|
||
|
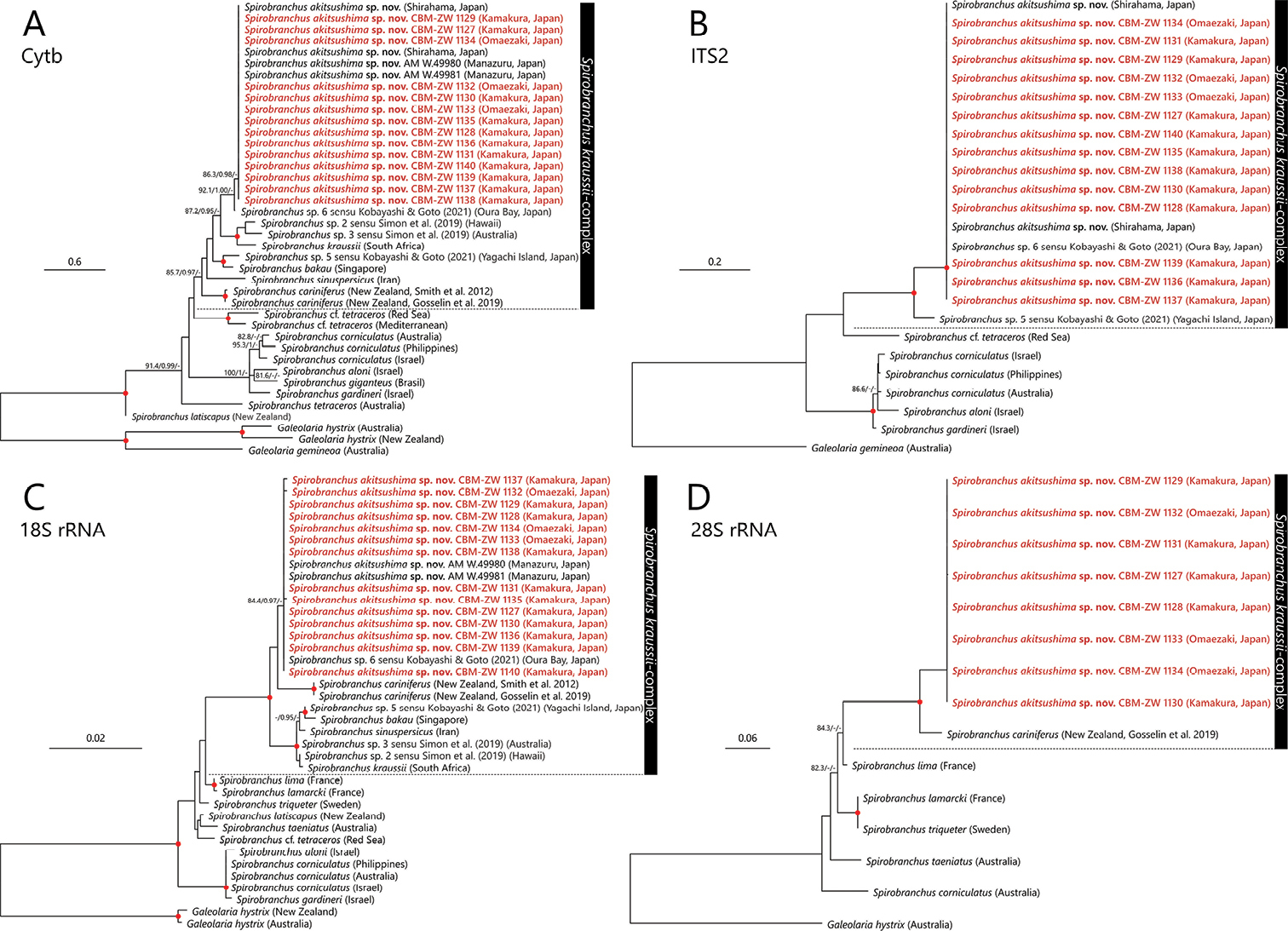
Maximum Likelihood tree of Spirobranchus species inferred from mitochondrial cytb (A), nuclear ITS2 (B), 18S (C), and 28S rRNA (D) gene/region sequences obtained from the present study and from DDBJ/EMBL/GenBank (Table 1). The gene sequences obtained in the present study are highlighted by red color. SH-aLRT/approximate Bayes support/ultrafast bootstrap support values of ≥ 80%, ≥ 0.95, ≥ 95%, respectively are given beside the respective nodes. Red circles at nodes indicate triple high support values of SH-aLRT ≥ 80%, approximate Bayes support ≥ 0.95, and ultrafast bootstrap support ≥ 95%. The scale bar represents the number of substitutions per site. Sequences of Galeolaria hystrix Mörch, 1863 and G. gemineoa Halt, Kupriyanova, Cooper & Rouse, 2009 from DDBJ/EMBL/GenBank were used for outgroup rooting. |