(C) 2010 Alberto López-Pancorbo. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
A new troglobitic species, Nesticus baeticus sp. n. (♂♀), inhabiting the karst landscapes of the high part of the Cazorla, Segura and Las Villas Natural Park (NE Jaén, Spain) where it has been found in 8 caves is diagnosed and described, its distribution and habitat are also analyzed.The new species belongs to the Iberian species group that includes Nesticus luquei, Nesticus lusitanicus and Nesticus murgis. Evolutionary relationships of the Iberian Nesticus species are discussed on the basis of morphological and molecular data (cox1 and rrnL).
Arachnida, Araneae, taxonomy, description, new species, caves, Iberian Peninsula, Mediterranean basin
The genus Nesticus
Thorell, 1869 is distributed worldwide except for south-eastern Asia
and Australia and comprises 125 species and 8 subspecies (
The first species described from Iberia was Nesticus obcaecatus
Simon, 1907, found only from a single locality: Cueva del Molino de
Aso (Huesca), on the southern slopes of the Central Pyrenees (
This work describes a new cavernicolous species whose distribution includes several karst landscapes of the different mountain formations that make up the Sistema Bético, the ridge in southern Iberian Peninsula.
Materials and methods TaxonomyAbbreviations used in the text are as follows: PL = prosoma length (from posterior edge of carapace to front edge of clypeus, measured at midline); PW = maximum prosoma width; OL = opisthosoma length (excluding the pedicel); OW = maximum opisthosoma width; MA = median apophysis; Mt = metatarsus; Tb = tibia; TTA = theridioid tegular apophysis; TTA p1 = process 1 of TTA; TTA p2 = process 2 of TTA; TL = total length (excluding the pedicel). Eyes: AM = anterior median; AL = anterior lateral; PM = posterior median; PL = posterior lateral; i = immature; sub = subadult; GEV = Grupo de Espeleología de Villacarrillo.
Female vulva was removed and treated with 30% KOH.
After observation and drawing, the vulva was washed in distilled water
and stored in 70% ethanol. Left male palps were illustrated in all
cases. We follow
A Nikon Coolpix 4500 digital camera attached to a stereomicroscope was used to capture images. Following
Taxonomic sampling. Taxa analyzed in the present study are listed in Appendix 1. All the Iberian species are included except Nesticus murgis due to impossibility to obtain fresh material for DNA analysis. Nesticus eremita Simon, 1879 from Croatia and Nesticus ionescui Dumitrescu, 1979 from Romania are also included to test the monophyly of Iberian species. Sequence from Nesticus sp. from China (
Sample Storage and DNA Extraction. Specimens were preserved in 95% or absolute ethanol and stored at 4°C. Total genomic DNA was extracted from legs of a single specimen using the QIamp® DNA Mini Kit (QIAGEN) following the manufacturer’s protocols. The approximate concentration and purity of the DNA obtained were verified using 1% agarose/TBE gel electrophoresis.
PCR Amplification and Sequencing. Two regions of the mitochondrial DNA corresponding to a fragment of the cytochrome oxidase I gene (cox1) and 16S rRNA (rrnL) were selectively amplified using PCR with the following primer pairs: for cox1 C1-J-1718 (5' GGAGGATTTGGAAATTGATTAGTTCC 3') with C1-N-2191 (5' CCCGGTAAAATTAAAATATAAACTTC 3') (
A Perkin-ElmerCetus Moldel 480 thermocycler was used to perform 35 iterations of the following cycle: 30s at 95°C, 45s at 45°C, and 1 min at 72°C, beginning with an additional step of 3 min at 95°C, and ending with another step of 10 min at 72°C. PCR results were visualized by means of a 1% agarose/TBE gel. Amplified products were purified using Microcon PCR columns following the manufacturer’s specifications. Purified products were directly cycle-sequenced from both strands using ABI BigDye (Applied Biosystems) chemistry, precipitated in DyeEx Spin kit (Qiagen, Chatsworth, CA) columns, and run out on ABI Prism 377 (Applied Biosystems) automated sequencers. Sequencing reactions were performed in our lab with the forward and reverse PCR primers. Resulting product were run and analyzed at the Serveis Científico-Tècnics of the Universitat de Barcelona.
Alignment. Raw sequences were compared against
chromatograms and complementary contigs built and edited using the
Geneious Pro 3.6.2 software (http://www.genious.com). Sequences were manipulated and preliminary manual alignments constructed using BioEdit V.7.0.5.3 (
Phylogenetic analyses. Parsimony analyses of the combined data matrices were conducted with the program Winclada v.1.00.08 (
Holotype: ♂ (1619-A25) Cueva de la Murcielaguina, Hornos, Jaén, Spain, 5.11.2006, GEV leg. Paratypes: 2♀♀ (1619-A25) same locality and data; 1♀ (1720-A29) same locality, 30.12.2007, GEV leg.; 1♀ (1530-A22) Sima HO-55, Hornos, Jaén, 14.8.2006, GEV leg. (drawings and description of the female are based on this specimen); 1♂ (1524-A21), 1♂sub., 1♀, 7i (1525-A22) same locality and data, GEV Leg.; 1♀ (3811–150) Sima de los Alhaurinos, Hornos, Jaén, 12.05.2002, GEV leg.; 1♀ (3812–150) same locality and data; 2i (3860-151) Sima del Campamento, Hornos, Jaén, 02.03.2003, GEV leg.; 2i, (5023-189) same locality, 27.08.2004, GEV leg.; 2♂sub., 1f, 5i (1157-A07) Sima del Laberinto, Hornos, Jaén, 04.02.2006 López, A. & Pérez, A. leg.; 1♀, 1♀sub. (1343-A14) Cueva SE-20, Santiago de la Espada - Pontones, Jaén, 30.04.2006, GEV leg.; 1i (1539-A22) Sima del Órgano (HO-25), Hornos, Jaén, 14.08.2006, GEV leg.; 1i (5014-189) Sima Irene, Hornos, Jaén, 15.02.2004, GEV leg.
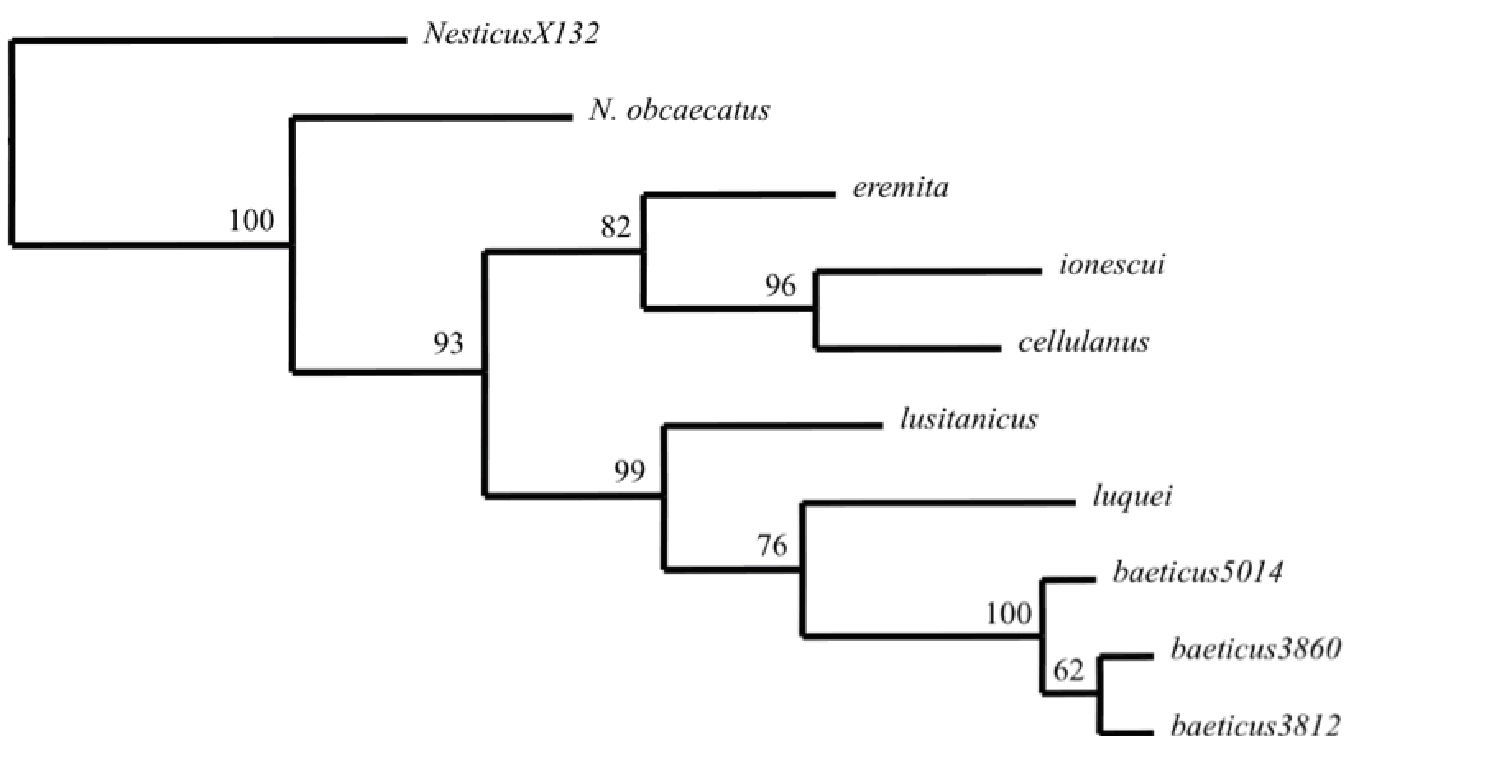
Single most parsimonious tree (L = 3454, CI = 0.447, RI = 0.769) found by MP analysis of the combined data set(cox1=472 bp, rrnL=456 bp and 22 gap characters) of Iberian Nesticus species (except Nesticus murgis), Nesticus ionescui from Romania and Nesticus eremita from Croatia. Numbers on nodes represent bootstrap support values. The outgroup Nesticus X130 is from China. Numbers on terminals refers to different localities (see material examined)
The Latin name ‘baeticus’ means ‘from Baetica’ (the south of Spain) and refers to the ‘Sistema Bético’, the ridge containing the karst landscapes from where the new species was collected.
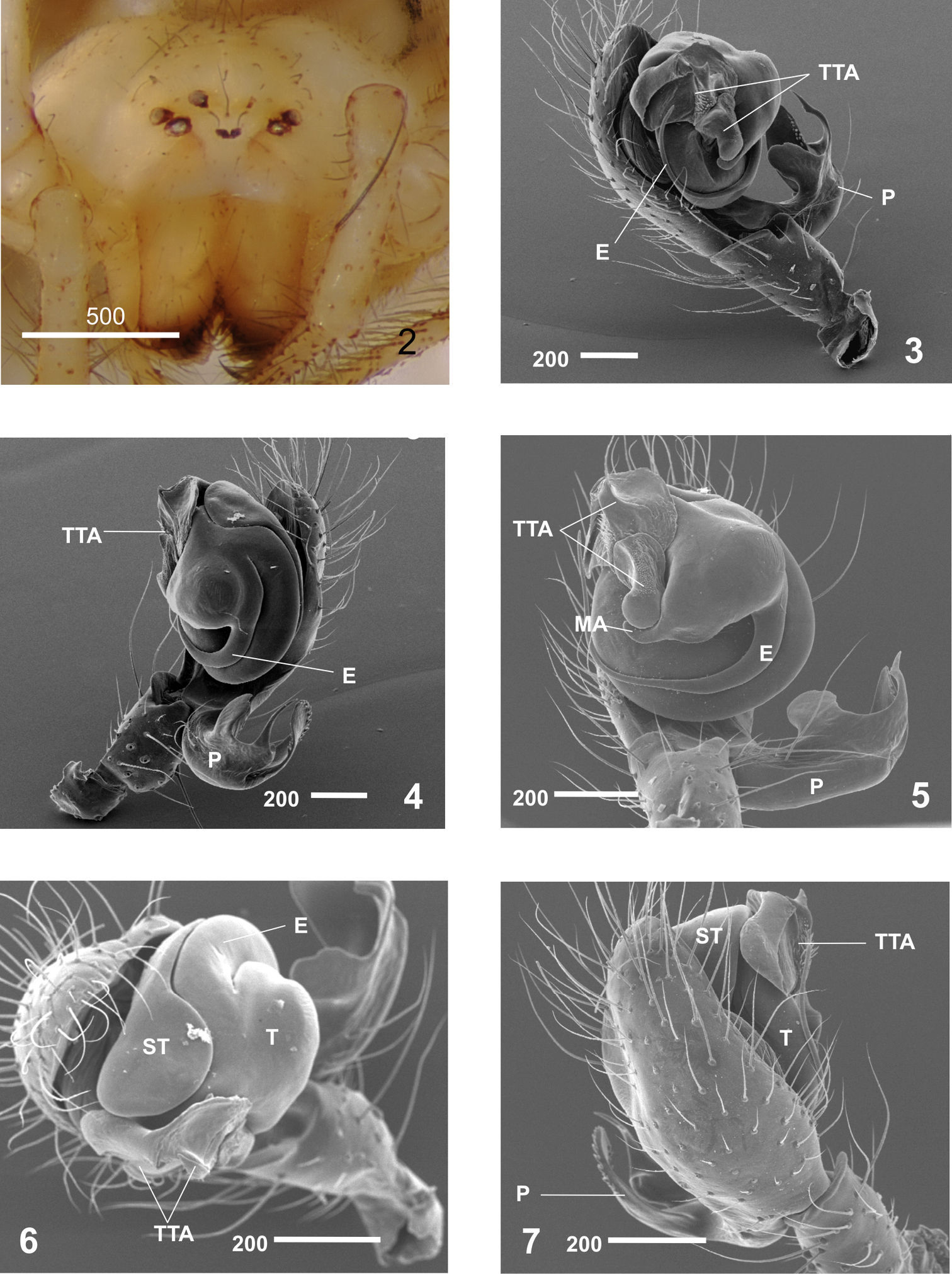
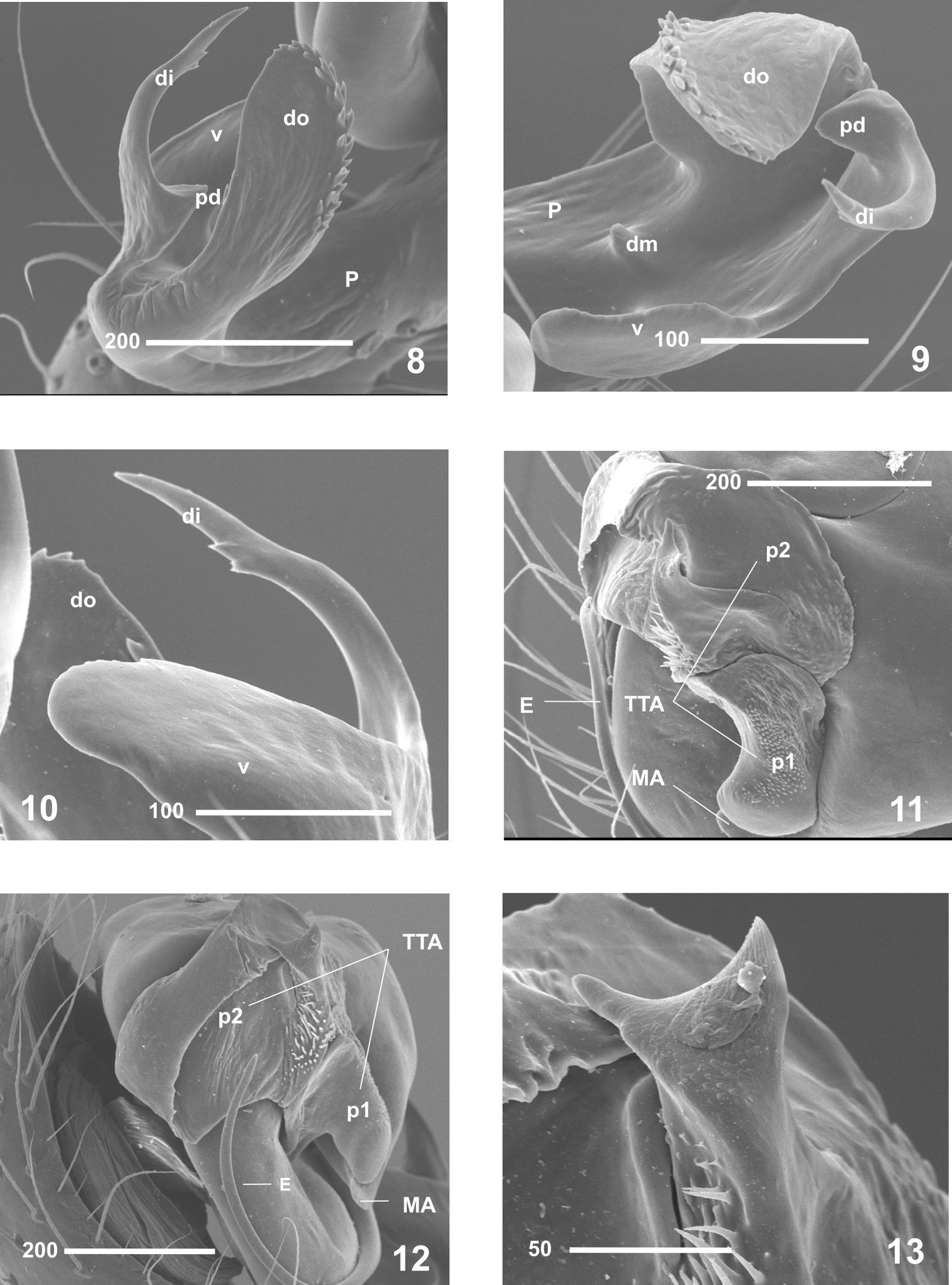
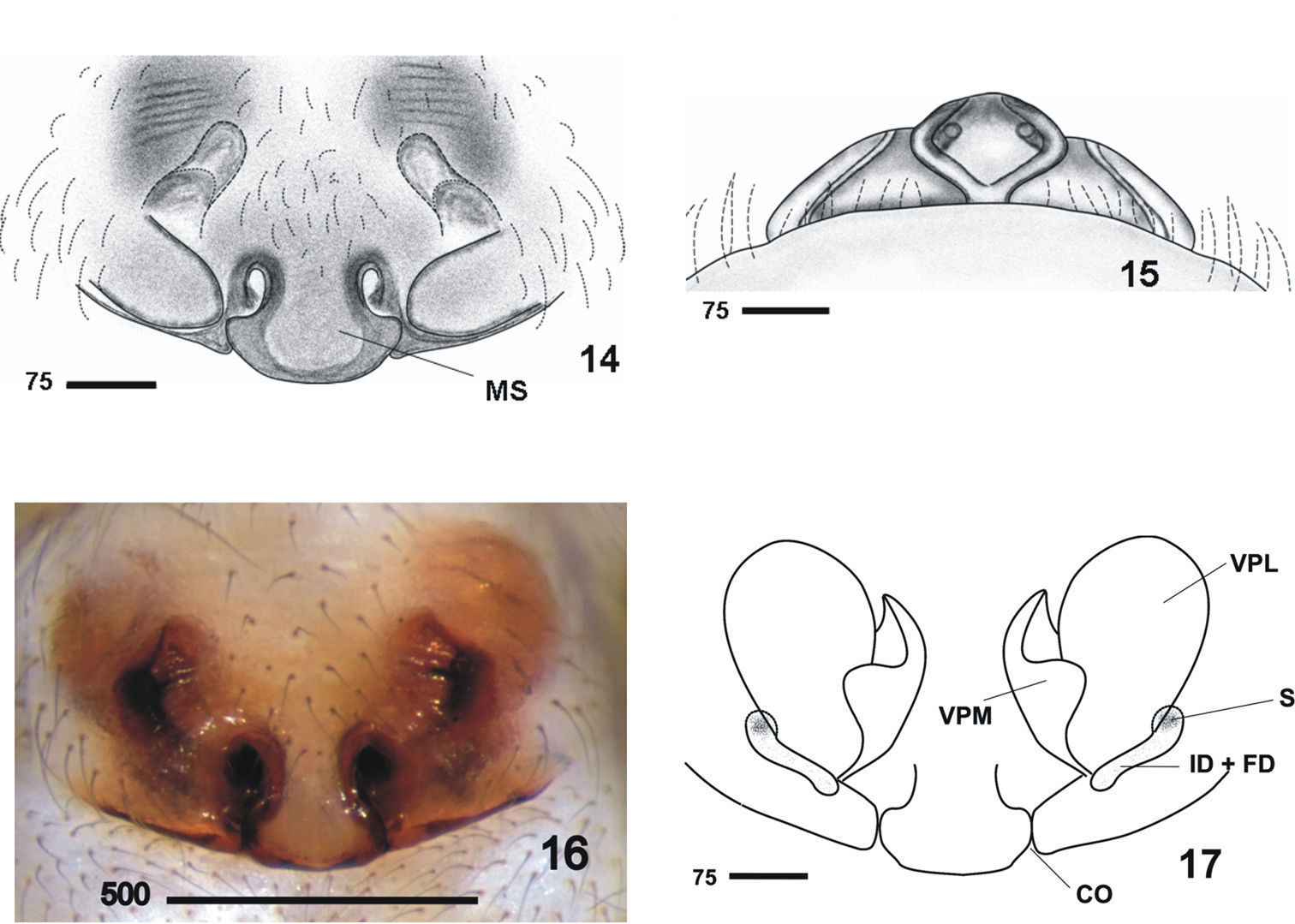
Males clearly differ from those of other Nesticus species in the shape of paracymbium (Figs 4–5, 8–10) and in the TTA structure (Figs 3–5, 11–12). In females, the development of the median septum of the vulva (Figs 14–16), the shape of the spermathecae and adjoining structures are also diagnostic (Fig. 17). The degree of ocular reduction of the AM eyes (Fig. 2) is also characteristic compared to other Iberian species.
Nesticus baeticus sp. n.: 2 female (1525) frontal view 3–6 male palp (Holotype). 3 retrolateral view 4 prolateral view 5 ventral view 6 frontal view 7 dorsal view. Abbreviations: E embolus MA median apophysis P paracymbium ST subtegulum T tegulum TTA theridioid tegular apophysis. Scale bar in μm.
On the basis of morphology, Nesticus baeticus sp. n. lies within the group including Nesticus murgis (known from Almería) and Nesticus luquei (an endemic to northwestern Spain). The shape and arrangement of the median apophysis (Figs 5, 11–12), the embolus (Figs 3–7, 11–12) and the paracymbial processes of male palp (Figs 8–10), plus the location and structure of the spermathecae and vulval glands of the female (Fig 17) are similar in all three species. Nesticus baeticus sp. n.differs more significantly from Nesticus lusitanicus, both in the morphology of the copulatory organs of both sexes.
Nesticus baeticus sp. n., male (holotype). 8–10 paracymbium 8 lateral view 9 frontal view 10 ventral view 11 median apophysis and theridioid tegular apophysis, ventral view 12 ditto, retrolateral view 13 apical protuberance of TTA p2. Abbreviations: di distal process dm dorsomedial apophysis do dorsal process E embolus P paracymbium pd paradistal apophysis MA median apophysis TTA theridioid tegular apophysis TTA p1 process 1 of TTA TTA p2 process 2 of TTA v ventral process. Sacale bar in μm.
The new species cannot be assigned to Carpathonesticus, Typhlonesticus or Canarionesticus, and differs from their representatives in having a different shape, ramification and modifications associated to the paracymbium, the general structure and arrangement of the embolous, as well as of the p1 and p2 TTA processes. Yet, the shape and disposition of the spermathecas and vulval glands shows markedly differences.
(Holotype). Coloration: carapace uniform
yellowish. Opisthosoma grayish, with some clearly-marked darker
patches. Appendages of the same colour as the carapace, slightly darker
around distal segments. Sternum yellowish, slightly paler than the
carapace. Carapace: approximately circular in dorsal view.
Cephalic region not raised but differentiated from the rest of the
prosoma. Fovea and thoracic grooves clearly visible. Significantly
reduced eyes, more evident in the AM (Fig. 1).
Eye size and interocular distances: AM = 0.03; AL = 0.07; PM = 0.06; PL
= 0.07; AM - AL = 0.14 AL; AM - AM = 0.03; PM - PL = 0.14; PM - PM =
0.18 PM; PL - AL almost touching. Opisthosoma: sub-elliptical in dorsal view. Appendages: prolateral margin of the chelicerae with 3 teeth, the two distal ones larger. Male palp(Figs 3–7). Paracymbium large (Figs 3–5) with well-developed dorsal and ventral processes (Figs 8–10). Broad, translucent dorsal process with a saw-toothed upper edge (Figs 8–10). Dorsomedial apophysis small and pointed (Fig. 9). Ventral region apically notched (Fig. 10). Short paradistal region, almost conical (Figs 8–9). Distal apophysis long, acuminate and slightly curved (Figs 8–10). Poorly developed MA, reduced to a small fingerlike process fused to the tegulum (Figs 5, 11–12). Conductor absent. TTA with two processes, TTA p1 and TTA p2 (Figs 11–12) (homologous to p1-p6 processes of conductor complex in
| Leg | coxa | troc. | femur | patella | tibia | meta. | tarsus | total |
|---|---|---|---|---|---|---|---|---|
| I | 1.1 | 0.7 | 9.4 | 1.4 | 9.1 | 9.7 | 2.9 | 34.3 |
| II | 1.0 | 0.4 | 6.9 | 1.4 | 6.6 | 6.9 | 2.6 | 25.8 |
| III | 0.9 | 0.6 | 5.1 | 1.1 | 4.0 | 4.6 | 1.9 | 18.2 |
| IV | 1.1 | 0.6 | 8.3 | 1.4 | 6.3 | 6.6 | 2.3 | 26.6 |
All characters as in male except: cephalic region scarcely differentiated, much less marked than in male. Fovea visible and thoracic grooves not clearly marked. Epigynum and vulva. Epigynum wide and convex (Figs 14–16). Median septum wide and prominent, caudally projected by a bell-shaped flap. Vulva (Fig. 17, drawing is based on specimen 3812-150) with well-developed lateral pockets divided by a ventral fold in two, the lateral and medial part. Measurements: PL: 2.9; PW: 2.9; OL: 3.4; OW: 2.6; total length = 6.3. Leg I>leg IV>leg II> leg III.
Nesticus baeticus sp. n., female. 14 epigynum (1530) ventral view 15 ditto, caudal view 16 epigynum (1619) ventral view 17 vulva ventral view. Abbreviations: CO copulatory orifice ID + FD insemination duct + fertilization duct MS median septum S spermathecae VPL vulval pocket lateral VPM vulval pocket medial. Scale bar in μm.
| Leg | coxa | troc. | femur | patella | tibia | meta. | tarsus | total |
|---|---|---|---|---|---|---|---|---|
| I | 1.1 | 0.7 | 8.9 | 1.1 | 8.9 | 8.6 | 3.4 | 32.7 |
| II | 1.0 | 0.6 | 8.0 | 1.1 | 6.0 | 6.3 | 1.4 | 24.4 |
| III | 0.9 | 0.4 | 6.3 | 1.0 | 3.4 | 4.0 | 1.7 | 17.7 |
| IV | 1.3 | 0.9 | 7.4 | 1.0 | 6.0 | 6.3 | 1.7 | 24.6 |
Nesticus baeticus sp. n. inhabits the karst landscapes of the high part of the Cazorla, Segura and Las Villas Natural Park (NE Jaén, Spain) where it has been found in 8 caves. Most of the material studied comes from the area surrounding the Tranco’s Reservoir, in Hornos, Jaén. The area is calcareous, lush and quite humid, with numerous, medium-sized caves, both horizontal and vertical. The specimens were generally located within the first few meters of the dark zone, their presence reaching towards the cave interiors, which were sampled more intensively.
Specimens and sequences, with corresponding Genbank accession numbers, analyzed in the present study, are listed in Appendix 1. Alignments of two mitochondrial genes and gap scores as presence/absence were merged resulting in a combined data matrix of 950 characters (cox1=472, rrnL=456 and 22 gap characters). Uncorrected cox1 genetic divergences among terminal taxa are summarized in Appendix 2. Parsimony analyses of the combined data matrix yielded a single most-parsimonious tree of 705 steps (CI = 73 and RI = 62) (Fig. 1).
The results show that the Iberian species do not constitute a monophyletic group. Nesticus luquei, Nesticus lusitanicus and Nesticus baeticus sp. n. form a clade with high bootstrap support (99%), while Nesticus cellulanus is nested within a clade that also includes Nesticus ionescui, from Romania, and Nesticus eremita, from Croatia. Nesticus obcaecatus is the sister group of the remaining species of the ingroup. This topology, along the high genetic divergences observed between Nesticus obcaecatus, Nesticus cellulanus and the remaining Iberian species suggest the existence of tree independent colonization to the Iberian Peninsula. Preliminary results of a more extensive phylogenetic analysis, including almost all the Mediterranean species of Nesticidae (our work in progress), support this hypothesis.
The morphology of both the male and female copulatory organs of this Iberian group of species (Nesticus luquei, Nesticus lusitanicus, Nesticus murgis and Nesticus baeticus sp. n.) shows important differences as compared to Nesticus cellulanus, the type species of the genus Nesticus, as well as to the Carpathonesticus species. Thus, the absence of paracymbial ramification, the shape and size of the median apophysis, plus clear differences in size and arrangement of the TTA processes constitute the major differences in the males. The number of spermathecae is the most noticeable character in the females.
With regards to Nesticus obcaecatus, significant differences in the shape and structure of the paracymbium, the median apophysis and the TTA (see
Nesticus obcaecatus shows highly troglomorphic characters, such as complete depigmentation, reduction of the eye size and their number (only six eyes), and is known from a single cave. These data alongside its phylogenetic uniqueness (basal position and a deep genetic distance from other Iberian congeners) suggest that this species may be considered a relict representative of an old colonization to Iberia, and should be a candidate for protection.
Climatic relict hypothesis assume that adaptation and
speciation to caves are mainly driven by climatic factors. The
Pleistocene glacial cycles has been identified as de main driver of the
evolution of cave-dwelling fauna in the Paleartic (
We would like to thank representatives of the caving groups who gave us support: Grupo de espeleología de Villacarrillo (GEV) and Grupo de Espeleología de Priego (GESP). Dr. Manuel Baena and Toni Pérez Fernández are thanked for providing us with most of the studied specimens, guiding us to some caves and for useful comments regarding the new species. We also want to thank Dr. M.A. Arnedo and Dr. S. Carranza for their helpful comments about the first draft. The Spanish Ministry of Education and Science grants CGL2004-05771 and CGL2006-13374/BOS supported this research. Alberto Lopez was supported by a grant (BES-2005-9591) from the Ministerio de Ciencia y Tecnología, Spain
Species included in the cladistic analysis and GenBank accession numbers for the cox1 and rrnL. All accession numbers starting with EU are new sequences obtained in the present study.
| species | Locality | cox1 | rrnL |
|---|---|---|---|
| Nesticus obcaecatus | Cueva del Molino de Aso, Valle de Añisclo, Huesca, Spain | EU746428 | EU746437 |
| Nesticus luquei | Cueva de la Picona, San Pedro de Carmona, Cabuerniga, Cantabria, Spain | EU746430 | EU746439 |
| Nesticus lusitanicus | Algar de Marradinhas II Concelho de Alcanena, Portugal | EU746429 | EU746438 |
| Nesticus baeticus | Sima Irene, Hornos, Jaén, Spain | EU746431 | EU746440 |
| Nesticus baeticus | Sima del Campamento, Hornos, Jaén, Spain | EU746432 | EU746441 |
| Nesticus baeticus | Sima de los Alhaurinos, Hornos, Jaén, Spain | EU746433 | EU746442 |
| Nesticus ionescui | Pestera Tismana, Tismana, Romania | EU746434 | EU746443 |
| Nesticus cellulanus | Manantiales Monte Castro, Sueras, Castellón, Spain. | EU746435 | EU746444 |
| Nuesticus eremita | Cave Pishurka (=Paganetijeva Pécina), Korcula Is., Croatia. | EU746436 | EU746445 |
| “Nesticus” X130 | China | AY231024 | AY230941 |
Uncorrected genetic distances of cox 1 gene between terminal taxa analyzed in the present study. Numbers on terminals refers to different localities (see material examined)
| obcaeca | lusitan | luquei | baet5014 | baet3860 | baet3812 | ionescui | cellula | eremita | |
|---|---|---|---|---|---|---|---|---|---|
| lusitanicus | 0.173 | ||||||||
| luquei | 0.180 | 0.143 | |||||||
| baeticus5014 | 0.197 | 0.144 | 0.153 | ||||||
| baeticus3860 | 0.199 | 0.143 | 0.155 | 0.002 | |||||
| baeticus3812 | 0.199 | 0.143 | 0.155 | 0.002 | 0.000 | ||||
| ionescui | 0.159 | 0.164 | 0.170 | 0.160 | 0.161 | 0.161 | |||
| cellulanus | 0.153 | 0.168 | 0.176 | 0.168 | 0.169 | 0.169 | 0.110 | ||
| eremita | 0.159 | 0.155 | 0.161 | 0.153 | 0.151 | 0.151 | 0.115 | 0.117 | |
| NesticusX130 | 0.169 | 0.193 | 0.195 | 0.204 | 0.206 | 0.206 | 0.174 | 0.184 | 0.170 |