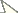(C) 2010 Wolfgang Arthofer. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Phylogenetic studies based on mtDNA become increasingly questioned because of potential pitfalls due to mitochondrial pseudogenes and mitochondrial selective sweeps. While the inclusion of nuclear markers should preferentially be considered for future studies, there is no need to abandon mtDNA as long as tests for the known mtDNA artefacts are performed. In this study we presentadditionaldata and test previous phylogeographical studies of Pityogenes chalcographus. We did not detect nuclear copies (numts) of the previously used mitochondrial markers by performing a combined long range/nested PCR of the COI gene and by an in silico analysis of the COI sequence data. This confirms the robustness of our previous phylogenetic study of Pityogenes chalcographus. Results of an in-situ hybridization of Wolbachia in Pityogenes chalcographus confirm the presence of this endosysmbiont in this species. However, we did not detect a correlation between infection status, geographical region and mtDNA haplotypes. The hybridisation data also support a previous hypothesis that infections do not result from parasitoids or parasitic nematodes, insect surface or laboratory contaminations and are hence a true infection of Pityogenes chalcographus. We conclude that the deep structure found in mitochondrial populations of Pityogenes chalcographus indeed represents the evolutionary history of European populations.
Wolbachia, Scolytinae, pseudogenes, numts, mtDNA, phylogeny, phylogeography
In the last two decades several phylogeographic (e.g.
Mitochondria originated from the endosymbiosis of
α-proteobacteria in ancestral eukaryotic cells. Mitochondrial genomes
contain fewer genes than those of free-living α-proteobacteria, due to a
loss of genes during their evolutionary history. This gene loss is
explained by (1) the functional redundancy of mitochondrial genes with
pre-existing nuclear genes and (2) the functional transfer of
mitochondrial genes to the nucleus. The transfer of mtDNA derived
sequences to the nucleus is an ongoing process in eukaryotes and
mitochondrial pseudogenes have been identified in the nuclear genome of
many species (
A set of strategies is available in order to avoid numt based errors, including in silico
analysis of sequences to detect an eventual increased number of
non-synonymous base substitutions, frameshifts, additional stop codons
and reduced transition/transversion ratios (
A specific feature of mtDNA is its strict maternal
inheritance in most insects. Due to this asymmetrical inheritance within
a species the marker only reflects the female part of the species’
genealogy. Hence, mtDNA transmission will be influenced by any
selection for maternally transmitted genes or other maternally selective
traits. Several maternally transmitted endosymbionts are well known in
invertebrates, with Wolbachia as the most prominent one (
While some Wolbachia
infections do not alter host physiology and reproduction, such effects
have been found in others. Reproductive fitness traits range from
cytoplasmatic incompatibility (CI) to male-killing, feminisation and
the induction of thelytokous parthenogenesis (see
In this study we show that numts do not influence the phylogenetic pattern of Pityogenes chalcographus (
Numt search
Mitogenomic sequences of the coleopteran species Pyrocoelia rufa (Lampyridae), Tribolium castanaeum (Tenebrionidae) and Crioceris duodecimpunctata (Chrysomelidae) were obtained from GeneBank (for accession numbers see table 1) and aligned using Clustal X (
Primer sequences of Met/F and CO2/R for Pityogenes chalcographus amplifying 3463bp: alignments and GenBank accession numbers.
| Met/F | 5' | g | c | t | w | h | t | g | g | g | t | t | c | a | t | a | c | c | c | 3' | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crioceris duodecimpunctata | NC_003372 | . | . | . | a | t | . | . | . | . | . | . | . | . | . | . | . | . | . | ||
| Pyrocoelia rufa | NC_003970 | . | . | . | t | t | . | . | . | . | . | . | . | . | . | . | . | . | . | ||
| Tribolium castaneum | NC_003081 | . | . | . | a | t | . | a | . | . | . | . | . | . | . | . | . | . | . | ||
| Apis mellifera ligustica | NC_001566 | . | . | . | a | a | c | a | . | . | . | . | . | . | . | . | . | . | . | ||
| Bombyx mori | NC_002355 | . | . | . | a | t | . | . | . | . | c | . | . | . | . | . | . | . | . | ||
| Drosophila simulans | NC_005781 | . | . | . | a | c | . | . | . | . | . | . | . | . | . | . | . | . | . | ||
| CO2/R | 5' | c | a | a | a | t | t | t | c | t | g | a | a | c | a | t | t | g | 3' | ||
| Crioceris duodecimpunctata | NC_003372 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||
| Pyrocoelia rufa | NC_003970 | . | g | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||
| Tribolium castaneum | NC_003081 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||
| Apis mellifera ligustica | NC_001566 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||
| Bombyx mori | NC_002355 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||
| Drosophila simulans | NC_005781 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||
Fourteen DNA extracts of Pityogenes chalcographus
representing all clades were selected for analysis. Thermocycling was
performed in a Primus 25 advanced thermocycler (peqlab, Germany). Full
length PCR was performed in 10 µl reactions using 0.4 µM of each Met/F
and CO2/R primer, 6 mM magnesium sulphate, 200 µM dNTPs, 0.4 U Taq DNA polymerase (Sigma, USA), 0.01 U Sawady Pwo polymerase (peqlab) and 1 µl DNA template in the buffer provided with the Pwo
polymerase. Cycling conditions were 3 min initial denaturation at 94° C
followed by 32 cycles of 94° C (30 sec), 55° C (1 min) and 68° C (2.5
min) and a final extension step at 68° C (10 min). Products were diluted
1:10, 000 with sterile distilled water and 1 µl diluted amplicon was
used as template for the nested PCR. Dilution series were carried out to
prove that the carry over of genomic DNA from the full length to the
nested PCR reaction was small enough to avoid detectable amounts of
amplicon. Nested PCR was done in 25 μl reactions containing 3.75 mM
magnesium chloride, 125 μM dNTPs (Fermentas, Lithuania), 0.5 μM of
each K698 (
An in-silco analysis was performed on 262 sequences of the original study (
In silico analysis of CO1 mutations of data presented in
| Total | Relative (%) | Expected value for mthNA a | |
|---|---|---|---|
| Single base substitutions | 125 | 100.0 | - |
| 1st codon position substitutions | 15 | 12.0 | 14.9 ± 9.4% b |
| 2nd codon position substitutions | 2 | 1.6 | 4.5 ± 3.5% b |
| 3rd codon position substitutions | 108 | 86.4 | 80.6 ± 21% b |
| Nonsynonymous substitutions | 13 | 10.4 | 7.47 ± 5.4% c |
| C › T substitutions | 25 | 20.0 | - |
| GC › GT substitutions | 3 | 12.0 d | 25 ± 14.0% e |
| Insertions | 0 | 0 | none f |
| Deletions | 0 | 0 | none f |
| Additional stop-codons | 0 | 0 | none f |
| Transitions (3rd codon position) | 95 | 88.0 g | 84.9% ± 18.1% h |
| Transversions (3rd codon position) | 13 | 12.0 g | 15.1 ± 7.6% h |
| Trasition-transversion ratio | 7.31 | - | - |
| GC content | - | 34.6 | 28.66 ± 10.5% i |
a expected relative values as given in reference ± χ
2 confidence interval at α=0.05 (
Identification of Wolbachia infections by in situ hybridization
In situ hybridization followed a slightly modified protocol of
Phylogeographic analysis of European Pityogenes chalcographus
populations revealed a deep genetic structure between the most diverged
haplotypes with three major clades and an estimated divergence time of
100, 000 years before present (
Long/nested PCR and in silico analysis for presence of numts
Alignment of mitochondrial genomes of three
coleopteran and three non-coleopteran insect species resulted in six
candidate primers (data not shown), of which one primer pair (Table 1), after extensive optimization of PCR conditions, amplified a clear band from Pityogenes chalcographus
DNA extracts. Dilution series of genomic DNA gave no visible bands in
dilutions of more than 1:1, 000, ensuring that all amplicons produced
in the nested PCR originated solely from the full length PCR product and
not from genomic carry-over (data not shown). After nested PCR
extensive products of the expected size could be obtained from almost
all haplotypes of Pityogenes chalcographus
examined. Even templates without visible amplification in the full
length PCR had formed enough product to be amplified in the subsequent
nested reaction. Comparison of the NJ trees derived from direct PCR
sequences (
PCR conditions were chosen to remove any numt shorter than 3.4 kb, i.e. three times longer than the largest numts ever observed in insects. Both direct and long/nested PCR sequences were identical, and so were the phylogenetic trees. With our test, co-amplification of numts in the direct PCR approach would have led to discrepancies in tree topology between direct and long PCR sequences.
In order to extend numt screening to 262 individual sequences representing 58 different haplotypes, an in silico analysis was performed targeting characteristic differences between mtDNA and numt sequence composition. Eleven numerical traits were analyzed independently and all of them resulted in values within 5% confidence intervals for authentic mtDNA (Table 2). Thus, presence of numts in the analyzed populations of Pityogenes chalcographus can be excluded.
Several strategies to avoid numt co-amplifications
are known. The purification of mtDNA by caesium chloride gradient
centrifugation (
While we consider the long/nested PCR approach as very reliable to exclude any numt from a genetic analysis, it requires additional handling time, costs for PCR consumables and high quality DNA allowing the amplification of >3kb products. Especially the latter condition will not be given when long term stored specimens have to be analyzed that might have degraded DNA. The in silico approach presented here can be readily applied to individual haplotypes within any mtDNA alignment and does not require additional manipulations in the laboratory. It is thus suitable for a re-check of existing mtDNA based phylogenies.
Detection of Wolbachia by in situ hybridization
The principial functionality of a modified protocol for Wolbachia detection by in situ hybridization with DIG labelled probes was tested using ovarial tissue of Wolbachia free Drospohila simulans STC and Drospohila simulans flies infected with wRi. Differences in colouration were clearly distiguishable between infected and uninfected Drospohila simulans (Fig. 1 A, B).
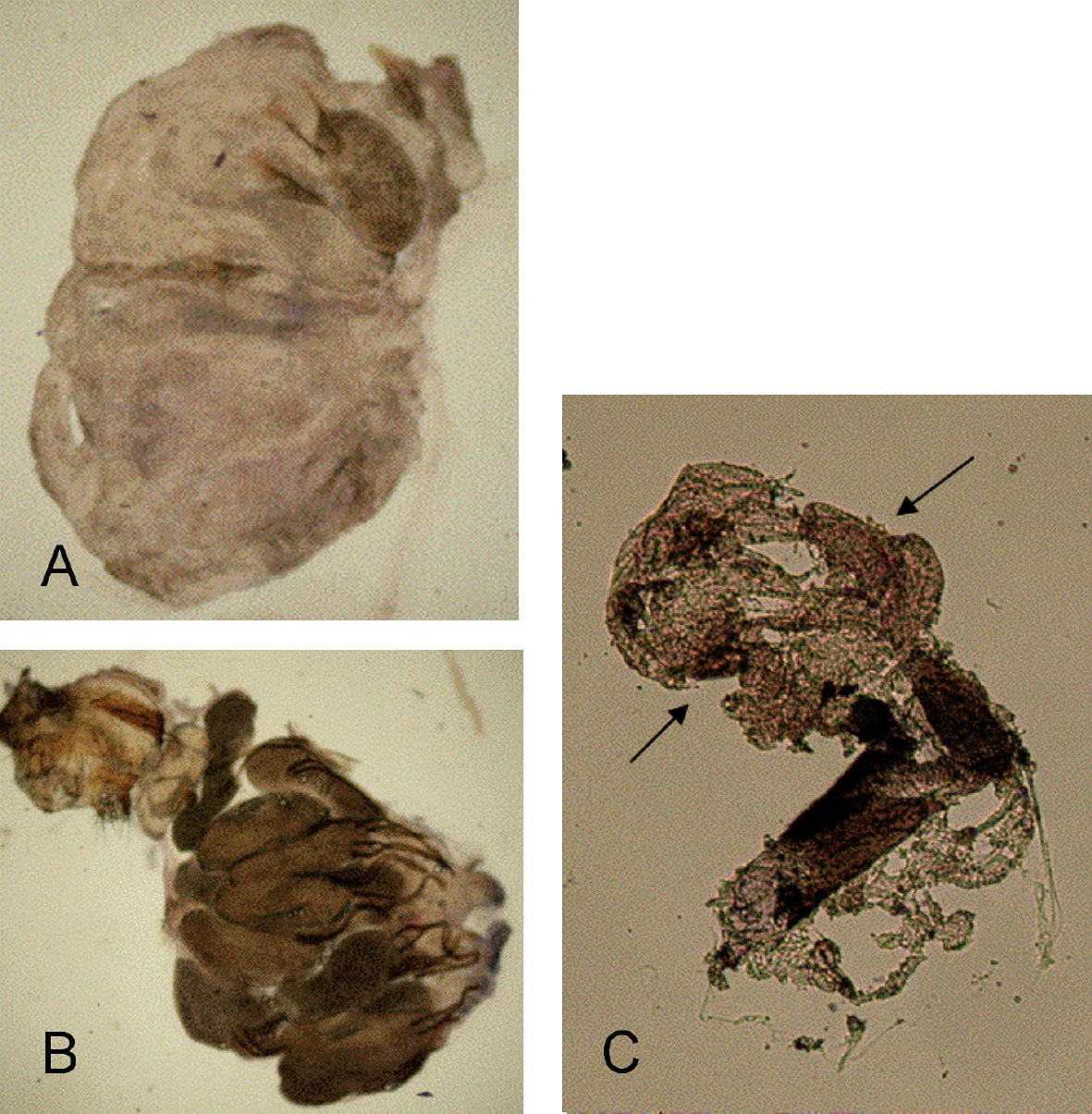
Compared to wRi in Drospohila simulans, Wolbachia titre in Pityogenes chalcographus was low, and in average only 35.5% of the individuals were infected (
in situ hybridization with wsp specific probe and staining with NBT/BCIP solution on uninfected A and Wolbachia infected Drosophila simulans B. An accumulation of dark color is observed only in ovarioles of Wolbachia infected Drospohila simulans. C Results of in situ hybridization of ovarial tissue excised from one Pityogenes chalcographus individual with accumulation of dark color (arrows). Three specimens were analysed. All ictures taken with 40-fold magnification.
Evidence of a range of selective forces on mtDNA markers
make phylogenetic studies that are purely based on mtDNA less reliable.
While the inclusion of nuclear markers like microsatellites or AFLP
should preferentially be considered for future studies, there is no
need to completely abandon mtDNA as long as tests for the potential
manipulation of mtDNA sequences are performed. Such tests should also be
included in ongoing efforts to barcode the tree of life based on mtDNA (
Furthermore, we have detected Wolbachia in Pityogenes chalcographus cells in low titre by in situ hybridisation. Our results confirm earlier work that used a highly sensitive PCR method (
We wish to thank Wolfgang Miller for providing Drosophila strains, Andrea Stradner for her help in the Wolbachia work and the Austrian Science Foundation (FWF) for financial support.