ZooKeys 176: 123–131, doi: 10.3897/zookeys.176.2284
Widespread
Wolbachia infection in terrestrial isopods and other crustaceans
Richard Cordaux 1, Samuel Pichon 1,3, Houda Ben Afia Hatira 2, Vincent Doublet 1,4, Pierre Grève 1, Isabelle Marcadé 1, Christine Braquart-Varnier 1, Catherine Souty-Grosset 1, Faouzia Charfi-Cheikhrouha 2, Didier Bouchon 1
1 Université de Poitiers, CNRS UMR 6556, Laboratoire Ecologie Evolution Symbiose, 40 Avenue du Recteur Pineau, 86022 Poitiers, France
2 Faculté des Sciences de Tunis, Unité de Recherche de Bioécologie et Systématique Evolutive, 2092 Université Tunis Manar, Tunisia
3 Present address: Zoological Institute, University of Basel, Vesalgasse 1, 4051 Basel, Switzerland
4 Present address: Institut für Biologie, Martin-Luther-Universität Halle-Wittenberg, Hoher Weg 8, 06120 Halle (Saale), Germany
Abstract
Wolbachia bacteria are obligate intracellular alpha-Proteobacteria of arthropods and nematodes. Although widespread among isopod crustaceans, they have seldom been found in non-isopod crustacean species. Here, we report Wolbachia infection in fourteen new crustacean species. Our results extend the range of Wolbachia infections in terrestrial isopods and amphipods (class Malacostraca). We report the occurrence of two different Wolbachia strains in two host species (a terrestrial isopod and an amphipod). Moreover, the discovery of Wolbachia in the goose barnacle Lepas anatifera (subclass Thecostraca) establishes Wolbachia infection in class Maxillopoda. The new bacterial strains are closely related to B-supergroup Wolbachia strains previously reported from crustacean hosts. Our results suggest that Wolbachia infection may be much more widespread in crustaceans than previously thought. The presence of related Wolbachia strains in highly divergent crustacean hosts suggests that Wolbachia endosymbionts can naturally adapt to a wide range of crustacean hosts. Given the ability of isopod Wolbachia strains to induce feminization of genetic males or cytoplasmic incompatibility, we speculate that manipulation of crustacean-borne Wolbachia bacteria might represent potential tools for controlling crustacean species of commercial interest and crustacean or insect disease vectors.
KeywordsWolbachia, endosymbiont, Crustacea, Maxillopoda, terrestrial isopod, distribution, adaptation
Introduction
Wolbachia pipientis (hereafter Wolbachia) bacteria are obligate intracellular alpha-Proteobacteria of arthropods and nematodes (O’Neill et al. 1997; Bourtzis and Miller 2003). These maternally-inherited bacteria are often referred to as reproductive parasites because they are able to manipulate the reproduction of their hosts to increase their own transmission, via mechanisms such as cytoplasmic incompatibility, male killing, thelytokous parthenogenesis and feminization of genetic males (O’Neill et al. 1997; Bourtzis and Miller 2003; Cordaux et al. 2011). In addition to vertical transmission, Wolbachia bacteria are occasionally transmitted horizontally (Werren et al. 1995; Vavre et al. 1999; Cordaux et al. 2001). These transmission patterns probably explain, at least partly, why Wolbachia bacteria are found in a highly diverse range of hosts.
Wolbachia bacteria are particularly frequent in arthropods. Hence, it has been estimated that 20–75% of insect species may be infected by Wolbachia (Werren and Windsor 2000; Hilgenboecker et al. 2008). These bacteria have also been reported in chelicerates such as mites (Breeuwer and Jacobs 1996), spiders (Cordaux et al. 2001) and scorpions (Baldo et al. 2007). In crustaceans, Wolbachia have long been known to infect the terrestrial isopod Armadillidium vulgare (Rousset et al. 1992) (order Isopoda, suborder Oniscidea; classification from Martin and Davis (2001)), in which Wolbachia induce functional feminization of genetic males (Rigaud et al. 1997; Cordaux et al. 2004; Bouchon et al. 2008; Cordaux et al. 2011). A systematic search in 80 crustacean species suggested that Wolbachia infection was restricted to isopods, with a prevalence of 46% in terrestrial isopod species (Bouchon et al. 1998). Based on additional screenings, this figure has recently been updated to 61% (Bouchon et al. 2008). The initial screening of 80 crustacean species also provided molecular evidence for Wolbachia infection in two other isopod suborders (Asellotta and Flabellifera) (Bouchon et al. 1998).
Despite multiple screenings of crustacean groups, Wolbachia bacteria have seldom been found in non-isopod crustacean species (Bouchon et al. 1998; Fitzsimmons and Innes 2005; Maniatsi et al. 2010). To date, only two amphipod and two non-marine ostracod crustacean species outside of the Isopoda order have been reported to be infected by Wolbachia (Cordaux et al. 2001; Baltanas et al. 2007). The extent to which this uneven Wolbachia distribution among crustacean species reflects host spectrum specificity or biased sampling remains to be clarified. In this study, we report Wolbachia infections in fourteen new crustacean species. Our results extend the range of Wolbachia infection within terrestrial isopods and amphipods (class Malacostraca). Moreover, the discovery of Wolbachia in the goose barnacle Lepas anatifera (subclass Thecostraca) establishes Wolbachia infection in class Maxillopoda. Molecular characterization indicates that the new strains are closely related to B-supergroup Wolbachia strains previously reported from crustacean hosts (Bouchon et al. 1998; Cordaux et al. 2001). The identification of closely related Wolbachia strains in highly divergent crustacean hosts suggests that this group of Wolbachia endosymbionts can naturally adapt to a wide range of crustacean hosts, not just isopods.
Materials and methods
Wild-caught individuals belonging to fourteen crustacean species were studied (Table 1). Seven species have been previously detected in a survey of Wolbachia infection of woodlice fauna in Tunisia (Ben Afia Hatira et al. 2007). For these species, Wolbachia prevalence (ranging from 40% to 100%) is available in the original publication. The seven remaining species were included as part of an ongoing effort in our laboratory to sample and test novel crustacean species for Wolbachia infection. For these species, one or two individuals were collected. Therefore, no information on prevalence is available for these species. Genomic DNA from single individuals was extracted as previously described (Bouchon et al. 1998). Wolbachia infection status of each individual was tested using a PCR assay based on the standard wsp marker. We used primers 81f and 691r (Braig et al. 1998) and previously described PCR conditions (Cordaux et al. 2001). Purified PCR products were directly sequenced in both directions, as previously described (Cordaux et al. 2001). The wsp sequences generated in this study were deposited in GenBank under accession numbers HE616815-HE616830 (Table 1).
Table 1.
Novel crustacean species infected by Wolbachia bacteria reported in this study.
|
Class
|
Infraclass (I) or Order (O)
|
Species
|
Sampling location
|
GenBank accession number for wsp
|
|
Maxillopoda
|
Cirripedia (I)
|
Lepas anatifera
|
La Rochelle, France |
HE616817
|
|
Malacostraca
|
Amphipoda (O)
|
Talitrus saltator
|
La Rochelle, France |
HE616820 and HE616821
|
|
Isopoda (O)
|
Armadillidium granulatum
|
Sidi Massaoud Khniss, Tunisia |
HE616828
|
|
Armadillidium pelagicum
|
Ras Jbel, Tunisia |
HE616829
|
|
Armadillidium sulcatum
|
Natural Reserve Mhibes, Tunisia |
HE616827
|
|
Cubaris murina
|
Baie Mahault, Guadeloupe, France |
HE616815
|
|
Hemilepistus reaumuri
|
Metbasta, Tunisia |
HE616816
|
|
Platyarthrus hoffmansegghi
|
Liniers, France |
HE616818
|
|
Porcellio albinus
|
Kibili, Tunisia |
HE616830
|
|
Porcellio buddelundi
|
Skhira cliff, Tunisia |
HE616823 and HE616824
|
|
Porcellio lamellatus
|
Menzel Jmil, Tunisia |
HE616825
|
|
Porcellio variabilis
|
Jbel Ouest, Tunisia |
HE616826
|
|
Porcellionides cingendus
|
Archigny, France |
HE616819
|
|
Trachelipus rathkei
|
Cosne Cours sur Loire, France |
HE616822
|
Sequences were aligned using ClustalW as implemented in the software BioEdit ver. 7.0 (Hall 1999), followed by manual adjustments. Representative sequences from the B supergroup of Wolbachia diversity were included for comparison and two of the A supergroup as an outgroup, as previously described (Cordaux et al. 2001; Cordaux et al. 2004). There was a total of 618 positions in the dataset. Hypervariable regions were deleted because they could not be aligned with confidence. The resulting alignment included 512 positions of which 185 were considered informative by parsimony criteria. Recombination analyses were performed using Recombination Detection Program (RDP) v3.41 (Martin et al. 2010). Parameters were set as follows: sequences were considered linear, the highest acceptable P value cutoff was 0.01, a Bonferroni correction was applied, consensus daughter sequences were found, gaps were included, different window sizes of variable sites were tested (10, 20, and 30 VI), and 1, 000 permutations were performed. Phylogenetic analyses were conducted using the Minimum Evolution (ME) and Neighbor-Joining (NJ) methods, as implemented in MEGA ver 4.0 (Tamura et al. 2007). Evolutionary distances were computed using the Kimura 2-parameter substitution model. The ME tree was searched using the close-neighbor-interchange algorithm at a search level of 1 and the NJ algorithm was used to generate the initial tree. All positions containing alignment gaps and missing data were eliminated in pairwise sequence comparisons (pairwise deletion option). Bootstrap analyses were carried out with 1000 replicates.
Results and discussion
Our findings extend the range of Wolbachia infections among crustacean hosts to twelve additional terrestrial isopod species, one amphipod species and one cirriped species (class Maxillopoda) (Table 1). We performed a molecular characterization of the Wolbachia strains based on the wsp gene, a Wolbachia-specific genetic marker (Braig et al. 1998) which has been shown to be highly informative for the analysis of crustacean Wolbachia strains (Cordaux et al. 2001). In total, we identified sixteen different Wolbachia strains: two strains in the amphipod Talitrus saltator and the terrestrial isopod Porcellio buddelundi, and one strain in each of the other crustacean host species. This is the third report of multiple Wolbachia strains being harbored by a single terrestrial isopod host species and the first case reported in amphipods. Indeed three different Wolbachia strains have previously been identified in Armadillidium vulgare (Cordaux et al. 2004; Verne et al. 2007) and Porcellionides pruinosus (Michel-Salzat et al. 2001). As in Armadillidium vulgare and Porcellionides pruinosus, the Wolbachia strains found in Talitrus saltator and Porcellio buddelundi were identified in different individuals, and no multiple infection of single individuals have been reported so far in crustaceans, in contrast to what has been widely documented in insects (Vautrin et al. 2007).
Querying GenBank with the sixteen wsp sequences through BLASTN searches resulted in best matches to other Wolbachia strains with 98.0–100% nucleotide similarity. This analysis revealed that all novel Wolbachia strains belong to the B supergroup of Wolbachia diversity, as do all isopod and amphipod Wolbachia strains characterized to date (Rousset et al. 1992; Bouchon et al. 1998; Cordaux et al. 2001; Michel-Salzat et al. 2001; Cordaux et al. 2004).
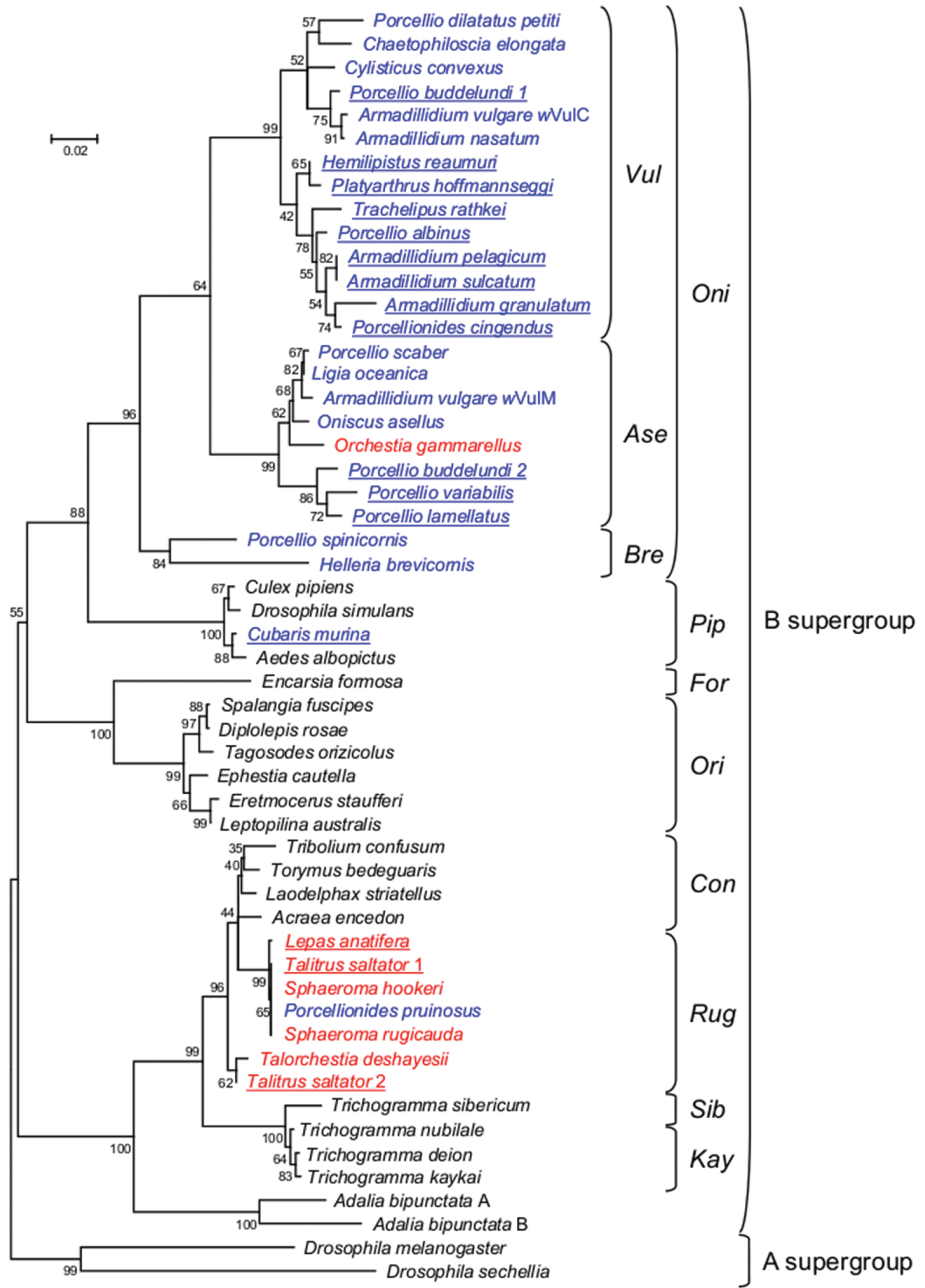
To further characterize the novel crustacean Wolbachia strains, we performed a phylogenetic analysis of B supergroup Wolbachia strains. To facilitate comparisons with previous analyses of crustacean Wolbachia strains, we added the sixteen strains reported in this study to the set of representative B supergroup strains previously used in Cordaux et al. (2001). No significant support for recombination was detected with RDP and the topologies of the ME and NJ trees (see Materials and Methods) reconstructed in this study were highly similar, as they were to the phylogenetic inferences reported previously (Cordaux et al. 2001). We also emphasize that, as far as crustacean Wolbachia strains are concerned, we have shown previously that a wsp-based phylogeny yielded essentially the same relationships between taxa as phylogenies based on other Wolbachia genes such as 16S rRNA, ftsZ and GroE (Bouchon et al. 1998; Cordaux et al. 2001; Wiwatanaratanabutr et al. 2009). Because our updated phylogeny of crustacean Wolbachia strains is in agreement with phylogenetic results obtained in previous studies using various genes, we are reasonably confident that the phylogenetic analysis we present in the manuscript is reliable. Figure 1 shows the tree inferred from the ME analysis. All but one crustacean Wolbachia strains clustered together in one of the two groups Oni and Rug, previously shown to encompass isopod and amphipod Wolbachia strains (Bouchon et al. 1998; Cordaux et al. 2001; Wiwatanaratanabutr et al. 2009). Our extended dataset shows a newly emerging trend in the diversity of crustacean Wolbachia strains, in that the Oni group mostly contains strains isolated from terrestrial isopods, whereas the Rug group mostly contains strains isolated from non terrestrial isopod crustaceans. We speculate that this might reflect two major ancestral Wolbachia acquisitions in crustaceans, in terrestrial and aquatic environments. Ecosystems in the transition zones from land to sea, as estuaries and wetlands, are highly productive and therefore attract a multitude of wildlife. Moreover these ecosystems are in a continuous state of change. The finding of closely related symbionts in the Rug group suggesting recent symbiont dispersal across divergent hosts sharing the same habitats suggests that horizontal transfers of Wolbachia could be facilitated in such ecosystems.
The only exception to the clustering of crustacean Wolbachia strains in the Oni or Rug groups is the Wolbachia strain isolated from the terrestrial isopod Cubaris murina (Fig. 1). Indeed, the latter strain falls within the Pip group, closely related to Wolbachia strains from mosquitoes and drosophila. This result suggests the possibility of a Wolbachia horizontal transfer between isopod and insect hosts, as previously proposed to explain the similarity between strains of the Rug group with other insect Wolbachia strains (Bouchon et al. 1998; Cordaux et al. 2001). In any event, it is noteworthy that Cubaris murina is a terrestrial isopod with a pan-tropical distribution (Schmalfuss 2003). Interestingly, a similar result has been obtained by Wiwatanaratanabutr et al. (2009) who showed that the Wolbachia strain isolated from an endemic isopod sampled in Thailand also belongs to the Pip group. This is in contrast to most terrestrial isopods screened so far for Wolbachia infection, which were generally sampled in temperate regions (Bouchon et al. 1998; Bouchon et al. 2008). This suggests that screenings of terrestrial isopods and other crustaceans in regions that have not been extensively studied so far may have the potential to uncover a tremendous and unexpected Wolbachia diversity that we are only beginning to realize.
Figure 1.
Phylogenetic tree of B-supergroup Wolbachia strains based on wsp sequences, using Minimum Evolution analysis. The tree is rooted with two A-supergroup Wolbachia strains. Bootstrap values inferred from 1000 replicates are shown as percentages. Strains are identified by the host species from which they were isolated. Wolbachia strains from terrestrial isopods and non terrestrial isopod crustaceans are shown in blue and red, respectively. New crustacean Wolbachia infections reported in this study are underlined. Wolbachia strains from insects are shown in black. Names assigned to groups of Wolbachia strains are shown on the right, following Cordaux et al. (2001).
An important result of this study is the discovery of Wolbachia in the goose barnacle Lepas anatifera. This is the first report of Wolbachia infection in the class Maxillopoda, a major crustacean group comprising ~15, 000 species, that is, more than one quarter of all described crustacean species (Martin and Davis 2001). Interestingly, the Maxillopoda Wolbachia strain is closely related to other crustacean Wolbachia strains in the Rug group, being 99.8% similar to the Wolbachia strains from the terrestrial isopod Porcellionides pruinosus, the intertidal isopods Sphaeroma rugicauda and Sphaeroma hookeri and the amphipod Talitrus saltator, based on the wsp marker (Fig. 1). This result thus indicates that highly divergent crustacean hosts can harbour highly similar Wolbachia endosymbionts. By implication, our results suggest that this group of Wolbachia endosymbionts can naturally adapt to a wide range of crustacean hosts. Given the demonstrated ability of terrestrial isopod Wolbachia strains to induce feminization of genetic males or cytoplasmic incompatibility between infected males and uninfected females (Legrand and Juchault 1986; Rigaud et al. 1997; Bouchon et al. 1998; Moret et al. 2001; Cordaux et al. 2004; Bouchon et al. 2008; Cordaux et al. 2011), we speculate that manipulation of crustacean-borne Wolbachia bacteria might represent potential tools for controlling crustacean species of commercial interest and crustacean or insect disease vectors. For example, it has been shown that freshwater crustaceans could be used as predators to control immature forms of the mosquito Aedes aegypti, the vector of the Dengue fever, one of the major infectious diseases in several tropical and subtropical countries in Asia, Africa, and The Americas (Kosiyachinda et al. 2003). The finding of Wolbachia infection in a wide range of crustacean species, including freshwater crustaceans, may open new opportunities in the biological control of insect disease vectors via a Wolbachia-based strategy. This might allow researchers to manipulate the population dynamics of crustacean predators of insects, to enhance their efficacy as biological control agents.
Acknowledgements
We thank Hajer Khemaissia and Karima Nasri-Ammar for assistance with sample collection. This research was funded by a European Research Council Starting Grant (FP7/2007-2013 grant 260729 EndoSexDet) to RC and a Comité Mixte de Coopération Universitaire Franco-Tunisien grant to DB and FCC.
ReferencesBaldo L, Prendini L, Corthals A, Werren JH (2007)
Wolbachia are present in southern african scorpions and cluster with supergroup f. Curr Microbiol 55: 367-373. doi:
10.1007/s00284-007-9009-4
Baltanas A, Zabal-Aguirre M, Pita M, Lopez-Fernandez C (2007)
Wolbachia identified in a new crustacean host: an explanation of the prevalence of asexual reproduction in non-marine ostracods? Fundamental and Applied Limnology 169: 217–221. doi:
10.1127/1863-9135/2007/0169-0217
Bouchon D, Rigaud T, Juchault P (1998) Evidence for widespread
Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc Biol Sci 265: 1081-1090. doi:
10.1098/rspb.1998.0402
Bouchon D, Cordaux R, Grève P (2008) Feminizing
Wolbachia and the evolution of sex determination in isopods. In: Bourtzis K, Miller T (Eds). Insect Symbiosis, Volume 3. Taylor and Francis Group LLC, Boca Raton, FL: 273-294. doi:
10.1201/9781420064117.ch12
Bourtzis K, Miller TA (2003) Insect symbiosis. CRC Press, Boca Raton.
Braig HR, Zhou W, Dobson SL, O’Neill SL (1998) Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont
Wolbachia pipientis. J Bacteriol 180: 2373-2378.
Breeuwer JA, Jacobs G (1996)
Wolbachia: intracellular manipulators of mite reproduction. Exp Appl Acarol 20: 421-434. doi:
10.1007/BF00053306
Cordaux R, Michel-Salzat A, Bouchon D (2001)
Wolbachia infection in crustaceans: novel hosts and potential routes for horizontal transmission. J Evol Biol 14: 237-243. doi:
10.1046/j.1420-9101.2001.00279.x
Cordaux R, Michel-Salzat A, Frelon-Raimond M, Rigaud T, Bouchon D (2004) Evidence for a new feminizing
Wolbachia strain in the isopod Armadillidium vulgare: evolutionary implications. Heredity 93: 78-84. doi:
10.1038/sj.hdy.6800482
Cordaux R, Bouchon D, Greve P (2011) The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet 27: 332-341. doi:
10.1016/j.tig.2011.05.002
Fitzsimmons J, Innes D (2005) No evidence of
Wolbachia among Great Lakes area populations of Daphnia pulex (Crustacea: Cladocera). J Plankton Res 27: 121-124. doi:
10.1093/plankt/fbh145
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95-98.
Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH (2008) How many species are infected with
Wolbachia?–A statistical analysis of current data. FEMS Microbiol Lett 281: 215-220. doi:
10.1111/j.1574-6968.2008.01110.x
Legrand JJ, Juchault P (1986) Rôle de bactéries symbiotiques dans l’intersexualité, la monogénie et la spéciation chez des crustacés oniscoïdes. Boll Zool 53: 161-172. doi:
10.1080/11250008609355500
Maniatsi S, Bourtzis K, Abatzopoulos TJ (2010) May parthenogenesis in Artemia be attributed to
Wolbachia? Hydrobiologia 651: 317–322. doi:
10.1007/s10750-010-0306-8
Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26: 2462-2463. doi:
10.1093/bioinformatics/btq467
Martin JW, Davis GE (2001) An updated classification of the recent crustacea. Natural History Museum of Los Angeles County, Science Series 39: 1-124.
Michel-Salzat A, Cordaux R, Bouchon D (2001)
Wolbachia diversity in the Porcellionides pruinosus complex of species (Crustacea: Oniscidea): evidence for host-dependent patterns of infection. Heredity 87: 428-434. doi:
10.1046/j.1365-2540.2001.00920.x
Moret Y, Juchault P, Rigaud T (2001)
Wolbachia endosymbiont responsible for cytoplasmic incompatibility in a terrestrial crustacean: effects in natural and foreign hosts. Heredity 86: 325-332. doi:
10.1046/j.1365-2540.2001.00831.x
O’Neill SL, Hoffmann AA, Werren JH (1997) Influential passengers: inherited microorganisms and arthropod reproduction. Oxford University Press, New York.
Rigaud T, Juchault P, Mocquard JP (1997) The evolution of sex determination in isopod crustaceans. Bioessays 19: 409-416. doi:
10.1002/bies.950190508
Rousset F, Bouchon D, Pintureau B, Juchault P, Solignac M (1992)
Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc Biol Sci 250: 91-98. doi:
10.1098/rspb.1992.0135
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596-1599. doi:
10.1093/molbev/msm092
Vavre F, Fleury F, Lepetit D, Fouillet P, Bouletreau M (1999) Phylogenetic evidence for horizontal transmission of
Wolbachia in host-parasitoid associations. Mol Biol Evol 16: 1711-1723.
Werren JH, Zhang W, Guo LR (1995) Evolution and phylogeny of
Wolbachia: reproductive parasites of arthropods. Proc Biol Sci 261: 55-63. doi:
10.1098/rspb.1995.0117
Werren JH, Windsor DM (2000)
Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc Biol Sci 267: 1277–1285. doi:
10.1098/rspb.2000.1139
Wiwatanaratanabutr I, Kittayapong P, Caubet Y, Bouchon D (2009) Molecular phylogeny of
Wolbachia strains in arthropod hosts based on groE-homologous gene sequences. Zoolog Sci 26: 171-177. doi:
10.2108/zsj.26.171