|
||
|
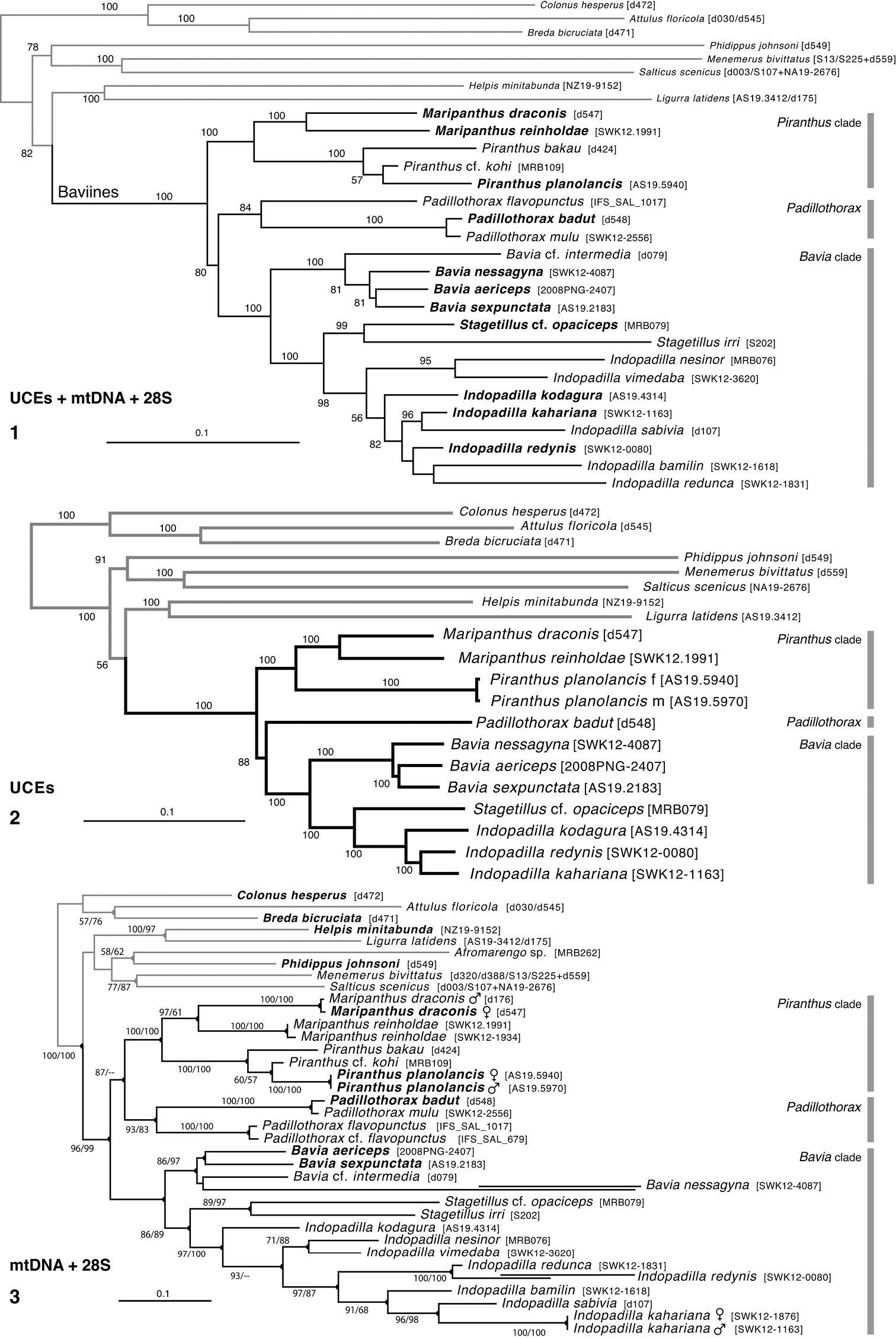
Phylogeny. 1 Maximum Likelihood tree (best of 20 replicates) from combined data set of 1313 UCE loci, plus mitochondrial 16SND1 and COI regions, plus 28S (dataset #4 in Methods). Baviines whose names are in bold, and all outgroups, have UCE data. Numbers are percentage of 500 bootstrap replicates showing the clade 2 Maximum Likelihood tree (best of 50 replicates) from concatenated data from 1313 UCE loci (dataset #1 in Methods). Numbers are percentage of 1000 bootstrap replicates showing the clade 3 Maximum Likelihood tree (best of 50 replicates) from concatenated data from mitochondrial data and 28S (dataset #2 in Methods). Taxa in bold have data from the entire mitochondrial genome, or nearly so. Numbers are percentage of 1000 bootstrap replicates showing the clade for the full dataset (#2), followed by the bootstrap percentage for the restricted mtDNA+28S dataset (#3 in Methods). Spots at nodes show those clades that also appear in the ML tree in the restricted dataset. The branches to B. nessagyna and I. redynis are long, compacted visually by cutting and sliding part of the length over itself; the actual length therefore should be seen as longer by the length of the overlap. |