|
||
|
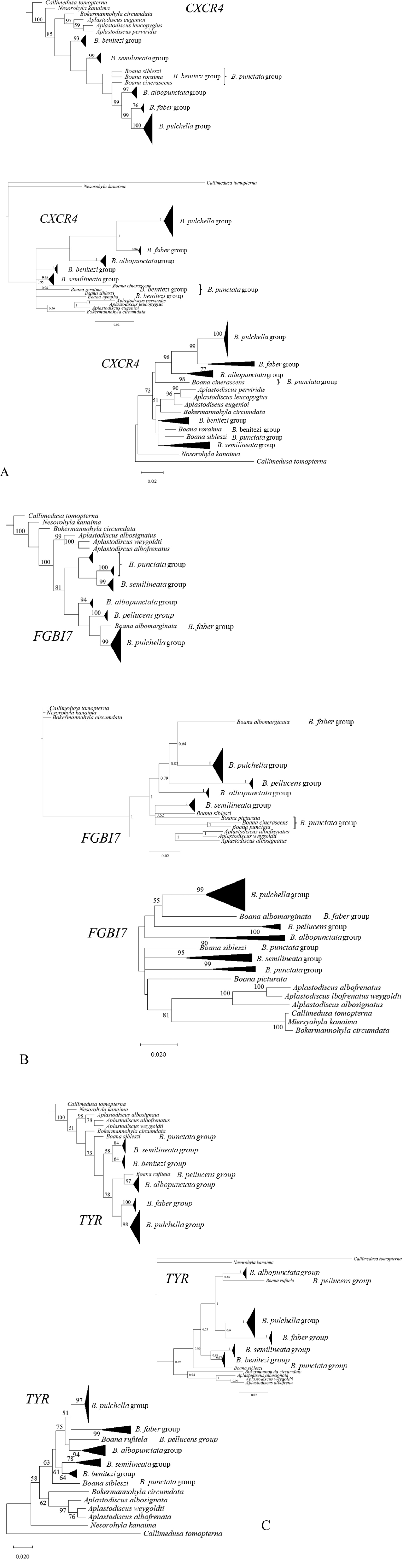
Phylogenetic trees corresponding to the studied markers provided more sensitive and highly informative performance (A CXCR4 B FGBI7 C TYR), and the methods used–MP, MB, and ML, corresponding to the first, second, and third trees for each marker, respectively). For Jackknife support values from the MP method, and bootstrap support values for the ML method, values below 50% were not presented. |