(C) 2011 Anu Veijalainen. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Correct species identification is the basis of ecological studies. Nevertheless, morphological examination alone may not be enough to tell species apart. Here, our integrated molecular and morphological studies demonstrate that the relatively widespread and common neotropical parasitoid wasp Pimpla croceipes Cresson, 1874 (Hymenoptera: Ichneumonidae: Pimplinae) actually consists of two distinct species. The name Pimpla molesta (Smith, 1879), stat. rev. is available for the second species. The two species were identified by DNA barcoding and minor differences in morphology and colouration. Our results support the previous notions that DNA barcoding can complement morphological identification and aid the discovery of cryptic species complexes.
cryptic species, integrative taxonomy, Neotropics, parasitoid wasp, Pimplinae
In the midst of global biodiversity loss and substantial
taxonomic shortcomings, improved identification methods for
hyperdiverse and poorly known invertebrate groups are good news.
Integrating molecular methods with morphological species identification
can accelerate biodiversity inventories and facilitate spotting cryptic
species (i.e. two or more distinct species classified as one due to
morphological similarity; e.g.
The parasitoid wasp family Ichneumonidae (Hymenoptera)
may well be the largest animal family on earth, but it is considered
taxonomically challenging and poorly known (
When working through neotropical ichneumonid samples, we encountered a number of specimens that we identified as Pimpla croceipes (hereafter Pimpla croceipes sensu lato) according to the keys in
The data consisted of specimens collected by the LLAMA project in Guatemala and Honduras (see below), currently on loan to the Zoological Museum, University of Turku (ZMUT) and later to be deposited in the collaborative institutions of the LLAMA project, and the collections of the Natural History Museum, London, UK (BMNH). The LLAMA specimens were studied using both molecular and morphological species identification methods, the BMNH specimens focusing exclusively on morphology. The images were taken in ZMUT using an Olympus SZX16 stereomicroscope attached to an Olympus E520 digital camera. The layer photos were combined using the programmes Deep Focus 3.1, Quick PHOTO CAMERA 2.3 and Combine ZP.
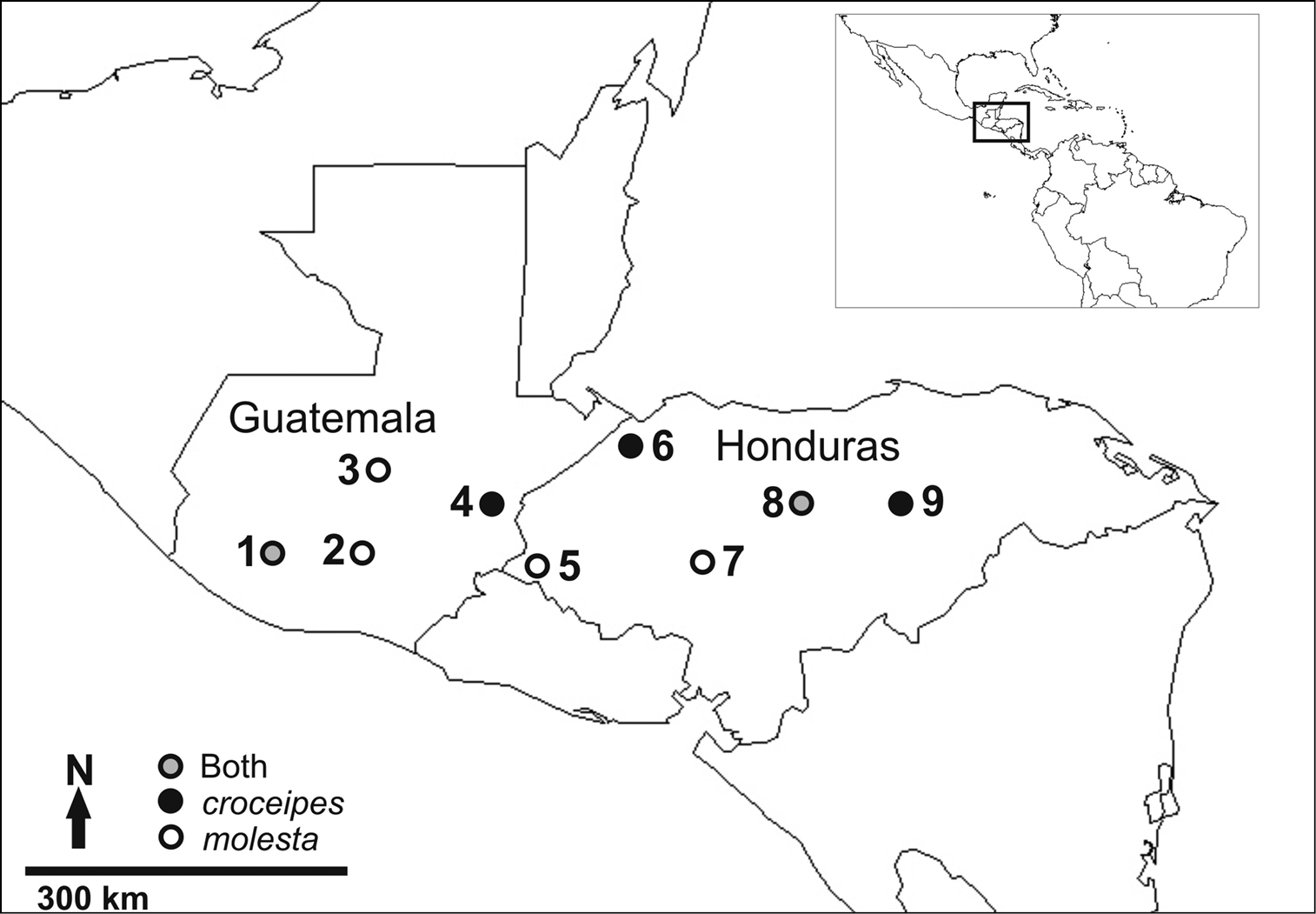
LLAMAIn the LLAMA samples, there were 97 specimens of Pimpla croceipes s.l. The specimens were collected by Malaise traps as a part of the Leaf Litter Arthropods of Mesoamerica project (LLAMA; http://llama.evergreen.edu) led by JTL in Guatemala and Honduras from May to June 2009 and 2010, respectively (see online Supplementary material for geospatial information and sampling periods). The LLAMA project applied similar Malaise sampling effort at 17 study sites, ranging from 50–2400 m asl. Four or five Malaise traps were set for four days at each site. Pimpla croceipes s.l. specimens were collected at nine of the 17 sites (Suchitepequez: 4 km S volcano Atitlán, Sacatepequez: 5 km SE Antigua, Baja Vera Paz: Biotopo El Quetzal, Zacapa: 2 km SE La Unión, Ocotepeque: 13 km E Nueva Ocotepeque, Cortés: Parque Nacional Cusuco, Comayagua: 10 km E Comayagua, Olancho: Parque Nacional La Muralla, and Olancho: 9–11 km N Catacamas; Figs 3, 5). All nine sites were at mid to high elevation, 1200–2335 m asl. Habitats were all mature wet forest, typically diverse mesophyll cloud forest, but some sites with variable densities of pine, oak, and Liquidambar. Traps were generally located on forest edges or in small clearings. Sampling took place during the transition from dry season to wet season.
DNA barcodingWe extracted the DNA of all the LLAMA specimens of Pimpla croceipes
s.l.using the DNeasy® Blood & Tissue Kit (QIAGEN) and following the
standard bench protocol for animal tissue in DNeasy Blood & Tissue
Handbook 07/2006 (the samples were also incubated at 70° C for 10
minutes after adding the Buffer AL and vortexing). Next, we amplified
the approximately 650-base fragment of the 5’ end of the mitochondrial
gene cytochrome c oxidase I(cox1, COI) well known as the
barcode region for animals. Each PCR was done in a 20 µl volume
consisting of 1 µl of DNA extract and 19 µl of master mix (12.5 µl dH2O,
2.0 µl 10× PCR Gold Buffer, 2.0 µl MgCl2 solution, 1.0 µl primer
LCO, 1.0 µl primer HCO, 0.4 µl dNTP, 0.1 µl Ampli Taq Gold). The PCRs
were run for 40 cycles with an annealing temperature of 50°C. The
succesful PCRs were cleaned and sequenced by Macrogen (South Korea),
after which we edited and aligned the sequences and constructed the
neighbour-joining tree based on genetic distance calculated with the K2P
model using MEGA v4 (
In the Natural History Museum, there were 237 Costa Rican specimens, 24 Mexican specimens, 2 from Panama and 1 from Venezuela, plus the holotype of Pimpla molesta, from Costa Rica (Supplementary material). These were sorted into Pimpla croceipes and Pimpla molesta based on their morphological characters (Table 1).
Pimpla croceipes female, lateral view.
Pimpla molesta female, lateral view.
The LLAMA study sites where specimens of either Pimpla croceipes, Pimpla molesta, or both, were collected 1 Suchitepequez 2 Sacatepequez 3 Baja Vera Paz 4 Zacapa 5 Ocotepeque 6 Cortés 7 Comayagua 8 Olancho: “La Muralla” 9 Olancho: “Catacamas”. See text for more specific descriptions of site locations.
A comparison of the diagnostic morphological characters of Pimpla croceipes and Pimpla molesta.
| Sex | Character | Pimpla croceipes (Fig. 1) | Pimpla molesta (Fig. 2) |
|---|---|---|---|
| F | Subalar prominence | Yellow/white | Black |
| F | Fore coxa | Yellow | Black or black and yellow |
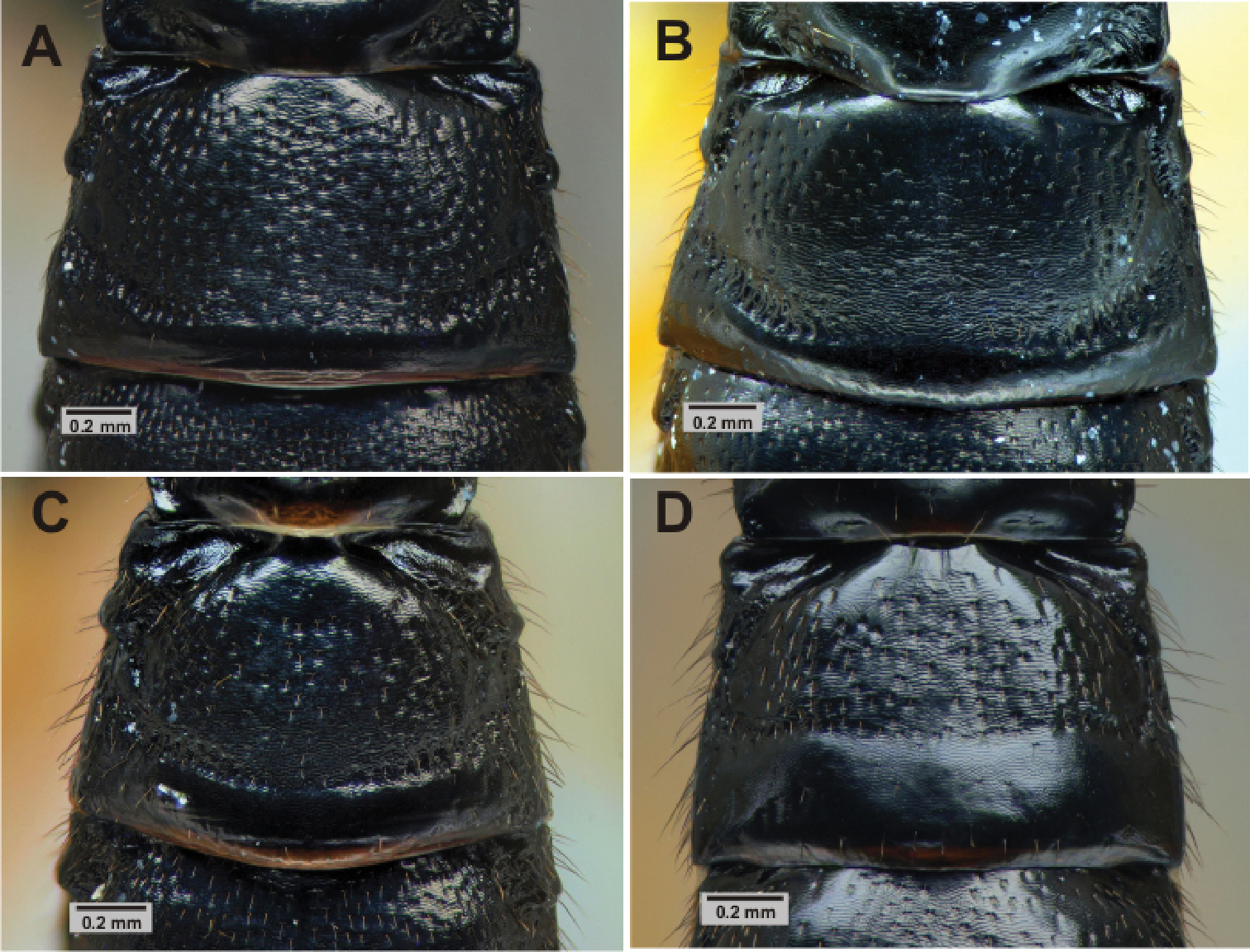
| F | 2nd tergite | More closely punctate(Fig. 6A) | More sparsely punctate (Fig. 6B) |
| M | 2nd tergite | Transverse groove at or behind posterior 0.65 (Fig. 6C), curved; tergite usually more closely punctate | Transverse groove at posterior 0.55 (Fig. 6D), almost straight; tergite more sparsely punctate |
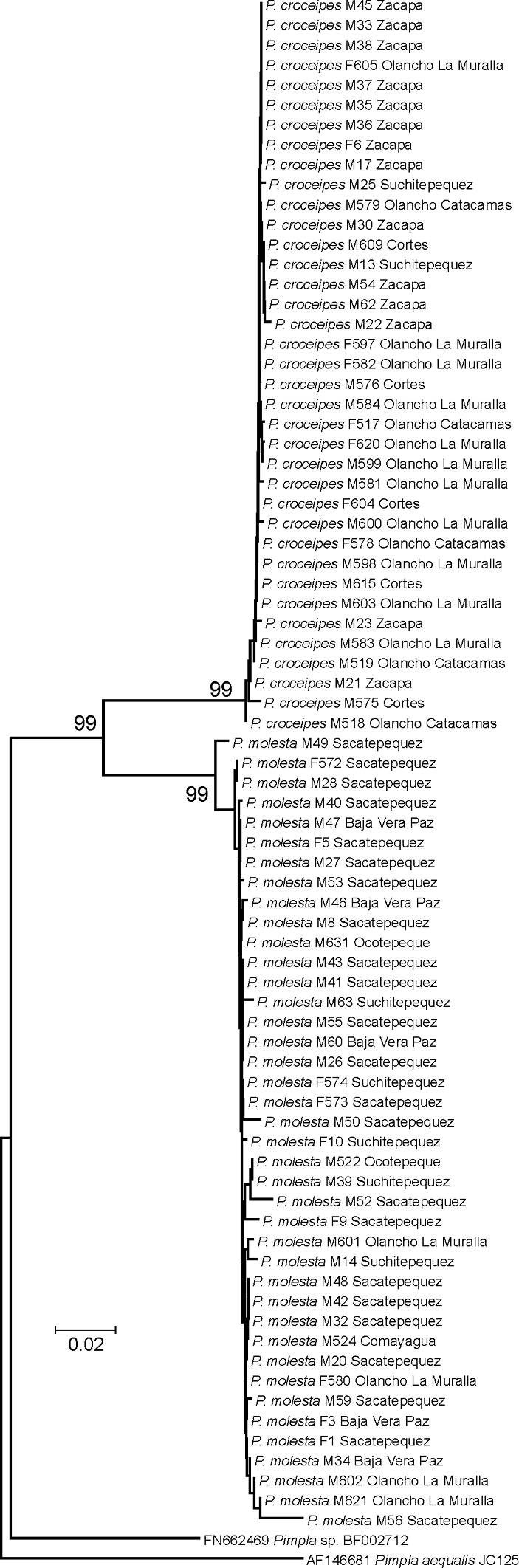
We obtained COI sequences for 77 specimens and the DNA barcoding results clearly group the specimens into two distinct species (Fig. 4).
The two species differed from each other by 9.9% (K2P distance), and
had virtually no intraspecific variation (0–0.5% variation). The
molecular results, enabling us to preliminarily divide the successfully
barcoded LLAMA specimens into two groups, greatly facilitated the
morphological identification process, and we also found interspecific
differences in the specimens’ colouration and morphology (Table 1). Careful morphological examination and comparison with type material revealed that the two species each have available names: Pimpla croceipes and Pimpla molesta
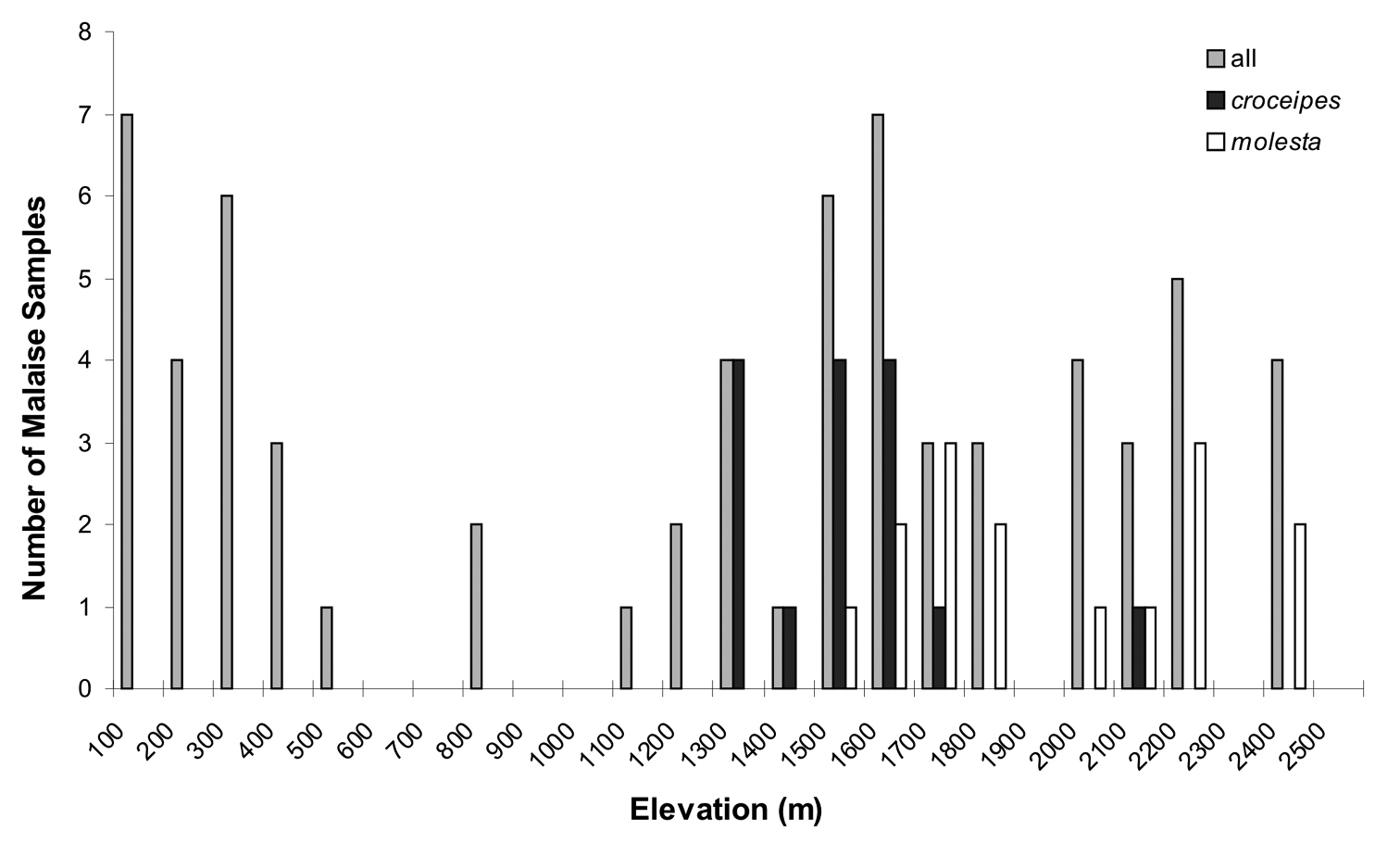
(stat. rev.). The ecological information that was available (e.g.
altitude, habitat) could not be used to entirely predict the identity
of the specimens as the distribution areas of the two species overlap in
the total LLAMA dataset and both species could occur in the same traps
(Supplementary material). This is in accordance with
We examined the holotype female of Pimpla molesta in BMNH and images of the lectotype female of Pimpla croceipes (deposited in Philadelphia Academy of Sciences). Fortuitously, these two types correspond to our two morphotypes, designated on the basis of the molecular separation. We were therefore able to assign all of our specimens of Pimpla croceipes s.l.to either Pimpla croceipes or Pimpla molesta. The species can be identified according to the diagnosis below and separated with the character differences summarized in Table 1 and illustrated in Figures 1, 2 and 6.
Males are relatively straightforward to separate on the
basis of the sculpture on the second metasomal tergite, although colour
pattern appears to offer no differences. Females are more difficult to
separate but do show small differences in colour and metasomal
sculpture. Table 1 should serve to separate almost all individuals. Both key easily to Pimpla croceipes using
In total, we studied 361 specimens (excluding the holotypes) and finally assigned them into 175 individuals of Pimpla croceipes and 186 of Pimpla molesta (Supplementary material). The two species were equally abundant in the LLAMA samples (Pimpla croceipes: 48, Pimpla molesta: 49 individuals). In BMNH, there are Pimpla croceipes specimens from Costa Rica and Venezuela (Las Mercedes) and Pimpla molesta specimens from Costa Rica, Mexico (several sites in Guerrero State) and Panama (Chiriquí).
Neighbour-joining tree of the successfully DNA barcoded LLAMA specimens (species; sex; DNA voucher code; location). Numbers above the branches are bootstrap proportions. The two species Pimpla croceipes and Pimpla molesta (stat. rev.) are clearly separated into two well-supported clusters. The results were further supported by morphological examination.
A frequency histogram of elevations of Malaise samples, for 1) all LLAMA samples, 2) those with Pimpla croceipes, and 3) those with Pimpla molesta.
We confirm with morphological and molecular evidence and occurrence of sympatry that Pimpla croceipes and Pimpla molesta are two separate species.
When cryptic species are discovered, linking previously compiled species data to the correct “new” species may be difficult (
We identify at least three sources of error that should be kept in mind while interpreting the presence of the two Pimpla species in specific study localities. First, the sampling efficiency of Malaise traps may be influenced by the precise positioning of the trap in the sampling locality. Second, many species of ichneumonids are often rare in samples. For this reason, their presence or absence in a Malaise trap sample may be largely a coincidence. Third, pimplines are large parasitoids which are normally strong fliers. Thus, a Malaise trap may sample individuals that are just passing a forest patch instead of actually being resident there.
We have shown that the Pimpla croceipes
s.l., previously thought to be one species, is in fact two
morphologically very similar but molecularly clearly different species
which are both common and co-occur in Central America. As with other
studies on neotropical parasitoids (
Metasoma. 2nd tergite sculpture of Pimpla croceipes female A, Pimpla molesta female B, Pimpla croceipes male C, and Pimpla molesta male D.
Lissonota sp. male (Ichneumonidae: Banchinae) collected at Comayagua (Honduras) showing a similar mimicry pattern to Pimpla croceipes and Pimpla molesta.
Project LLAMA was supported by National Science Foundation grant DEB-0640015. The Finnish Society of Forest Science, Jenny and Antti Wihuri Foundation, Smithsonian Institution Graduate Student Fellowship, and SYNTHESYS provided personal funding and travel grants to Anu Veijalainen. The lab work expenses were covered by The Academy of Finland grant 129811 to Niklas Wahlberg, and Kone Foundation grant (Biodiversity and multiple trophic interactions) to Ilari E. Sääksjärvi. Matthew Buffington (USDA) and Michael Sharkey (University of Kentucky) arranged the distribution of specimens, and the assistance from the TEGLab staff (University of Turku) was of great help. Thank you to Jason Weintraub (Academy of Natural Sciences, Philadelphia) for sending images of the lectotype of Pimpla croceipes. Norman Johnson, Jose Fernandez-Triana and one anonymous referee gave valuable comments on the manuscript.