Research Article |
|
Corresponding author: Amelie Höcherl ( amelie.hoecherl@gmail.com ) Academic editor: Erinn Fagan-Jeffries
© 2024 Amelie Höcherl, Mark R. Shaw, Caroline Boudreault, Dominik Rabl, Gerhard Haszprunar, Michael J. Raupach, Stefan Schmidt, Viktor Baranov, José Fernández-Triana.
This is an open access article distributed under the terms of the CC0 Public Domain Dedication.
Citation:
Höcherl A, Shaw MR, Boudreault C, Rabl D, Haszprunar G, Raupach MJ, Schmidt S, Baranov V, Fernández-Triana J (2024) Scratching the tip of the iceberg: integrative taxonomy reveals 30 new species records of Microgastrinae (Braconidae) parasitoid wasps for Germany, including new Holarctic distributions. ZooKeys 1188: 305-386. https://doi.org/10.3897/zookeys.1188.112516
|
Abstract
Substantial parts of the European and German insect fauna still remain largely unexplored, the so-called “dark taxa”. In particular, midges (Diptera) and parasitoid wasps (Hymenoptera) are abundant and species-rich throughout Europe, yet are often neglected in biodiversity research. One such dark taxon is Microgastrinae wasps (Hymenoptera: Braconidae), a group of parasitoids of lepidopteran caterpillars with 252 species reported in Germany so far. As part of the German Barcode of Life Project GBOL III: Dark Taxa, reverse DNA barcoding and integrative taxonomic approaches were used to shed some light on the German Fauna of Microgastrinae wasps. In our workflow, DNA barcoding was used for molecular clustering of our specimens in a first step, morphological examination of the voucher specimens in a second step, and host data compared in a third step. Here, 30 species are reported for the first time in Germany, adding more than 10% to the known German fauna. Information for four species is provided in a new Holarctic context, reporting them for the Nearctic or, respectively, Palaearctic region, and 26 additional country records are added from sequenced material available in the collections accessible to us. Molecular clusters that show signs of discrepancies are discussed. Results show that we are just scratching the tip of the iceberg of the unexplored Microgastrinae diversity in Germany.
Key words
Dark taxa, DNA barcoding, faunistics, host-parasitoid associations, morphology, parasitoid biology
Introduction
With approximately 105,000 insect species documented (
However, substantial parts of the German insect fauna remain largely unexplored: the so-called “dark taxa” (
As part of the German Barcode of Life Initiative, GBOL III: Dark Taxa project, we aim to change this situation by applying a reverse DNA barcoding and integrative taxonomic approach. In contrast to traditional DNA barcoding workflows, we first performed molecular analyses, tested these results via morphological comparison in a second step, and compared available host data in a third step. Our molecular work relies on DNA barcoding (
In the last two decades, DNA barcoding and integrative taxonomy have revolutionised the study of Microgastrinae parasitoid wasps (e.g.,
Materials and methods
Specimens were collected in southern Germany (Fig.
Molecular workflow
We manually size-fractioned Malaise trap bulk samples by sieving and then sorted into first-glance morphotypes, of which subsamples were chosen for sequencing. We used legs as tissue samples, depending on size of the specimen one to three legs. COI-sequencing was done at the CCDB (Canadian Centre for DNA Barcoding) using their at-the-time standard sequencing protocols and primers, which can be reviewed for each sequence in the BOLD database (www.boldsystems.org). Sequences were analysed and clustered using the BOLD workbench and database (
Morphological workflow
We chemically dried our voucher specimens from Germany using a modified Hexamethyldisilazane (HMDS) protocol (
Biology (host information)
We checked original descriptions for collecting any host information related to type material. The supposed host data compiled by literature abstraction, such as in the database Taxapad 2016 (
Additional information, abbreviations, and terminology
New information is marked by an asterisk (*). All original descriptions of the species we report here are cited in the References section. Specimens were photographed using a Keyence VHX-6000 digital microscope and panorama stacks were computed using the built-in software of the microscope. Subsequent processing and construction of image plates and figures was done using Photoshop and Inkscape. Maps were done using QGIS and Inkscape. Voucher codes that are referred to in the notes and material examined sections refer to the “SampleID” in BOLD, more information about these specimens can be retrieved from the supplementary material or from BOLD. We have, however, added MS, MRS, or MRS_JFT voucher codes for barcoded specimens housed in
Results
Species recorded for Germany and other regions for the first time
Apanteles galleriae
Material examined
Germany: Baden-Württemberg, Malsch, Hansjakobstr. 7, Urban Garden, 48.884, 8.32, 120 m, Malaise trap, 27.ix.2020, leg. D. Doczkal,
Geographical distribution
AFR, AUS, NEA, NEO, OTL, PAL.
AFR- Mauritius, Réunion; AUS- Hawaiian Islands, New Zealand; NEA- Canada (BC), United States (GA, NC, OH, SC); NEO- Argentina, Brazil (SP); OTL- China (FJ, GD, GX, GZ, HI, HN, JX, TW, ZJ), India, Pakistan; PAL- Armenia, Bulgaria, France, Greece, Germany*, Hungary, Iran, Italy, Japan, Malta, Romania, Russia (PRI), Spain, Turkey, United Kingdom.
Molecular data
BIN: BOLD:AAG1400.
Host information
Pyralidae: type reared from Galleria mellonella (Linnaeus, 1758) (
Notes
Specimens in BIN BOLD:AAG1400 morphologically identified as Apanteles galleriae were reared from both Galleria mellonella (CNCHYM45392=MRS_JFT 0107 ex. beehive with that species) and Achroia grisella (MRS_JFT0380). Additional host species recorded for non-type specimens are based on literature (e.g.,
Apanteles kubensis
Material examined
Germany: Baden-Württemberg: Malsch, Hansjakobstr. 7, Urban Garden, 48.884, 8.32, 120 m, Malaise trap, 16.viii.2020, leg. D. Doczkal,
Geographical distribution
PAL.
PAL- Azerbaijan, Germany*, Hungary, Iran, Korea, Moldova, Mongolia, Russia (NC, S), Turkey, Ukraine*.
Molecular data
BIN: BOLD:AAH1340.
Host information
Host of type unknown (
Notes
The German specimens were compared with a specimen from Ukraine (CNCHYM 00136=CNC280641) which had been identified by Kotenko in 1981 and donated to the
Choeras ciscaucasicus
Material examined
Czech Republic: South Moravia, Obora Soutok, Lanzhot, 48.69, 16.945, 165 m, ex. Sterrhopterix fusca, 09.v.2014, leg. P. Drozd, BC-
Geographical distribution
PAL.
PAL- Czech Republic*, Germany*, Lithuania, Russia (AD, PRI).
Molecular data
BIN: BOLD:ACU3996.
Host information
Host of type unknown; also Psychidae*: Sterrhopterix fusca* (Haworth, 1809).
Notes
This species is morphologically very distinct from all other Palaearctic species of Choeras and can be identified by the combination of the following characters: T1 strongly narrowing; ovipositor sheaths short, ~ ½ length of metatibia; propodeum smooth and shiny; T1 and T2 smooth, with only slight wrinkles on the posterior half of T1. Our German specimens keyed out as Choeras ciscaucasicus in all keys that we used (
Choeras gnarus
Material examined
Germany: Baden-Württemberg: Gaggenau, Michelbach, 48.821, 8.388, 340 m, Malaise trap, 9.vii.2011, leg. D. Doczkal,
Geographical distribution
PAL.
PAL- Belarus, Germany*, Moldova*, Russia (NC, C), Sweden*, Ukraine.
Molecular data
BIN: BOLD:AAU6216.
Host information
Unknown.
Notes
The German specimens were compared with a specimen from Moldova (CNCHYM 00280) which had been identified by Kotenko in 1986 and donated to the
Cotesia coryphe
Material examined
Austria: Hinterreit, Gemeinde Großgmain, 47.75, 12.946, 493 m, ex. Hemaris fuciformis, 22.vi.2021, leg. W. Langer,
Geographical distribution
PAL.
PAL: Austria*, Germany*, Netherlands*, United Kingdom.
Molecular data
BIN: partially BOLD:AAA7143.
Host information
Sphingidae: type reared from Hemaris fuciformis (Linnaeus, 1758); also Hemaris tityus* (Linnaeus, 1758).
Notes
The German specimens were identified using
Cotesia eunomiae
Material examined
Belgium: Luxembourg, Pisserotte, ex. Boloria eunomia, vi.2004, leg. J. Choutt, individuals from separate gregarious broods, MS 106; MS 107; MS 108; Finland: Janakkala, ex. Boloria eunomia, 14.vi.1992, leg. M. R. Shaw, MRS-JFT 0655; MRS-JFT 0656; France: Pyrénées-Orientales, Porte, ex. Boloria eunomia, 30.v.2001, leg. T. Lafranchis, MRS_JFT 0118; Germany: Bavaria: Rhön, Hausen, Kleines Moor, 50.487, 10.039, 890 m, Malaise trap, 11.vii.2018, leg. D. Doczkal,
Geographical distribution
PAL.
PAL- Belgium, Finland*, France*, Germany*.
Molecular data
BIN: BOLD:AAV9098.
Host information
Nymphalidae: type reared from Boloria eunomia (Esper, 1800).
Notes
Our specimen from Germany matches the original description and clusters very closely (max. p-dist 0.34%) with six specimens (MRS-JFT 0655, MRS-JFT 0656, MS 106, MS 107, MS 108, MRS_JFT0118) reared from Boloria eunomia, the host of the holotype (
Cotesia inducta
Material examined
Germany: Bavaria: Oberstdorf, Oytal, rubble cone east of Gleitweg, 47.389, 10.348, 1200 m, Malaise trap, 16.vi.2014, leg. D. Doczkal, S. Schmidt, J. Voith,
Geographical distribution
PAL.
PAL- Bulgaria, Germany*, Hungary, Ireland, Israel, Korea, Moldova, Russia (KDA, PRI), Slovakia, Spain, Turkey, Ukraine, United Kingdom, Uzbekistan.
Molecular data
BIN: BOLD:AAV9096.
Host information
Host of type unknown; also Lycaenidae: Callophrys avis Chapman, 1909, Celastrina argiolus (Linnaeus, 1758), Glaucopsyche melanops (Boisduval, 1828), Leptotes pirithous* (Linnaeus, 1767), Satyrium w-album (Knoch, 1782), Tomares ballus (Fabricius, 1787).
Notes
German specimens were compared with the description and keys in
Cotesia mendicae
Material examined
Austria: Lower Austria, Raglitz, ex. Phragmatobia fuliginosa, 06.viii.2006, leg. J. Connell, MS 055; Germany: Bavaria: Balderschwang, Leiterberg, 47.489, 10.088, 1600 m, Malaise trap, 21.ix.2017, leg. D. Doczkal, J. Voith,
Geographical distribution
PAL.
PAL: Austria*, Germany*, Kazakhstan, Russia (VOR).
Molecular data
BIN: partially BOLD:AAA7143.
Host information
Host of (para-)type Diaphora mendica (Clerck, 1759); also Erebidae: Phragmatobia fuliginosa* (Linnaeus, 1758).
Notes
Specimens were compared with the information provided in
Cotesia risilis
Material examined
Finland: Uusimaa: Helsinki, Kaisaniemi Botanic Garden, 60.175700, 24.944700, Malaise trap, 29.viii-5.ix.2018, leg. J. Paukkunen, CNC1182785; France: Var, Callas, ex. Satyrium w-album, 5.v.2015, leg. P. Kan, B. Kan, MRS-JFT 0604; Germany: Baden-Württemberg: Malsch, Luderbusch, 48.913, 8.332, 117 m, Malaise trap, 26.vii.2020, leg. D. Doczkal, K. Grabow,
Geographical distribution
PAL.
PAL- Finland*, France, Germany*, Greece, Hungary, Iran, Italy, Mongolia, Montenegro, Netherlands, Romania, Slovakia, Spain, Sweden, Turkey, United Kingdom.
Molecular data
BIN: partially BOLD:AAA6099.
Host information
Pieridae: type reared from Gonepteryx rhamni (Linnaeus, 1758); also Lycaenidae: Satyrium w-album (Knoch, 1782).
Notes
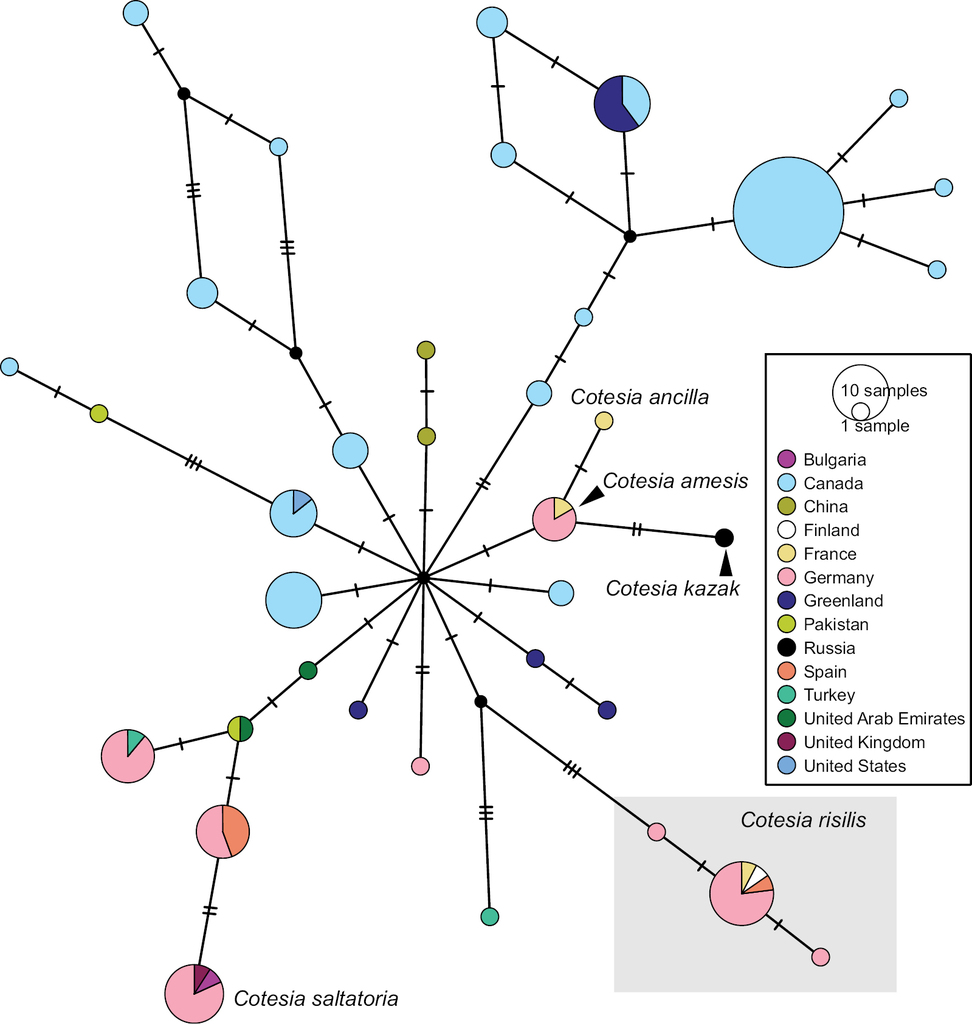
Barcoding cluster BIN BOLD:AAA6099 currently includes 155 sequences which have been assigned seven species names: Cotesia risilis, C. saltatoria (+ C. cf. saltatoria), C. amesis, C. ancilla, C. cyaniridis, C. kazak, and C. flaviconchae. The barcoding cluster also includes a large number of specimens from the Nearctic currently labelled as “Cotesia jft09”. Many of these species names are represented by reared material and, based on morphology and biology, clearly represent different species. They are all parasitoids of Pieridae and Lycaenidae with the exception of C. kazak (which might have been a misidentification) and possibly C. flaviconchae.
We performed a Haplotype Network analysis including most sequences in this BIN (excluding specimens CNCHYM00406 (C. cyaniridis) and DQ538819 (C. flaviconchae) due to the sequences being significantly shorter than the other available sequences and with incomplete collection data). Our German material clusters in eight different haplotypes (Fig.
TCS haplotype network of BIN BOLD:AAA6099, sequence length for analysis: 504 bp. The haplotypes morphologically identified as Cotesia risilis as part of this project are marked by a box. Each hatch mark in the network represents a single mutational change; small black dots at nodes indicate missing haplotypes. The diameter of the circles is proportional to the number of haplotypes sampled and the countries are colour-coded. The aligned sequences and traits can be reviewed in Suppl. materials
Cotesia selenevora
Material examined
Belgium: Luxembourg: Libin, ex. Boloria selene, 01.vi.2008, leg. J. Choutt, MRS-Cot-cal [paratype]; Germany: Bavaria: Lkr. Kelheim, Siegenburg, Bombodrom, 48.755, 11.791, 411 m, Malaise trap, 8.ix.2017, leg. D. Doczkal, J. Voith,
Geographical distribution
PAL.
PAL- Belgium, Finland, Germany*, Sweden.
Molecular data
BIN: partially BOLD:AAA7143.
Host information
Nymphalidae: type reared from Boloria selene (Denis & Schiffermüller, 1775).
Notes
Our sequences of this species were formerly part of BIN BOLD:AAA9381 and merged into BOLD:AAA7143 in February 2023. This BIN includes many clearly different species of Cotesia; see discussion below about this “megaBIN”. ASAP clustering resolves this species as a single cluster, including a specimen reared from Boloria selene (Denis & Schiffermüller, 1775) and part of the type series (MRS-Cot-cal=MS 075). Our German specimen matches the barcode of the paratype 100% and also matches the species morphologically, based on
Cotesia subordinaria
Material examined
Germany: Bavaria: Plattling, Isarmündung, renat. gravel bar, 48.781, 12.906, 317 m, Malaise trap, 30.vi.2021, leg. GBOL3, R. Albrecht,
Geographical distribution
PAL.
PAL: Azerbaijan, Georgia, Germany*, Netherlands, Poland*, Russia (NC), United Kingdom.
Molecular data
BIN: partially BOLD:ACO3220.
Host information
Host of type unknown; also Erebidae: Rivula sericealis (Scopoli, 1763).
Notes
German specimens were identified using the keys of
Deuterixys plugarui
Material examined
Georgia: Kakheti: Lagodekhi reserve, Mt Kudigora, 41.855850, 46.292733, 847 m, Malaise trap, 25.viii-4.ix.2014, leg. G. Japoshvili, CNC506818; Germany: Bavaria: Bad Windsheim, Rappenau, 49.482, 10.468, 382 m, canopy fogging, 9.vii.2020, leg. B. Leroy,
Geographical distribution
PAL.
PAL- Georgia*, Germany*, Hungary, Moldova, Netherlands*, Russia (S), Ukraine, United Kingdom.
Molecular data
BIN: BOLD:AEJ7518.
Host information
Bucculatricidae: type reared from Bucculatrix ulmella Zeller, 1848.
Notes
German specimens were identified using the keys and information provided in
Dolichogenidea cerialis
Material examined
Germany: Baden-Württemberg: Malsch, Hansjakobstr. 7, Urban Garden, 48.884, 8.32, 120 m, Malaise trap, 13.ix.2020, leg. D. Doczkal,
Geographical distribution
PAL.
PAL- Bulgaria, Germany*, Hungary, Israel, Italy, Kazakhstan, Russia (S), Spain, Ukraine*.
Molecular data
BIN: BOLD:AAZ9570.
Host information
Host of type unknown.
Notes
The sequences of our German specimens match that of a specimen from Ukraine, identified by Kotenko and stored in the
Dolichogenidea cheles
Material examined
Germany: Baden-Württemberg: Malsch, Hansjakobstr. 7, Urban Garden, 48.884, 8.32, 120 m, Malaise trap, 19.vii.2020, leg. D. Doczkal,
Geographical distribution
PAL.
PAL- Finland, Germany*, Hungary, Poland, Russia (NW), Sweden, Turkey.
Molecular data
BIN: BOLD:ACQ9527.
Host information
Host of type unknown. Other host associations in need of verification.
Notes
German specimens were compared with the original description (
Dolichogenidea coleophorae
Material examined
Canada: Newfoundland and Labrador: 2 miles west of Gambo, 48.789478, -54.261043, 19.vi.1975, leg. A. G. Raske, J. D. Rowe, ex. Coleophora serratella, CNCHYM 01020; Gambo, 48.786481, -54.215467, 27.vii.1985, CNCHYM 01019; Germany: Baden-Württemberg: Malsch, Luderbusch, 48.912, 8.332, 112 m, Malaise trap, 24.v.2020, leg. D. Doczkal, K. Grabow,
Geographical distribution
NEA, PAL.
NEA: Canada (NL); PAL: Azerbaijan, Finland, Germany*, Hungary, Poland, Romania, Russia (KHA, VOR, YAR), Slovakia, Switzerland, Tajikistan, Tunisia, Turkey, United Kingdom, Uzbekistan.
Molecular data
Host information
Coleophoridae: type reared from Coleophora serratella (Linnaeus, 1761); also possibly Coleophora ? ibipennella Zeller, 1849; Coleophora ? lusciniaepennella (Treitschke, 1833); Coleophora ? obducta (Meyrick, 1931); Coleophora ? tadzhikiella Danilevsky, 1955.
Notes
Several Canadian (CNCHYM 01019, CNCHYM 01020) and European specimens at the
Glyptapanteles indiensis
Material examined
Czech Republic: South Moravia: Obora Soutok, Lanžhot, 48.69, 16.945, 165 m, 14.v.2013, leg. P. Drozd, BC-
Geographical distribution
NEA, OTL, PAL.
NEA: USA (PA), OTL: India, PAL: Czech Republic*, Germany*, Japan*.
Molecular data
Host information
Erebidae: type reared from Lymantria obfuscata (Marsh, 1979); also Lymantria dispar (Linnaeus, 1758); Geometridae*: Operophtera brumata* (Linnaeus, 1758).
Notes
We record G. indiensis for the first time in the Palaearctic region, based on specimens from Germany, Japan and Czech Republic. This species is morphologically similar, especially in habitus, to several Glyptapanteles species. Our identification was therefore based on a careful study (detailed below) which included a combination of morphology (see Figs
Neighbour-joining topology of the COI barcoding region of Glyptapanteles indiensis, G. popovi and morphologically similar species, based on Kimura 2-parameter distances. Numbers next to nodes represent non-parametric bootstrap values > 90% (1,000 replicates). The aligned sequences and N-J topology can be reviewed in Suppl. materials
Other species of Glyptapanteles parasitising Lymantria dispar are Glyptapanteles liparidis (BOLD:AAV2164, including several reared specimens from this host such as WAM 0445=MRS_JFT 0028, BC-
The known hosts of G. mygdonia include Operophtera brumata and the multiple hosts recorded for G. vitripennis in the literature (many of them likely incorrect) include both O. brumata and L. dispar. The many available sequences of G. mygdonia (BOLD:AAU5027) and G. vitripennis (BOLD:AAA7148) are also very distinctive and far apart from those of indiensis [The sequences of G. vitripennis and G. liparidis are relatively very close (2.13% p-distance); furthermore, G. vitripennis seems to include a complex of species that remains unresolved, but that is beyond the scope of the present paper]. There are also morphological differences between these species and G. indiensis (
The last species we compared to G. indiensis was G. popovi, which is much less understood. Until now, G. popovi was only known from Turkey and Turkmenistan (
Glyptapanteles popovi
Material examined
Armenia: [translated and transcribed from Russian] Khosrov Forest State Reserve, Vediiskii reservoir sector (of reserve), montane forest, 30.vi.1981, CNCHYM 01335; Germany: Bavaria: Garmisch-Partenkirchen, Zugspitze, Platt, 47.406, 11.009, 1965 m, Malaise trap, 11.ix.2018, leg. D. Doczkal, J. Voith,
Geographical distribution
PAL.
PAL- Armenia*, Germany*, Turkey, Turkmenistan.
Molecular data
BIN: BOLD:AEJ4298.
Host information
Host unknown.
Notes
Our specimens were identified morphologically using keys and information in
Illidops cloelia
Material examined
Germany: Bavaria: Garmisch-Partenkirchen, Zugspitze, Platt, 47.407, 11.008, 2005 m, Malaise trap, 2.viii.2018, leg. D. Doczkal, J. Voith,
Geographical distribution
PAL.
PAL- Austria, Germany*, Hungary, Korea, Russia (E, NC), Slovakia, Switzerland, Tajikistan, former Yugoslavia.
Molecular data
BIN: BOLD:AEO8223.
Host information
Host unknown.
Notes
The German specimen was identified by comparison with the keys and details from the works of
Illidops splendidus
Material examined
Germany: Bavaria: Lkr. Kelheim Siegenburg, Bombodrom, 48.755, 11.791, 411 m, Malaise trap, 26.v.2017, leg. D. Doczkal, J. Voith,
Geographical distribution
PAL.
PAL- Germany*, Hungary, Russia (C).
Molecular data
BIN: BOLD:AEJ7519.
Host information
Host unknown.
Notes
The German specimen was identified by comparison with the keys of
Microgaster arctostaphylica
Material examined
Germany: Bavaria: Bad Tölz, forest close to Isarstausee, 47.77, 11.547, 652 m, Malaise trap, 16.vii.2019, leg. J. Müller,
Geographical distribution
PAL.
PAL- Germany*, Sweden*, United Kingdom.
Molecular data
BIN: BOLD:AAH1039.
Host information
Tortricidae: type reared from Argyroploce arbutella (Linnaeus, 1758); also Epinotia nemorivaga (Tengström, 1848), Stictea mygindiana (Denis & Schiffermüller, 1775).
Notes
German specimens were identified by comparison with the original description and specimens from that paper (
Microgaster caris
Material examined
Germany: Bavaria: Allgäu, Oberstdorf, Oytal Magerweide östlich Oytalhaus, 47.388, 10.344, 1056 m, 1.vi.2014, leg. D. Doczkal, S. Schmidt, J. Voith, BC-
Geographical distribution
PAL.
PAL- Austria, China (JL), Czech Republic, Germany*, Hungary, Russia (C, PR), Slovakia, Switzerland.
Molecular data
BIN: BOLD:ACN6851.
Host information
Host of type unknown. Other host associations in need of verification.
Notes
The German specimens were identified by comparison with the original description (
Microgaster nervosae
Material examined
Germany: Baden-Württemberg: Malsch, Luderbusch, 48.912, 8.332, 114 m, Malaise trap, 12.iv.2020, leg. D. Doczkal, K. Grabow,
Geographical distribution
PAL.
PAL- Germany*, United Kingdom.
Molecular data
BIN: BOLD:ACR4142.
Host information
Depressariidae: type reared from Agonopterix nervosa (Haworth, 1811); also Agonopterix umbellana (Fabricius, 1794).
Notes
This species was very recently described from Britain (
Microgaster nixalebion
Material examined
Austria: Lower Austria: Opponitz, ex. Patania ruralis, vi.2007, leg. J. Connell, MRS_JFT0934; Germany: Baden-Württemberg: Malsch, Hansjakobstr. 7, Urban Garden, 48.884, 8.32, 120 m, Malaise trap, 13.ix.2020, leg. D. Doczkal,
Geographical distribution
PAL.
PAL- Austria*, Belgium, France, Germany*, Greece, Serbia*, Spain*, United Kingdom.
Molecular data
BIN: BOLD:ABY6385.
Host information
Choerutidae: type reared from Anthophila fabriciana (Linnaeus, 1767); also Prochoreutis myllerana (Fabricius, 1794); Nymphalidae: Aglais urticae (Linnaeus, 1758), Vanessa atalanta (Linnaeus, 1758); Vanessa cardui* (Linnaeus, 1758); Pyralidae: Patania ruralis (Scopoli, 1763).
Notes
German specimens were identified by comparing with the information in the original description (
TCS haplotype network of BIN BOLD:ABY6385, sequence length for analysis: 392 bp to accommodate MRS_JFT0342=MARKB109-21 ex. Anthophila fabriciana from the United Kingdom. The haplotypes morphologically identified as Microgaster nixalebion as part of this project are marked by a grey background. Each hatch mark in the network represents a single mutational change; small black dots at nodes indicate missing haplotypes. The diameter of the circles is proportional to the number of haplotypes sampled (see legend). The aligned sequences and traits can be reviewed in Suppl. materials
Microgaster raschkiellae
Material examined
Germany: Bavaria: Rhön Hausen, Eisgraben, basalt block heap at forest edge, 50.503, 10.09, 735 m, Malaise trap, 23.vii.2018, leg. D. Doczkal,
Geographical distribution
NEA, PAL.
NEA: Canada (MB); PAL: Germany*, United Kingdom.
Molecular data
Host information
Momphidae: type reared from Mompha raschkiella (Zeller, 1839).
Notes
German specimens were identified by comparing to the original description (
TCS haplotype network of BIN BOLD:AAC9130, the haplotypes morphologically identified as Microgaster raschkiellae as part of this project are in cluster A and marked by a grey background. Each hatch mark in the network represents a single mutational change; small black dots at nodes indicate missing haplotypes. The diameter of the circles is proportional to the number of haplotypes sampled (see legend). The aligned sequences and traits can be reviewed in Suppl. materials
Microplitis coactus
Material examined
Canada: Newfoundland and Labrador: Saglek, Torngat Mountains NP, Base Camp south of park, 58.451, -62.798, 5 m, 01.viii.2014, leg. D. Whitaker, BIOUG18647-F03; Nunavut: Ellesmere Island, Hazen Camp, 81.816667, -71.300000, [date unknown, leg. unknown], CNC497575; Germany: Bavaria: Atzmannsberg, Hessenreuther and Atzmannsberger Forst, 49.825, 11.963, 550 m, Malaise trap, 11.vii.2019, leg. J. Müller,
Geographical distribution
NEA, PAL.
NEA- Canada (NL*, NU), Greenland; PAL- Germany*, Iceland.
Molecular data
BIN: BOLD:ACA4555.
Host information
Host of type unknown; also Noctuidae.
Notes
German specimens were identified by comparison with many specimens at the
Microplitis kewleyi
Material examined
Germany: Bavaria: Ammergebirge Halblech, Im Laich, gravel bar, 47.606, 10.841, 904 m, Malaise trap, 16.ix.2016, leg. D. Doczkal, J. Voith,
Geographical distribution
NEA, PAL.
NEA- Canada (AB, MB, NB, NL, NS, ON, PE, QC), United States (CA, DC, IA, MD, MI, NJ, NY, WI); PAL*- Germany*.
Molecular data
BIN: BOLD:AAB8493.
Host information
Noctuidae: type reared from Euxoa sp.; also Agrotis ipsilon (Hufnagel, 1766), Euxoa ochrogaster (Guenée, 1852), ?Pseudohermonassa bicarnea (Guenée, 1852).
Notes
German specimens were identified by comparison with many specimens in the
Microplitis naenia
Material examined
Czech Republic: South Moravia: Obora Soutok, Lanžhot, 48.69, 16.945, 165 m, ex. Orthosia cruda, 14.v.2013, leg. P. Drozd, BC-
Geographical distribution
PAL.
PAL- Czech Republic*, Germany*, Hungary, Russia (C, NW), Slovakia, Turkey, United Kingdom.
Molecular data
BINs: BOLD:ABV9098, BOLD:AEK2564.
Host information
Host of type unknown; also Noctuidae: Cosmia trapezina (Linnaeus, 1758), Conistra vaccinii (Linnaeus, 1761), Eupsilia transversa (Hufnagel, 1766), Orthosia cruda (Denis & Schiffermüller, 1775), Orthosia cerasi (Fabricius, 1775), Rileyiana fovea (Treitschke, 1825).
Notes
The morphology of our material (see Fig.
Pholetesor bedelliae
Material examined
Canada: New Brunswick: Fredericton, 45.963487, -66.6442, 9.vii.1970, leg. C. M. Yoshimoto, CNCHYM 03145 [paratype]; Germany: Bavaria: Bayreuth, Gemein, Trebgast, 49.989, 11.603, 348 m, Malaise trap, 11.vii.2019, leg. J. Müller,
Geographical distribution
AUS, NEA, NEO, PAL.
AUS: Hawaiian Islands; NEA: Canada (AB, BC, MB, NB, NS, ON, QC, SK), USA (AK, AZ, AR, CA, CT, DC, FL, IL, IA, KA, LA, MO, NJ, NY, OR, VA); NEO: Bermuda, Peru; PAL: Finland, Germany*.
Molecular data
BIN: BOLD:AAA9172.
Host information
Types reared from Bedellia sp. At least 21 host species within seven families of Lepidoptera have been recorded as hosts of this wasp species (
Notes
We compared our specimens to a paratype and many other Nearctic specimens stored at the
Protapanteles endemus
Material examined
France: Jura, Ounans, ex. Thyatira batis, 25.vii.2013, leg. M. R. Shaw, MRS_JFT0355; Germany: Baden-Württemberg: Malsch, Hansjakobstr. 7, Urban Garden, 48.884, 8.32, 120 m, Malaise trap, 30.viii.2020, leg. D. Doczkal,
Geographical distribution
PAL.
PAL: France, Germany*, Hungary, Kazakhstan, Poland*, Russia (ZAB, SPE), Switzerland, Ukraine, United Kingdom.
Molecular data
BIN: BOLD:AEI1558.
Host information
Geometridae: Type reared from Abraxas grossulariata (Linnaeus, 1758); also Drepanidae*: Thyatira batis* (Linnaeus, 1758).
Notes
The German specimens were identified based on information from the original description (
Rasivalva desueta
Material examined
Germany: Bavaria: Bayernwald National Park, Schöberg, ex. ?Eilema depressa, 16.vi.2015, leg. M. R. Shaw, MRS-JFT 0582; Berchtesgaden National Park, Königssee, Rinnkendlsteig, 47.555, 12.965, 750 m, Malaise trap, 21.viii.2017, leg. D. Doczkal, J. Voith,
Geographical distribution
PAL.
PAL- Germany*, Sweden*, Switzerland.
Molecular data
BIN: BOLD:ADE2589.
Host information
Host of type unknown. Other host associations in need of verification.
Notes
The non-reared German specimen was identified as Rasivalva desueta by comparing it with the detailed original description (
Additional species
Cotesia eulipis
Material examined
Canada: British Columbia: Winfield, 50.061, -119.431, [450m,] ex. Operophtera bruceata, 17.v.2001, leg. K. Deglow, CNCHYM 00330; CNCHYM 00331; Prince Edward Island: Brackley Beach, Prince Edward Island National Park, 46.431111, -63.216111, ex. Rheumaptera hastata, 10.viii.1940, leg. G.S. Walley, CNC1447813; Yukon: Top of the World Highway km 82, 64.09, -140.951, 19.vii.2006, leg. H. Goulet, C. Boudreault, HYM00001784; Finland: Lapland: Utsjoki, Kevo, ex. Operophtera brumata, 28.vi.2010, leg. K. Ruohomaki, MRS 0049; MRS_JFT0049; Utsjoki, Vetsikko, ex. Operophtera brumata, 28.vi.2010, leg. K. Ruohomaki, MRS 0050; Germany: Bavaria; St. Oswald, National Park Bayerischer Wald, 48.951, 13.422, 842 m, Malaise trap, 25.vii.2012, leg. G. Sellmayer, BIOUG07768-G08; Norway: Rogaland, Hana, ex. Operophtera brumata, 28.vi.2010, leg. K. Ruohomaki, MRS 0048; United Kingdom: Scotland: Argyll, Scotnish Farm, ex. Rheumaptera hastata, 25.iv.1990, leg. K. P. Bland, CNCHYM49288.
Geographical distribution
NEA, PAL.
NEA*- Canada* (BC, PE, YT); PAL- Bulgaria, Finland, Germany, Greece, Hungary, Norway*, Sweden, United Kingdom.
Molecular data
BIN: BOLD:ACZ1254.
Host information
Geometridae: type reared from Rheumaptera hastata (Linnaeus, 1758); also Operophtera bruceata* (Hulst, 1886), Operophtera brumata* (Linnaeus, 1758).
Notes
Barcoding cluster BOLD:ACZ1254 includes sequences of specimens from Finland, Germany, Norway, United Kingdom, and Canada. Specimens in this cluster from both sides of the Atlantic were reared from Rheumaptera hastata (the host of the holotype): from the United Kingdom (CNCHYM49288=MRS_JFT 0154) and from Canada (CNC1447813). Morphological examination resulted in some differences in leg colouration between Canadian and European specimens available to us (see Figs
Cotesia tetrica
Material examined
Austria: Lower Austria, Raglitz, ex. Aphantopus hyperantus, 04.v.2011, leg. J. Connell, MRS 0054; Germany: Bavaria: Allgäu, Oberstdorf, Oytal, Schochen, alpine meadow, 47.392, 10.37, 1930 m, 6.viii.2014, leg. D. Doczkal, S. Schmidt, J. Voith, BC-
Geographical distribution
PAL.
PAL- Austria*, Germany, Montenegro, Serbia, United Kingdom.
Molecular data
BIN: BOLD:AAV9103.
Host information
Host of type unknown; also Nymphalidae: Satyrinae*: Lasiommata megera (Linnaeus, 1767), Maniola jurtina (Linnaeus, 1758); Aphantopus hyperantus* (Linnaeus, 1758).
Notes
This species was previously recorded from Germany (
Diolcogaster claritibia
Material examined
Canada: Ontario: Ottawa, Central Experimental Farm, DBM Field Cage Trials, 45.389959, -75.711949, 23.vi.2010, leg. P. Mason, S. Girardoz, CNCHYM 01692; CNCHYM 01693; CNCHYM 01694; CYPRUS: Amathus, 21-iv-1966, leg. Mavromoustakis, CNCHYM 00892; FRANCE: Languedoc-Roussillon: Baillargues, Herault, 43.662, 4.014, 3-vi-1995, leg. P. Mason, CNCH1127; Bel Air; Herault, 43.639, 3.75, 5-vi-1995, leg. P. Mason, CNCH1126; Germany: Baden-Württemberg: Malsch, Hansjakobstr. 7, Urban Garden, 48.884, 8.32, 120 m, Malaise trap, 19.vii.2020, leg. D. Doczkal,
Geographical distribution
NEA, PAL.
NEA- Canada (AB, MB, ON); PAL- Afghanistan, Armenia, Austria, Azerbaijan, Belarus, Cyprus, Finland, France, Georgia, Germany, Greece, Hungary, Italy, Iran, Jordan, Kazakhstan, Lithuania, Macedonia, Moldova, Netherlands, Russia (ZAB, KDA), Spain, Syria, Tunisia, Turkey, Turkmenistan, Ukraine, former Yugoslavia.
Molecular data
BIN: BOLD:AAH1034, BOLD:AEV8838.
Host information
Host of type unknown; also Plutellidae: Plutella armoraciae Busck, 1912, Plutella xylostella (Linnaeus, 1758).
Notes
German specimens were identified based on the detailed species concept in
Microgaster procera
Material examined
Canada: Prince Edward Island: Near Georgetown; Georgetown, 46.417, -62.667, 8 m, 31.vii.2005, leg. M. Sharkey, WMIC 0244; WMIC 0245; Germany: Bavaria: Berchtesgaden National Park, Königssee, Rinnkendlsteig, 47.553, 12.964, 775 m, Malaise trap, 30.vii.2017, leg. D. Doczkal, J. Voith,
Geographical distribution
NEA, PAL.
NEA*- Canada* (PE); PAL- Austria, Finland, Germany, Hungary, Ireland, Mongolia, Netherlands, Poland, Romania, Russia (SPE), Spain, Ukraine.
Molecular data
BIN: BOLD:AAA9548.
Host information
Host of type unknown.
Notes
This is not a new record for Germany, but morphological examination of German material and assignment of the species name to a barcoding cluster allow us to record the species in the Nearctic for the first time. The Canadian specimens were compared to the European material and all sequences in this barcoding cluster have a maximum pairwise-distance of just 0.15%. The host information for this species must be considered as mostly unreliable; based on a recent discussion (
Discussion
DNA Barcoding
Processing 5364 specimens in this reversive DNA barcoding approach combined with integrative taxonomy enabled us to gather many new distributions as well as much biological data in a relatively short time frame. We were able to report 30 species for Germany for the first time, four species as occurring in the Holarctic for the first time and, by also taking into account material already determined in
We used the framework of the BOLD workbench and database as it already includes more than 65,000 sequences of Microgastrinae worldwide (www.boldsystems.org, accessed on 20 Aug 2023) and has been used as a main molecular analysis tool in Microgastrinae taxonomy in the past (
We investigated BINs that had high within-BIN maximum p-distances (> 2.2%) (Ratnasingham and Hebert 2013) and BINs with low p-distances to their Nearest Neighbour (< 2.2%) (
BOLD:AAA7143 is an even more extreme case of BIN-sharing, it includes four species of Cotesia treated in this paper: C. coryphe, C. mendicae, C. selenevora, and C. subordinaria. When starting our analyses in 2022, each of these species was part of a separate BIN but merged into this “megaBIN” in early 2023. In Table
P-distances and member counts of the BINs treated in this paper, retrieved on 8 Aug 2023. Single asterisks (*) mark each species name that was matched to a BIN for the first time, Double asterisks (**) mark each species that was sequenced for the first time. A BIN in parentheses shows that the BIN represents more than one species and cannot be used for molecular identification of the species. A BIN in brackets shows the previous BIN before February 2023. If the minimum NN p-distance is < 2.2% or within BIN maximum p-distance > 2.2%, then the within-BIN max. p-distance and minimum NN-p-distance are in bold font.
| Species names | BIN(s) | Members (BIN) | BIN-compliant members | within- BIN average p-dist | within-BIN max. p-dist | min. NN p-dist |
|---|---|---|---|---|---|---|
| Apanteles galleriae | BOLD:AAG1400 | 25 | 13 | 0.38% | 1.17% | 2.36% |
| Apanteles kubensis | BOLD:AAH1340 | 7 | 5 | 0.14% | 0.32% | 5.61% |
| Choeras ciscaucasicus* | BOLD:ACU3996 | 5 | 1 | 0.22% | 0.48% | 6.78% |
| Choeras gnarus* | BOLD:AAU6216 | 41 | 12 | 0.08% | 0.52% | 2.23% |
| Cotesia coryphe | (BOLD:AAA7143) [BOLD:ABY6805] | 1772 [25] | 859 [5] | 3.85% [0.16%] | 7.61% [0.71%] | 2.21% [1.50%] |
| Cotesia eulipis | BOLD:ACZ1254 | 16 | 4 | 0.29% | 0.59% | 1.18% |
| Cotesia eunomiae | BOLD:AAV9098 | 6 | 0 | 0.18% | 0.34% | 7.68% |
| Cotesia inducta | BOLD:AAV9096 | 4 | 1 | 0.22% | 0.34% | 2.48% |
| Cotesia mendicae | (BOLD:AAA7143) [BOLD:ABY8119] | 1772 [N/A] | 859 [N/A] | 3.85% [N/A] | 7.61% [N/A] | 2.21% [N/A] |
| Cotesia risilis | (BOLD:AAA6099) | 182 | 105 | 1.25% | 2.87% | 1.12% |
| Cotesia selenevora | (BOLD:AAA7143) [BOLD:AAA9381] | 1772 [23] | 859 [6] | 3.85% [0.62%] | 7.61% [2.84%] | 2.21% [1.40%] |
| Cotesia subordinaria | (BOLD:AAA7143) [BOLD:ACO3220] | 1772 [5] | 859 [0] | 3.85% [0.07%] | 7.61% [0.19%] | 2.21% [1.44%] |
| Cotesia tetrica | BOLD:AAV9103 | 25 | 17 | 0.29% | 0.66% | 2.84% |
| Deuterixys plugarui** | BOLD:AEJ7518 | 9 | 0 | 0.54% | 0.96% | 5.83% |
| Diolcogaster claritibia | BOLD:AAH1034 | 95 | 73 | 0.34% | 2.28% | 2.14% |
| BOLD:AEV8838 | 16 | 8 | 0.66% | 2.64% | 2.14% | |
| Dolichogenidea cerialis | BOLD:AAZ9570 | 8 | 5 | 0.28% | 0.49% | 1.28% |
| Dolichogenidea cheles* | BOLD:ACQ9527 | 10 | 5 | 0.57% | 1.42% | 3.33% |
| Dolichogenidea coleophorae** | BOLD:AEO8197 | 2 | 0 | 0.00% | 0.00% | 2.72% |
| Glyptapanteles indiensis | BOLD:ABY2372 | 16 | 8 | 0.31% | 0.81% | 3.52% |
| Glyptapanteles popovi | BOLD:AEJ4298 | 7 | 0 | 0.23% | 0.64% | 3.25% |
| Illidops cloelia** | BOLD:AEO8223 | 2 | 0 | 0.00% | 0.00% | 4.81% |
| Illidops splendidus** | BOLD:AEJ7519 | 1 | 0 | N/A | N/A | 6.58% |
| Microgaster arctostaphylica | BOLD:AAH1039 | 12 | 7 | 0.93% | 2.25% | 2.68% |
| Microgaster caris | BOLD:ACN6851 | 66 | 4 | 0.17% | 0.72% | 1.60% |
| Microgaster nervosae | BOLD:ACR4142 | 11 | 3 | 0.35% | 0.82% | 2.72% |
| Microgaster nixalebion | BOLD:ABY6385 | 62 | 12 | 0.20% | 1.48% | 0.97% |
| Microgaster procera* | BOLD:AAA9548 | 8 | 3 | 0.04% | 0.16% | 5.33% |
| Microgaster raschkiellae | BOLD:AAC9130 | 36 | 25 | 0.91% | 2.26% | 1.93% |
| Microplitis coactus | BOLD:ACA4555 | 6 | 4 | 0.58% | 0.96% | 3.57% |
| Microplitis kewleyi | BOLD:AAB8493 | 127 | 53 | 0.03% | 0.80% | 5.77% |
| Microplitis naenia | BOLD:ABV9098 | 18 | 17 | 0.35% | 1.28% | 1.70% |
| BOLD:AEK2564 | 9 | 2 | 0.36% | 0.64% | 1.70% | |
| Pholetesor bedelliae | BOLD:AAA9172 | 74 | 55 | 0.46% | 1.62% | 3.29% |
| Protapanteles endemus* | BOLD:AEI1558 | 2 | 1 | 0.17% | 0.17% | 7.64% |
| Rasivalva desueta* | BOLD:ADE2589 | 4 | 2 | 0.08% | 0.17% | 3.04% |
Overall, a more general observation from discussing integrative species concepts in our dataset is that the distances between barcoding clusters can vary substantially between genera. We were able to observe several cases of lumping (or BIN-sharing) in Cotesia with p-distances of ~ 1.0–1.5% between morphologically and biologically well-established species. We had to exclude some species from this paper because the situation turned out to be more complex than previously expected (e.g., Cotesia euchloevora Shaw, 2020 and Cotesia pilicornis (Thomson, 1895); see discussion of these two species in
So far, barcoding has worked well for the majority of Microgastrinae species and has been an essential tool in advancing the study of Microgastrinae diversity (
Fig.
Neighbour-joining topology of the barcoding region of our dataset of the analysed species based on Kimura 2-parameter distances. Triangles show the relative number of individuals sampled (height) and sequence divergence (width). Pale blue colouration indicates species associated with BINs that have a maximum within-BIN distance > 2.2%. Dark blue colouration indicates species associated with BINs that have a higher within-BIN p-distance compared to NN p-distance. Orange colouration indicates species maximum intraspecific distances > 2.2% (cases of “BIN-sharing”). Numbers next to nodes represent non-parametric bootstrap values > 90% (1,000 replicates). Sequences shorter than 400 bp were excluded from this analysis. The aligned sequences and N-J topology can be reviewed in Suppl. materials
Distribution in Europe and the Holarctic region
The known distribution in Europe of some species is significantly expanded: Choeras ciscaucasicus was previously only known from Russia and Lithuania; in this case the new record from Germany expands this species’ range to the western part of Europe. Microgaster arctostaphylica and Microgaster nervosae, two recently described species (
A main result of this study is that four species were recorded for the first time for the Palaearctic (Glyptapanteles indiensis, Microplitis kewleyi) and Nearctic regions (Cotesia eulipis, Microgaster procera). According to the supplementary material of the world checklist published in 2020, only 5% (56) of the 1,178 Holarctic species of Microgastrinae are shared between the Nearctic and Palaearctic regions. Of these 56 species, 29 have an exclusively Holarctic distribution (
Distribution maps for the four newly recorded Holarctic species. Coloured countries/states show the known distribution as in
Acknowledgments
We would like to acknowledge Wolfgang Langer, Ingrid Langer, and Pavel Drozd for reared specimens, and Benjamin Leroy, Dieter Doczkal, Johannes Voith, Jörg Müller, and the Swedish Malaise Trap Project for numerous bulk samples. The GBOL3-Team at
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
Funding was provided by the Bundesministerium für Bildung und Forschung (BMBF) of Germany, through the project “German Barcode of Life III: Dark Taxa” (FKZ 16LI1901C).
Author contributions
Conceptualization: AH, JLFT. Data curation: AH, JLFT, MRS, CB, DR. Formal analysis: AH, JLFT, MRS. Funding acquisition: SS. Investigation: AH, JLFT, MRS. Project administration: AH. Resources: SS. Supervision: JLFT, GH, MJJR, SS. Visualization: AH, CB. Writing – original draft: AH, JLFT. Writing – review and editing: AH, JLFT, MRS, VAB, CB, MJJR, SS, GH.
Author ORCIDs
Amelie Höcherl https://orcid.org/0009-0007-4211-7468
Mark R. Shaw https://orcid.org/0000-0002-6651-8801
Caroline Boudreault https://orcid.org/0000-0002-4511-2626
Dominik Rabl https://orcid.org/0000-0002-0613-7804
Michael J. Raupach https://orcid.org/0000-0001-8299-6697
Stefan Schmidt https://orcid.org/0000-0001-5751-8706
Viktor Baranov https://orcid.org/0000-0003-1893-3215
José Fernández-Triana https://orcid.org/0000-0003-0425-0309
Data availability
All of the data that support the findings of this study are available in the main text or Supplementary Information.
References
- Abdinbekova A (1969) New braconid wasps (Hymenoptera, Braconidae) in the fauna of Azerbaidjan. Doklady Akademii Nauk Azerbaidzhanskoi SSR 25: 72–77.
- Abdoli P, Talebi A, Farahani S, Fernandez-Triana JL (2019) Three new species of the genus Choeras Mason, 1981 (Hymenoptera: Braconidae, Microgastrinae) from Iran. Zootaxa 4545(1): 77–92. https://doi.org/10.11646/zootaxa.4545.1.4
- Abram PK, Thiessen J, Clarke P, Gillespie DR, Fernández-Triana JL, Bennett AMR, Gibson GAP, Huber JT, Mason PG, Landry J-F (2022) Natural History of Plutella armoraciae Busck, 1912, A Sympatric Congener of the Diamondback Moth, Plutella xylostella (L., 1758), in Southwestern Canada. Journal of the Lepidopterists Society 76(1): 25–39. https://doi.org/10.18473/lepi.76i1.a4
- Belokobylskij SA, Taeger A, van Achterberg C, Haeselbarth E, Riedel M (2003) Checklist of the Braconidae of Germany (Hymenoptera). Beiträge zur Entomologie = Contributions to Entomology 53: 341–435. https://doi.org/10.21248/contrib.entomol.53.2.341-435
- Brown BV (2005) Malaise trap catches and the crisis in neotropical dipterology. American Entomologist 51(3): 180–183. https://doi.org/10.1093/ae/51.3.180
- Brown BV, Borkent A, Adler PH, Amorim D de S, Barber K, Bickel D, Boucher S, Brooks SE, Burger J, Burington ZL, Capellari RS, Costa DNR, Cumming JM, Curler G, Dick CW, Epler JH, Fisher E, Gaimari SD, Gelhaus J, Grimaldi DA, Hash J, Hauser M, Hippa H, Ibáñez-Bernal S, Jaschhof M, Kameneva EP, Kerr PH, Korneyev V, Korytkowski CA, Kung G-A, Kvifte GM, Lonsdale O, Marshall SA, Mathis W, Michelsen V, Naglis S, Norrbom AL, Paiero S, Pape T, Pereira-Colavite A, Pollet M, Rochefort S, Rung A, Runyon JB, Savage J, Silva VC, Sinclair BJ, Skevington JH, Stireman III JO, Swann J, Thompson FC, Vilkamaa P, Wheeler T, Whitworth T, Wong M, Wood DM, Woodley N, Yau T, Zavortink TJ, Zumbado MA (2018) Comprehensive inventory of true flies (Diptera) at a tropical site. Communications Biology 1(1): 1–8. https://doi.org/10.1038/s42003-018-0022-x
- Capek M (1972) [Verzeichnis der aus Schädlingsinsekten erzogenen Parasiten. Teil V. – Die Brackwespen (Braconidae, Hymenoptera)]. Entomologické problémy 10: 125–140.
- Capek M, Hladil J, Sedivy J (1982) [Verzeichnis der aus verschiedenen Insekten erzogenen parasitschen Hymenopteren – Teil VI.]. Entomologické problémy 17: 325–371.
- Chimeno C, Hausmann A, Schmidt S, Raupach MJ, Doczkal D, Baranov V, Hübner J, Höcherl A, Albrecht R, Jaschhof M, Haszprunar G, Hebert PDN (2022) Peering into the darkness: DNA barcoding reveals surprisingly high diversity of unknown species of Diptera (Insecta) in Germany. Insects 13(1): 1–82. https://doi.org/10.3390/insects13010082
- Fagan-Jeffries EP, Cooper SJB, Bertozzi T, Bradford TM, Austin AD (2018) DNA barcoding of microgastrine parasitoid wasps (Hymenoptera: Braconidae) using high-throughput methods more than doubles the number of species known for Australia. Molecular Ecology Resources 18(5): 1132–1143. https://doi.org/10.1111/1755-0998.12904
- Fernandez-Triana JL (2022) Turbo taxonomy approaches: Lessons from the past and recommendations for the future based on the experience with Braconidae (Hymenoptera) parasitoid wasps. ZooKeys 1087: 199–220. https://doi.org/10.3897/zookeys.1087.76720
- Fernandez-Triana J, Shaw MR, Cardinal S, Mason P (2014a) Contributions to the study of the Holarctic fauna of Microgastrinae (Hymenoptera, Braconidae). I. Introduction and first results of transatlantic comparisons. Journal of Hymenoptera Research 37: 61–76. https://doi.org/10.3897/jhr.37.7186
- Fernandez-Triana J, Shaw MR, Cardinal S, Dosdall L, Mason P (2014b) First Nearctic record of Diolcogaster claritibia (Hymenoptera: Braconidae: Microgastrinae), with notes on taxonomic status and natural history. Canadian Entomologist 146(6): 609–620. https://doi.org/10.4039/tce.2014.16
- Fernandez-Triana J, Whitfield J, Rodriguez J, Smith MA, Janzen D, Hallwachs W, Hajibabaei M, Burns J, Solis A, Brown J, Cardinal S, Goulet H, Hebert P (2014c) Review of Apanteles sensu stricto (Hymenoptera, Braconidae, Microgastrinae) from Area de Conservación Guanacaste, northwestern Costa Rica, with keys to all described species from Mesoamerica. ZooKeys 383: 1–565. https://doi.org/10.3897/zookeys.383.6418
- Fernandez-Triana J, Shaw MR, Boudreault C, Beaudin M, Broad GR (2020) Annotated and illustrated world checklist of Microgastrinae parasitoid wasps (Hymenoptera, Braconidae). ZooKeys 920: 1–1089. https://doi.org/10.3897/zookeys.920.39128
- Fujie S, Japoshvili G, Fernandez-Triana J (2021) Review of the world species of Paroplitis Mason, 1981 (Hymenoptera, Braconidae, Microgastrinae), with description of three new species. Deutsche Entomologische Zeitschrift 68(1): 33–43. https://doi.org/10.3897/dez.68.59641
- Habel JC, Segerer A, Ulrich W, Torchyk O, Weisser WW, Schmitt T (2016) Butterfly community shifts over two centuries. Conservation Biology 30(4): 754–762. https://doi.org/10.1111/cobi.12656
- Hallmann CA, Sorg M, Jongejans E, Siepel H, Hofland N, Schwan H, Stenmans W, Müller A, Sumser H, Hörren T, Goulson D, de Kroon H (2017) More than 75 percent decline over 27 years in total flying insect biomass in protected areas. PLoS ONE 12(10): e0185809. https://doi.org/10.1371/journal.pone.0185809
- Hartop E, Srivathsan A, Ronquist F, Meier R (2022) Towards Large-Scale Integrative Taxonomy (LIT): Resolving the Data Conundrum for Dark Taxa. [Dayrat B (Ed. )] Systematic Biology 71(6): 1404–1422. [syac033] https://doi.org/10.1093/sysbio/syac033
- Hausmann A, Haszprunar G, Hebert PDN (2011a) DNA Barcoding the Geometrid Fauna of Bavaria (Lepidoptera): Successes, Surprises, and Questions. PLoS ONE 6(2): e17134. https://doi.org/10.1371/journal.pone.0017134
- Hausmann A, Haszprunar G, Segerer AH, Speidel W, Behounek G, Hebert PDN (2011b) Now DNA-barcoded: The butterflies and larger moths of Germany. Spixiana 34: 47–58.
- Hausmann A, Krogmann L, Peters R, Rduch V, Schmidt S (2020) GBOL III: Dark Taxa. iBOL Barcode Bulletin. https://ibol.org/barcodebulletin/research/gbol-iii-dark-taxa/ [August 25, 2022]
- Hawlitschek O, Morinière J, Lehmann GUC, Lehmann AW, Kropf M, Dunz A, Glaw F, Detcharoen M, Schmidt S, Hausmann A, Szucsich NU, Caetano-Wyler SA, Haszprunar G (2017) DNA barcoding of crickets, katydids and grasshoppers (Orthoptera) from Central Europe with focus on Austria, Germany and Switzerland. Molecular Ecology Resources 17(5): 1037–1053. https://doi.org/10.1111/1755-0998.12638
- Hebert PDN, Ratnasingham S, de Waard JR (2003a) Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London, Series B, Biological Sciences 270(suppl_1): S96–S99. https://doi.org/10.1098/rsbl.2003.0025
- Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003b) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London, Series B, Biological Sciences 270(1512): 313–321. https://doi.org/10.1098/rspb.2002.2218
- Hebert PDN, Bock DG, Prosser SWJ (2023) Interrogating 1000 insect genomes for NUMTs: A risk assessment for estimates of species richness. PLoS ONE 18(6): e0286620. https://doi.org/10.1371/journal.pone.0286620
- Hendrich L, Morinière J, Haszprunar G, Hebert PDN, Hausmann A, Köhler F, Balke M (2015) A comprehensive DNA barcode database for Central European beetles with a focus on Germany: Adding more than 3500 identified species to BOLD. Molecular Ecology Resources 15(4): 795–818. https://doi.org/10.1111/1755-0998.12354
- Heraty J, Hawks D (1998) Hexamethyldisilazane – A chemical alternative for drying Insects. Entomological News 109: 1–4.
- Jähnig SC, Baranov V, Altermatt F, Cranston P, Friedrichs-Manthey M, Geist J, He F, Heino J, Hering D, Hölker F, Jourdan J, Kalinkat G, Kiesel J, Leese F, Maasri A, Monaghan MT, Schäfer RB, Tockner K, Tonkin JD, Domisch S (2021) Revisiting global trends in freshwater insect biodiversity. WIREs. Water 8(2): e1506. https://doi.org/10.1002/wat2.1506
- Karlsson D, Hartop E, Forshage M, Jaschhof M, Ronquist F (2020) The swedish malaise trap project: a 15 year retrospective on a countrywide insect inventory. Biodiversity Data Journal 8: e47255. https://doi.org/10.3897/BDJ.8.e47255
- Kotenko A (2007) Microgastrinae. In: Lelej A (Ed.) Neuropteroidea, Mecoptera, Hymenoptera. Key to the Insects of Russia Far East. Dalnauka, Vladivostok, 134–192.
- Ku D, Belokobylskij S, Cha J (2001) Hymenoptera (Braconidae). Economic Insects of Korea 16. Insecta Koreana. Supplement 23. Junghaengsa, Suwon, 283 pp.
- Lazarević M, Stanković SS, van Achterberg C, Marczak D, Modic Š, Ilić Milošević M, Trajković A, Žikić V (2023) Morphological and genetic variability of Cotesia tibialis species complex (Hymenoptera: Braconidae: Microgastrinae). Zoologischer Anzeiger 302: 58–66. https://doi.org/10.1016/j.jcz.2022.10.007
- Leandro C, Jay-Robert P, Vergnes A (2017) Bias and perspectives in insect conservation: A European scale analysis. Biological Conservation 215: 213–224. https://doi.org/10.1016/j.biocon.2017.07.033
- Leigh JW, Bryant D (2015) PopART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution 6(9): 1110–1116. https://doi.org/10.1111/2041-210X.12410
- Liu Z, He J-H, Chen X-X, Gupta A (2020) The ater-group of the genus Apanteles Foerster (Hymenoptera, Braconidae, Microgastrinae) from China with the descriptions of forty-eight new species. Zootaxa 4807(1): 1–205. https://doi.org/10.11646/zootaxa.4807.1.1
- Lundbeck W (1896) Hymenoptera groenlandica. Videnskabelige Meddelelser fra den Naturhistoriske Forening i Kjøbenhaven 58: 220–251.
- Marczak P, Buszko J (1993) Braconid wasps (Hymenoptera, Braconidae) reared from mining Lepidoptera. Wiadomosci Entomologiczne 12: 259–272.
- Marsh PM (1979) The Braconid (Hymenoptera) Parasites of the Gypsy Moth, Lymantria dispar (Lepidoptera: Lymantriidae). Annals of the Entomological Society of America 72(6): 794–810. https://doi.org/10.1093/aesa/72.6.794
- Morinière J, Hendrich L, Balke M, Beermann AJ, König T, Hess M, Koch S, Müller R, Leese F, Hebert PDN, Hausmann A, Schubart CD, Haszprunar G (2017) A DNA barcode library for Germany′s mayflies, stoneflies and caddisflies (Ephemeroptera, Plecoptera and Trichoptera). Molecular Ecology Resources 17(6): 1293–1307. https://doi.org/10.1111/1755-0998.12683
- Muesebeck C (1922) A revision of the North American ichneumon-flies, belonging to the subfamilies Neoneurinae and Microgasterinae. Proceedings of the United States National Museum 61(2436): 1–76. https://doi.org/10.5479/si.00963801.61-2436.1
- Muesebeck C (1928) A new European species of Apanteles parasitic on the gipsy moth. Proceedings of the Entomological Society of Washington 30: 8–9.
- Mutanen M, Kivelä SM, Vos RA, Doorenweerd C, Ratnasingham S, Hausmann A, Huemer P, Dincă V, van Nieukerken EJ, Lopez-Vaamonde C, Vila R, Aarvik L, Decaëns T, Efetov KA, Hebert PDN, Johnsen A, Karsholt O, Pentinsaari M, Rougerie R, Segerer A, Tarmann G, Zahiri R, Godfray HCJ (2016) Species-level para- and polyphyly in DNA barcode gene trees: Strong operational bias in european Lepidoptera. Systematic Biology 65(6): 1024–1040. https://doi.org/10.1093/sysbio/syw044
- Nixon G (1965) A reclassification of the tribe Microgasterini (Hymenoptera: Braconidae). Bulletin of the British Museum (Natural History). Entomology 2(Supplement 2): 1–284. https://doi.org/10.5962/p.144036
- Nixon G (1968) A revision of the genus Microgaster Latreille (Hymenoptera: Braconidae). Bulletin of the British Museum (Natural History). Entomology 22: 31–72. https://doi.org/10.5962/bhl.part.9950
- Nixon G (1970) A revision of the N. W. European species of Microplitis Forster (Hymenoptera: Braconidae). Bulletin of the British Museum (Natural History). Entomology 25: 1–30.
- Nixon G (1972) A revision of the north-western European species of the laevigatus-group of Apanteles Förster (Hymenoptera, Braconidae). Bulletin of Entomological Research 61(4): 701–743. https://doi.org/10.1017/S0007485300047544
- Nixon G (1973) A revision of the north-western European species of the vitripennis, pallipes, octonarius, triangulator, fraternus, formosus, parasitellae, metacarpalis and circumscriptus-groups of Apanteles Förster (Hymenoptera, Braconidae). Bulletin of Entomological Research 63(2): 169–230. https://doi.org/10.1017/S0007485300039006
- Nixon GEJ (1974) A revision of the north-western European species of the glomeratus group of Apanteles Förster (Hymenoptera, Braconidae). Bulletin of Entomological Research 64(3): 453–524. https://doi.org/10.1017/S0007485300031333
- Nixon G (1976) A revision of the north-western European species of the merula, lacteus, vipio, ultor, ater, butalidis, popularis, carbonarius and validus-groups of Apanteles Förster (Hymenoptera, Braconidae). Bulletin of Entomological Research 65(4): 687–735. https://doi.org/10.1017/S0007485300006386
- O’Hara JE, Shima H, Zhang C (2009) Annotated catalogue of the Tachinidae (Insecta: Diptera) of China. Zootaxa 2190(1): 1–236. https://doi.org/10.11646/zootaxa.2190.1.1
- Okada I (1988) Three species of wax moths in Japan. Mitsubachi Kagaku 9: 145–149.
- Oltra-Moscardó MT, Jiménez-Peydró R (2005) The taxon Rasivalva (Hymenoptera: Braconidae) in the Palaearctic region and description of Rasivalva pyrenaica new species from Andorra. Journal of Entomological Science 40(4): 438–445. https://doi.org/10.18474/0749-8004-40.4.438
- Page RDM (2016) DNA barcoding and taxonomy: Dark taxa and dark texts. Philosophical Transactions of the Royal Society of London, Series B, Biological Sciences 371(1702): 1–7. https://doi.org/10.1098/rstb.2015.0334
- Papp J (1959) The Microgaster Latr., Microplitis Först., and Hygroplitis Thoms. species of the Carpathian Basin (Hymenoptera, Braconidae). Annales Historico-Naturales Musei Nationalis Hungarici 51: 397–413.
- Papp J (1973) New Apanteles Först. species from Hungary (Hymenoptera, Braconidae: Microgasterinae), II. Annales Historico-Naturales Musei Nationalis Hungarici 65: 287–304.
- Papp J (1974) New Apanteles Först. species from Hungary (Hymenoptera, Braconidae: Microgasterinae), III. Annales Historico-Naturales Musei Nationalis Hungarici 66: 325–337.
- Papp J (1976a) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgasterinae) I. The species-groups. Annales Historico-Naturales Musei Nationalis Hungarici 68: 251–274.
- Papp J (1976b) Key to the European Microgaster Latr. species, with a new species and taxonomical remarks (Hymenoptera: Braconidae; Microgasterinae). Acta Zoologica Academiae Scientiarum Hungaricae 22: 97–117.
- Papp J (1978) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgasterinae) II. The laevigatus-group, I. Annales Historico-Naturales Musei Nationalis Hungarici 70: 265–301.
- Papp J (1979) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgasterinae) III. The laevigatus-group, 2. Annales Historico-Naturales Musei Nationalis Hungarici 71: 235–250.
- Papp J (1980) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgasterinae) IV. The lineipes-, obscurus- and ater-group. Annales Historico-Naturales Musei Nationalis Hungarici 72: 241–272.
- Papp J (1981a) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgastrinae), V. The lacteus-, longipalpis-, ultor-, butalidis- and vipio-group. Annales Historico-Naturales Musei Nationalis Hungarici 73: 263–291.
- Papp J (1981b) Contributions to the Braconid fauna of Hungary, III. Opiinae and Microgasterinae (Hymenoptera: Braconidae). Folia Entomologica Hungarica 42: 127–141.
- Papp J (1982) A Survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgastrinae), VI. The laspeyresiella-, merula-, falcatus- and validus-group. Annales Historico-Naturales Musei Nationalis Hungarici 74: 255–267.
- Papp J (1983) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgastrinae), VII. The carbonarius-, circumscriptus-, fraternus-, pallipes-, parasitellae-, vitripennis-, liparidis-, octonarius- and thompsoni-group. Annales Historico-Naturales Musei Nationalis Hungarici 75: 247–283.
- Papp J (1984a) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgastrinae), VIII. The metacarpalis-, formosus-, popularis- and suevus-group. Annales Historico-Naturales Musei Nationalis Hungarici 76: 265–295.
- Papp J (1984b) Palaearctic species of Microgaster Latreille (= Microplitis Förster) with description of seven new species (Hymenoptera, Braconidae, Microgastrinae). Entomologische Abhandlungen 47: 95–140.
- Papp J (1986a) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgastrinae) IX. The glomeratus-group, 1. Annales Historico-Naturales Musei Nationalis Hungarici 78: 225–247.
- Papp J (1986b) First survey of the Glabromicroplitis Papp species of the Holarctic Region, with taxonomical remarks of three Microgaster Latreille species (Hymenoptera, Braconidae: Microgastrinae). Annales Historico-Naturales Musei Nationalis Hungarici 78: 249–253.
- Papp J (1987) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgastrinae), X. The glomeratus-group 2 and the cultellatus-group. Annales Historico-Naturales Musei Nationalis Hungarici 79: 207–258.
- Papp J (1988) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgastrinae) XI. “Homologization” of the species-group of Apanteles s. l. with Mason’s generic taxa. Checklist of genera. Parasitoid host list 1. Annales Historico-Naturales Musei Nationalis Hungarici 80: 145–175.
- Papp J (1989) Three new braconid species from Central Switzerland (Hymenoptera, Braconidae). Journal of the Swiss Entomological Society 62: 269–278. https://doi.org/10.5169/seals-402353
- Papp J (1990) A survey of the European species of Apanteles Först. (Hymenoptera, Braconidae: Microgastrinae) XII. Supplement to the key of the glomeratus-group. Parasitoid/host list 2. Annales Historico-Naturales Musei Nationalis Hungarici 81: 159–203.
- Pilotto F, Kühn I, Adrian R, Alber R, Alignier A, Andrews C, Bäck J, Barbaro L, Beaumont D, Beenaerts N, Benham S, Boukal DS, Bretagnolle V, Camatti E, Canullo R, Cardoso PG, Ens BJ, Everaert G, Evtimova V, Feuchtmayr H, García-González R, Gómez García D, Grandin U, Gutowski JM, Hadar L, Halada L, Halassy M, Hummel H, Huttunen K-L, Jaroszewicz B, Jensen TC, Kalivoda H, Schmidt IK, Kröncke I, Leinonen R, Martinho F, Meesenburg H, Meyer J, Minerbi S, Monteith D, Nikolov BP, Oro D, Ozoliņš D, Padedda BM, Pallett D, Pansera M, Pardal MÂ, Petriccione B, Pipan T, Pöyry J, Schäfer SM, Schaub M, Schneider SC, Skuja A, Soetaert K, Spriņģe G, Stanchev R, Stockan JA, Stoll S, Sundqvist L, Thimonier A, Van Hoey G, Van Ryckegem G, Visser ME, Vorhauser S, Haase P (2020) Meta-analysis of multidecadal biodiversity trends in Europe. Nature Communications 11(1): e3486. https://doi.org/10.1038/s41467-020-17171-y
- Puillandre N, Brouillet S, Achaz G (2021) ASAP: Assemble species by automatic partitioning. Molecular Ecology Resources 21(2): 609–620. https://doi.org/10.1111/1755-0998.13281
- Ratnasingham S, Hebert PDN (2007) BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Molecular Ecology Notes 7(3): 355–364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
- Ratnasingham S, Hebert PDN (2013a) A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 8(7): e66213. https://doi.org/10.1371/journal.pone.0066213
- Ratnasingham S, Hebert PDN (2013b) A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 8(7): e66213. https://doi.org/10.1371/journal.pone.0066213
- Raupach MJ, Hendrich L, Küchler SM, Deister F, Morinière J, Gossner MM (2014) Building-Up of a DNA Barcode Library for True Bugs (Insecta: Hemiptera: Heteroptera) of Germany Reveals Taxonomic Uncertainties and Surprises. PLoS ONE 9(9): e106940. https://doi.org/10.1371/journal.pone.0106940
- Raupach MJ, Hannig K, Morinière J, Hendrich L (2020) A DNA barcode library for ground beetles of Germany: The genus Pterostichus Bonelli, 1810 and allied taxa (Insecta, Coleoptera, Carabidae). ZooKeys 980: 93–117. https://doi.org/10.3897/zookeys.980.55979
- Reinhard H (1880) Beiträge zur Kenntniss einiger Braconiden-Gattungen. Fünftes Stück. XVI. Zur Gattung Microgaster, Latr. (Microgaster, Microplitis, Apanteles). Deutsche Entomologische Zeitschrift 24: 353–370. https://doi.org/10.1002/mmnd.4800240215
- Reinhardt R, Bolz R (2011) Rote Liste und Gesamtartenliste der Tagfalter (Rhopalocera) (Lepidoptera: Papilionoidea et Hesperioidea) Deutschlands. Naturschutz und Biologische Vielfalt 3: 167–194.
- Rennwald E, Rodeland J, Guggemoos T (2023) Lepiforum’s Checklist of European Lepidoptera. https://lepiforum.org/wiki/page/Downloads#Lepiforums-Europaliste_der_Schmetterlinge-Lepiforums_Checklist_of_European_Lepidoptera [July 21, 2023]
- Ronquist F, Forshage M, Häggqvist S, Karlsson D, Hovmöller R, Bergsten J, Holston K, Britton T, Abenius J, Andersson B, Buhl PN, Coulianos C-C, Fjellberg A, Gertsson C-A, Hellqvist S, Jaschhof M, Kjærandsen J, Klopfstein S, Kobro S, Liston A, Meier R, Pollet M, Riedel M, Roháček J, Schuppenhauer M, Stigenberg J, Struwe I, Taeger A, Ulefors S-O, Varga O, Withers P, Gärdenfors U (2020) Completing Linnaeus’s inventory of the Swedish insect fauna: Only 5,000 species left? PLoS ONE 15(3): e0228561. https://doi.org/10.1371/journal.pone.0228561
- Rumph JA, Turner WJ (1998) Alternative to critical point drying for soft-bodied insect larvae. Annals of the Entomological Society of America 91(5): 693–699. https://doi.org/10.1093/aesa/91.5.693
- Ruthe J (1860) Deutsche Braconiden. Erstes Stück. Deutsche Entomologische Zeitschrift 4: 105–160.
- Samin N, Chelav HS, Ahmad Z, Penteado-Dias AM, Samiuddin A (2020) A faunistic study on the family Braconidae (Hymenoptera: Ichneumonoidea) from Iran. Scientific Bulletin of the Uzhhorod University 48: 14–19. https://doi.org/10.24144/1998-6475.2020.48.14-19
- Schmid-Egger C, Straka J, Ljubomirov T, Blagoev GA, Morinière J, Schmidt S (2019) DNA barcodes identify 99 per cent of apoid wasp species (Hymenoptera: Ampulicidae, Crabronidae, Sphecidae) from the Western Palearctic. Molecular Ecology Resources 19(2): 476–484. https://doi.org/10.1111/1755-0998.12963
- Schmidt S, Schmid-Egger C, Morinière J, Haszprunar G, Hebert PDN (2015) DNA barcoding largely supports 250 years of classical taxonomy: Identifications for Central European bees (Hymenoptera, Apoidea partim). Molecular Ecology Resources 15(4): 985–1000. https://doi.org/10.1111/1755-0998.12363
- Schütte A, Stüben P, Astrin J (2023) Molecular Weevil Identification Project: A thoroughly curated barcode release of 1300 Western Palearctic weevil species (Coleoptera, Curculionoidea). Biodiversity Data Journal 11: e96438. https://doi.org/10.3897/BDJ.11.e96438
- Seibold S, Gossner MM, Simons NK, Blüthgen N, Müller J, Ambarlı D, Ammer C, Bauhus J, Fischer M, Habel JC, Linsenmair KE, Nauss T, Penone C, Prati D, Schall P, Schulze E-D, Vogt J, Wöllauer S, Weisser WW (2019) Arthropod decline in grasslands and forests is associated with landscape-level drivers. Nature 574(7780): 671–674. https://doi.org/10.1038/s41586-019-1684-3
- Shaw MR (1994) Parasitoid host ranges. In: Hawkins BA, Sheehan W (Eds) Parasitoid Community Ecology. Oxford University Press, Oxford, 111–144. https://doi.org/10.1093/oso/9780198540588.003.0007
- Shaw MR (2004) Microgaster alebion Nixon and its “var A”: Description of a new species and biological notes (Hymenoptera: Braconidae, Microgastrinae). Entomologist’s Gazette 55: 217–224.
- Shaw MR (2007) The species of Cotesia Cameron (Hymenoptera: Braconidae: Microgastrinae) parasitising Lycaenidae (Lepidoptera) in Britain. British Journal of Entomology and Natural History 20: 255–267.
- Shaw MR (2009) Cotesia Cameron (Hymenoptera: Braconidae: Microgastrinae) parasitoids of Heliconiinae (Lepidoptera: Nymphalidae) in Europe, with description of three new species. British Journal of Entomology and Natural History 22: 133–146.
- Shaw M (2012a) Notes on some European Microgastrinae (Hymenoptera: Braconidae) in the National Museums of Scotland, with twenty species new to Britain, new host data, taxonomic changes and remarks, and descriptions of two new species of Microgaster Latreille. Entomologist’s Gazette 63: 173–201.
- Shaw MR (2012b) Larval parasitoids of Rivula sericealis (Scopoli) (Lepidoptera: Noctuidae) in Britain, including notes on the biology of Cotesia subordinaria (Tobias) (Hymenoptera: Braconidae, Microgastrinae), a solitary-cum-gregarious parasitoid. Entomologist’s Gazette 63: 251–257.
- Shaw MR (2020) Discovery of the genus Venanides Mason 1981 (Hymenoptera: Braconidae, Microgastrinae) in Europe, with description of a new species parasitizing Carcina quercana (Fabricius) (Lepidoptera: Peleopodidae). Entomologist’s Gazette 71(1): 45–57. https://doi.org/10.31184/G00138894.711.1755
- Shaw MR (2022) Rearings of four European Microgastrinae (Hymenoptera: Braconidae), three new to Britain including a new species of Cotesia Cameron, 1891. Entomologist’s Gazette 73(3): 165–175. https://doi.org/10.31184/G00138894.733.1853
- Shaw MR (2023) Two new species of European Microgaster Latreille, 1804 (Hymenoptera: Braconidae, Microgastrinae), with host data on some further species. Entomologist’s Gazette 73(4): 219–232. https://doi.org/10.31184/G00138894.734.1867
- Shaw MR, Colom P (2023) Notes on the three species of Cotesia Cameron, 1891 (Hymenoptera: Braconidae, Microgastrinae) parasitizing Gonepteryx [Leach, 1815] species (Lepidoptera: Pieridae) in Europe, with description of a new species from the Balearic Islands. Entomologist’s Gazette 73(4): 253–260. https://doi.org/10.31184/G00138894.734.1872
- Shaw MR, Fernandez-Triana JL (2020) Two new European species of Cotesia Cameron, 1891 (Hymenoptera: Braconidae, Microgastrinae) parasitizing butterflies (Lepidoptera: Papilionoidea), and an unrelated synonymy in the genus. Entomologist’s Gazette 71(3): 181–195. https://doi.org/10.31184/G00138894.713.1768
- Shimamori K (1987) On the biology of Apanteles galleriae, a parasite of the two species of wax moths. Mitsubachi Kagaku 8: 107–112.
- Smith MA, Bertrand C, Crosby K, Eveleigh ES, Fernandez-Triana J, Fisher BL, Gibbs J, Hajibabaei M, Hallwachs W, Hind K, Hrcek J, Huang D-W, Janda M, Janzen DH, Li Y, Miller SE, Packer L, Quicke D, Ratnasingham S, Rodriguez J, Rougerie R, Shaw MR, Sheffield C, Stahlhut JK, Steinke D, Whitfield J, Wood M, Zhou X (2012) Wolbachia and DNA Barcoding Insects: Patterns, Potential, and Problems. PLoS ONE 7(5): e36514. https://doi.org/10.1371/journal.pone.0036514
- Smith AM, Fernández-Triana JL, Eveleigh E, Gómez J, Guclu C, Hallwachs W, Hebert PDN, Hrcek J, Huber JT, Janzen D, Mason PG, Miller S, Quicke DLJ, Rodriguez JJ, Rougerie R, Shaw MR, Várkonyi G, Ward DF, Whitfield JB, Zaldívar-Riverón A (2013) DNA barcoding and the taxonomy of Microgastrinae wasps (Hymenoptera, Braconidae): Impacts after 8 years and nearly 20 000 sequences. Molecular Ecology Resources 13(2): 168–176. https://doi.org/10.1111/1755-0998.12038
- Smith MA, Whitfield JB, Fernandez-Triana JL, Kula RR, Hallwachs W, Janzen DH (2015) Revision of the genera Microplitis and Snellenius (Hymenoptera, Braconidae, Microgastrinae) from Area de Conservacion Guanacaste, Costa Rica, with a key to all species previously described from Mesoamerica. Deutsche Entomologische Zeitschrift 62(2): 137–201. https://doi.org/10.3897/dez.62.5276
- Snell Q, Walker P, Posada D, Crandall K (2002) TCS: Estimating Gene Genealogies. International parallel and distributed processing symposium 2: 184–191.
- Telenga N (1955) Braconidae, subfamily Microgasterinae, subfamily Agathinae. Fauna USSR 5(4): 1–311.
- Thomas JA, Telfer MG, Roy DB, Preston CD, Greenwood JJD, Asher J, Fox R, Clarke RT, Lawton JH (2004) Comparative losses of british butterflies, birds, and plants and the global extinction crisis. Science 303(5665): 1879–1881. https://doi.org/10.1126/science.1095046
- Tobias V (1971) Review of the Braconidae (Hymenoptera) of the U.S.S.R. Trudy Vsesoyuznogo Entomologicheskogo Obshchestva 54: 156–268.
- Tobias V (1975) Two new species of braconids from the genus Apanteles Forst. (Hymenoptera, Braconidae) parasites of the moth Bucculatrix ulmella Z. Izvestiia Akademii nauk Moldavskoi SSR: seriia biologicheskikh i khimicheskikh nauk 3: 60–63.
- Tobias V (1976) Braconids of the Caucasus (Hymenoptera, Braconidae). Opred. Faune SSSR: 1–286.
- Tobias V (1986) Subfamily Microgasterinae. Key to the Insects of the European Part of the USSR III. 4. Insitute of Zoology, Academy of Sciences of the USSE, Leningrad, 605–815.
- Tobias V, Kotenko A (1984) Three new species of genus Apanteles Foerster of the parasitellae-group (Hymenoptera, Braconidae). Taxonomy and Zoogeography of Insects. Kiev, 61–66.
- van Achterberg C (2002) Apanteles (Choeras) gielisi spec. nov. (Hymenoptera: Braconidae: Microgastrinae) from The Netherlands and the first report of Trichoptera as host of Braconidae. Zoologische Mededelingen Leiden 72: 53–60.
- van Achterberg C (2006) The Braconidae (Hymenoptera) of Greenland. Zoölogische Mededeelingen 80: 1–179.
- Viereck HL (1911) Descriptions of six new genera and thirty-one new species of ichneumon flies. Proceedings of the United States National Museum 40(1812): 173–196. https://doi.org/10.5479/si.00963801.1812.173
- Voith J, Bräu M, Dolek M, Nunner A, Wolf W (2016) Rote Liste und Gesamtartenliste der Tagfalter (Lepidoptera: Rhopalocera) Bayerns.
- Völkl W, Blick T (2004) Die quantitative Erfassung der rezenten Fauna von Deutschland. BfN-Skripten 117: 1–85.
- Wagner DL (2020) Insect Declines in the Anthropocene. Annual Review of Entomology 65(1): 457–480. https://doi.org/10.1146/annurev-ento-011019-025151
- Watanabe C (1987) Occurrence of Apanteles galleriae (Hymenoptera, Braconidae), parasite of wax moth, in Japan. Kontyu 55: 3–94.
- Whitfield J (2006) Revision of the Nearctic species of the genus Pholetesor Mason (Hymenoptera: Braconidae). Zootaxa 1144(1): 3–94. https://doi.org/10.11646/zootaxa.1144.1.1
- Whitfield J, Cameron S (1993) Comparative notes on Hymenopteran parasitoids in bumble bee and honey bee colonies (Hymenoptera, Apidae) reared adjacently. Entomological News 104: 240–248.
- Whitfield JB, Oltra Moscardó MTO (2004) The neotropical species of Deuterixys Mason (Hymenoptera: Braconidae). Journal of Hymenoptera Research 13: 134–148.
- Wilkinson DS (1932) Four new Apanteles (Hym., Brac.). Proceedings of the Royal Entomological Society of London, Series B, Taxonomy 1(6): 139–144. https://doi.org/10.1111/j.1365-3113.1932.tb01372.x
- Wilkinson DS (1938) On a Further Two New Palaearctic Species of Apanteles (Hym. Brac.). Proceedings of the Royal Entomological Society of London, Series B, Taxonomy 7(10): 222–227. https://doi.org/10.1111/j.1365-3113.1938.tb01231.x
- Wilkinson D (1945) Description of Palaearctic species of Apanteles (Hymen., Braconidae). Transactions of the Royal Entomological Society of London 95(3): 35–226. https://doi.org/10.1111/j.1365-2311.1945.tb00436.x
- Wührl L, Pylatiuk C, Giersch M, Lapp F, von Rintelen T, Balke M, Schmidt S, Cerretti P, Meier R (2022) DiversityScanner: Robotic handling of small invertebrates with machine learning methods. Molecular Ecology Resources 22(4): 1626–1638. https://doi.org/10.1111/1755-0998.13567
- Yu D, van Achterberg C, Horstmann K (2016) Taxapad 2016, Ichneumonoidea 2015. Nepean, Ontario. http://www.taxapad.com
- Zeng J, He J-H, Chen X (2011) The genera Deuterixys Mason, 1981 and Wilkinsonellus Mason, 1981 (Hymenoptera, Braconidae, Microgastrinae) from China, with description of two new species. ZooKeys 120: 27–40. https://doi.org/10.3897/zookeys.120.891
- Žikić V, Lazarević M, Ilić Milošević M, Trajković A, Hric B, Stanković SS (2021) Diversity of the Genus Cotesia Cameron (Braconidae: Microgastrinae) in Serbia. Acta Entomologica Serbica 26: 27–35. https://doi.org/10.5281/zenodo.5704966
Supplementary materials
Specimen Data Spreadsheet (BOLD) for DS-MCGNRECS
Data type: xlsx
Explanation note: This table includes all relevant data associated with specimens that are included in our BOLD dataset (341 specimens).
COI-Sequences for DS-MCGNRECS
Data type: fas
Explanation note: Includes all COI sequences used in our interative species concepts.
Alignment used for NJ-topology of DS-MCGNRECS
Data type: fasta
Explanation note: Includes the alignment and all sequences used for the NJ-topology in Fig.
NJ-topology of DS-MCGNRECS
Data type: nwk
Coordinates for Holarctic distribution maps
Data type: csv
Explanation note: Includes the coordinates used for the distribution maps in Fig.
Glyptapanteles indiensis and related species sequences
Data type: fasta
Explanation note: All sequences used for NJ-topology in Fig.
Glyptapanteles indiensis and related species NJ-topology
Data type: nwk
Explanation note: NJ-topology in Fig.
BOLD:AAA6099 sequences
Data type: nexus
Explanation note: Sequences in BIN BOLD:AAA6099 used for haplotype network in Fig.
BOLD:AAA6099 traits
Data type: txt
Explanation note: Amelie Höcherl Traits (countries) for BIN BOLD:AAA6099 used for haplotype network in Fig.
BOLD:ABY6385 sequences
Data type: nexus
Explanation note: Sequences in BOLD:ABY6385 used for haplotype network in Fig.
BOLD:ABY6385 traits
Data type: txt
Explanation note: Traits (hosts) for BIN BOLD:ABY6385 used for haplotype network in Fig.
BOLD:AAC9130 sequences
Data type: nexus
Explanation note: Sequences in BIN BOLD:AAC9130 used for haplotype network in Fig.
BOLD:AAC9130 traits
Data type: txt
Explanation note: Traits (hosts) for BIN BOLD:AAC9130 used for haplotype network in Fig.
ASAP partition 1 of BOLD:AAA7143
Data type: csv
Explanation note: Sequences downloaded 7th of august, 2023, ASAP score 3.99.
ASAP partition 2 of BOLD:AAA7143
Data type: csv
Explanation note: Sequences downloaded 7th of august, 2023, ASAP score 6.50.