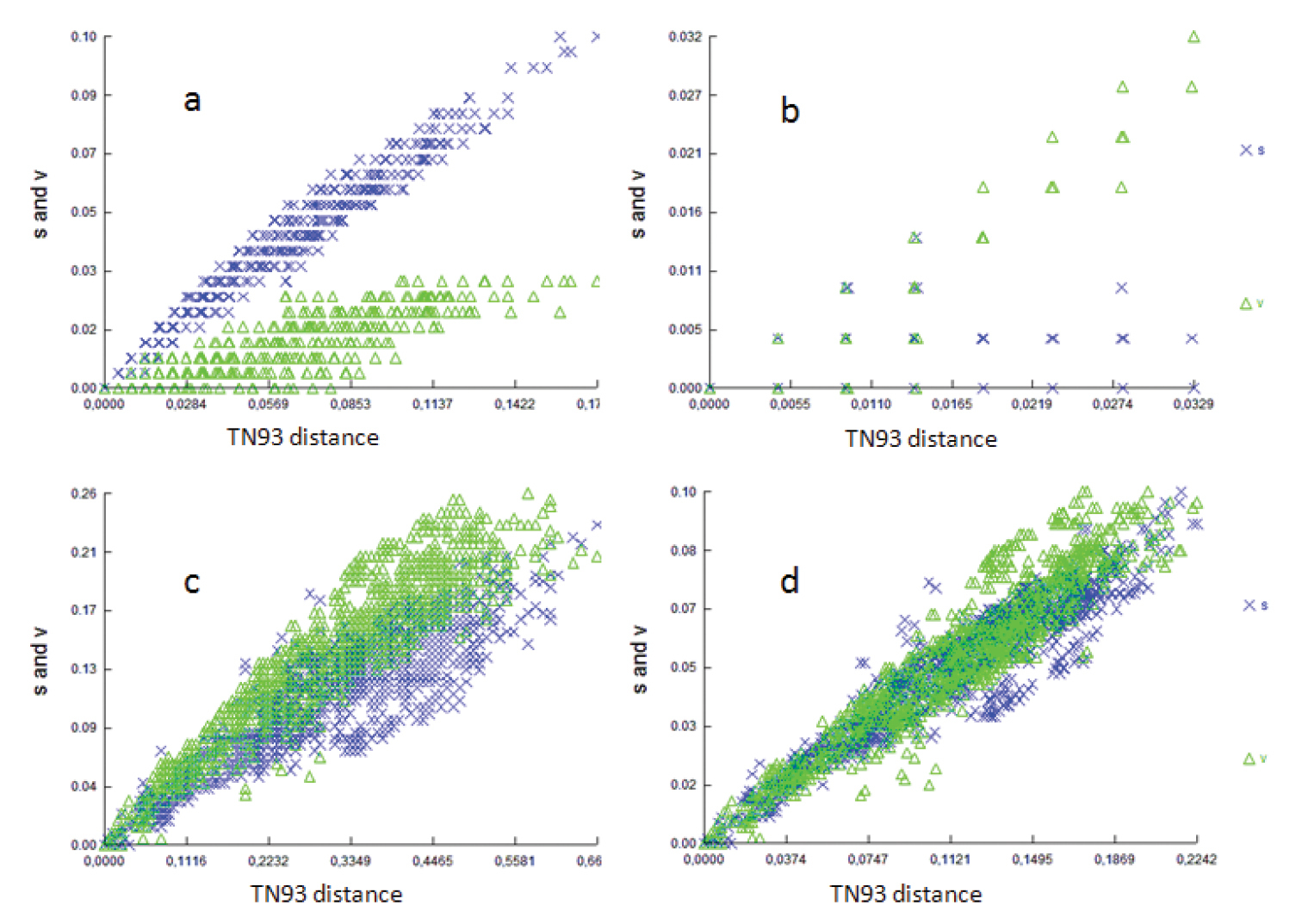
All three codon positions in the partial COI gene were analyzed for saturation, achieved by plotting the number of observed substitutions against the model TN93 genetic distance estimates. The scattergrams (Fig. 2a–c) showed that transitions and transversions for the first, second and third codons of the partial COI gene increased with the genetic distance, but considerable scattering was also observed. In addition, a similar plot of the third codon transition of the COI gene (Fig. 2d) suggested that saturation of transition occurred between certain pairs of the taxa, which may lead to higher levels of homoplasy (
Saturation plots of transversion and transition rates against JC69 distance at a first codon position b second codon position c third codon position, and d sum of data.
Result of likelihood mapping is presented in Fig. 3. High dichotomic phylogenetic signal was detected in the dataset. The percentage of the quartets suggesting a star- or network- like phylogeny is 9.9%, indicating that data are reliable for a dichotomic phylogenetic analysis (
Likelihood mapping analysis of the sequence alignments of COI gene present in the Cephinae. The regions at the corners of the triangles correspond to the three possible tree topologies for a quartet; the lateral regions to partly resolved trees and the central region to unresolved trees. The numbers indicate the percentage of quartets falling in each region.
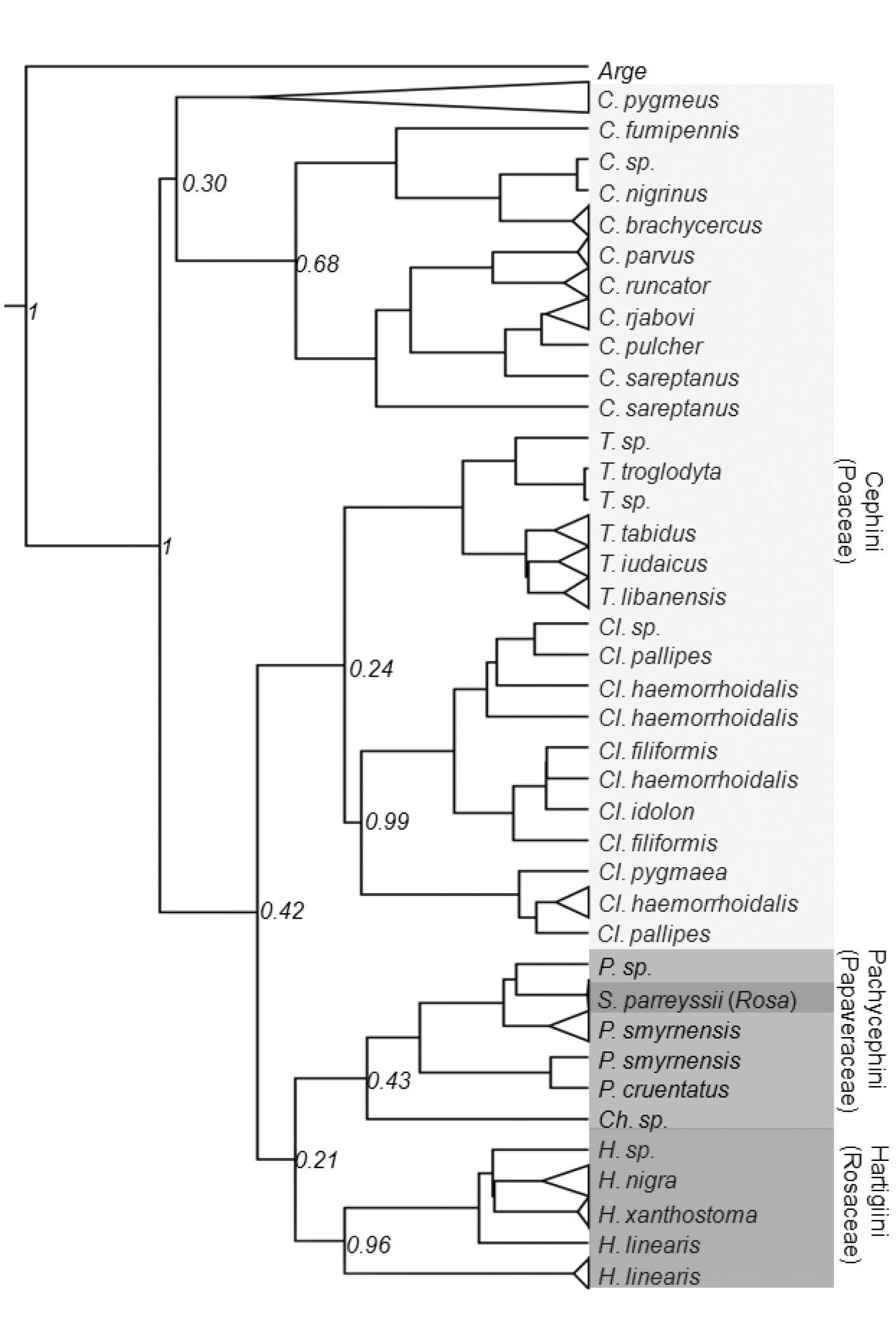
Bayesian interface tree based on the mitochondrial COI gene sequences of the Cephinae. Host plants are indicated in parentheses. Numbers at nodes indicate the posterior values.
Currently, the Cephinae is divided into three tribes based on morphology and feeding habits of larvae. The recovered mitochondrial gene trees substantially conflict with the current taxonomic arrangement, particularly the tribe level. Trees constructed under ML and BI methods supported monophyly of each genus except Pachycephus but failed to recover monophyly of any tribes. However, it should be noted that monophyly of most genera were supported by low posterior values (Fig. 4). This is probably due to the strongly biased nucleotide composition and the saturation at the third codon position (Fig. 2). The BI tree suggests that the most basal clade of Cephinae is the genus Cephus making the Cephini paraphyletic with respect to rest of Cephini and other tribes. Occurrence of Syrista within Pachycephini rather than Hartigiini makes both tribes polyphyletic and paraphyletic respectively (Fig. 4). Otherwise, Pachycephini and Hartigiini appear as sister groups. However, we do not propose a new classification as the present phylogeny is generated from a single gene fragment.
Evolution of phytophagy has occurred many times in insects, and is often accompanied by a significant increase in rates of speciation (
We thank Mahir Yildirim and Burcu Temel for accompanying us on some of our field trips and collecting specimens. This research was funded by Cumhuriyet University via research grants provided to the project CÜBAP F-224. We are grateful to Dr. Donald L.J. Quicke (Imperial College, London) and the other anonymous reviewer for their valuable comments on the early version of the manuscript.