|
||
|
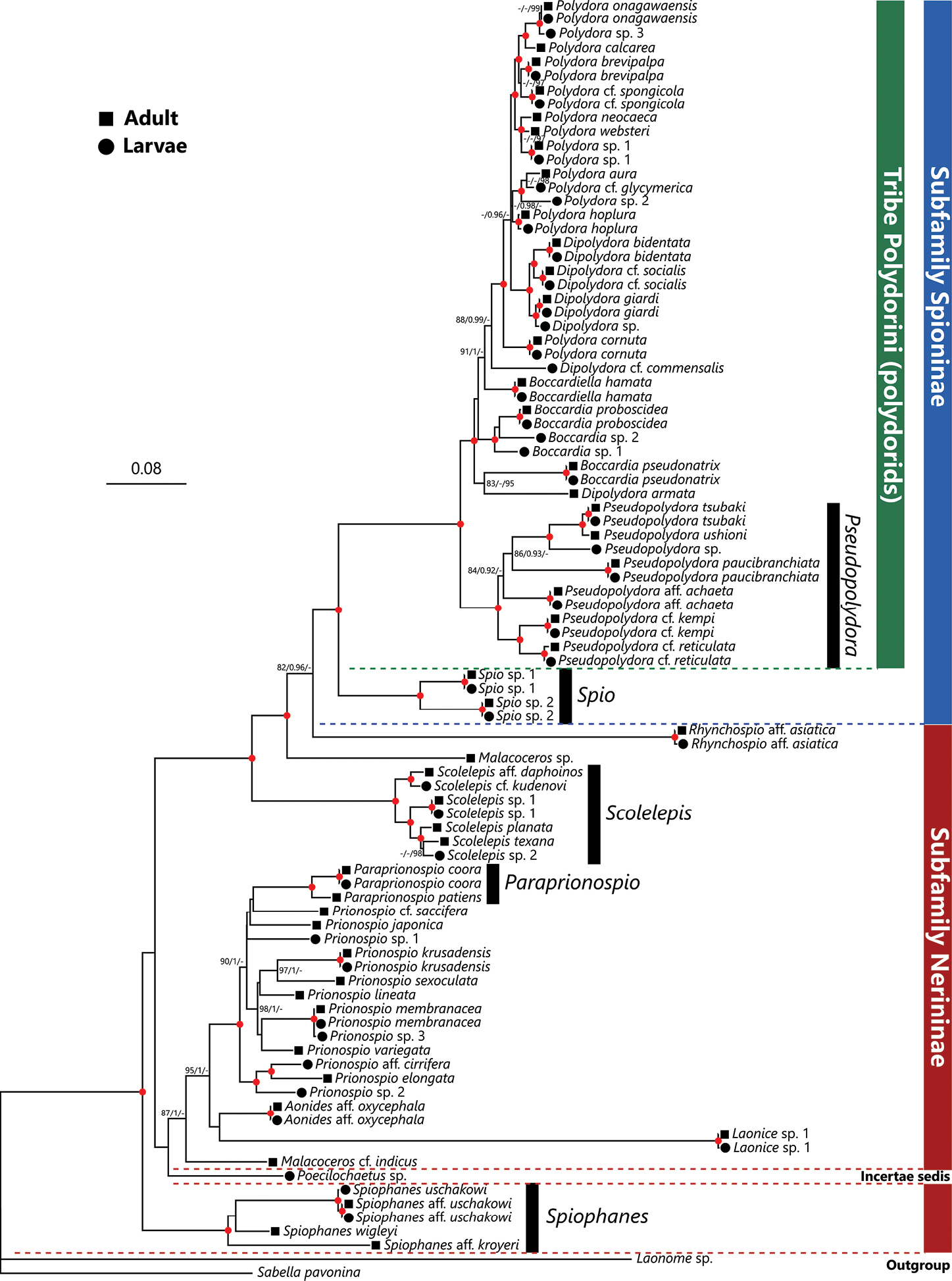
Maximum Likelihood tree inferred from nuclear 18S and mitochondrial 16S rRNA gene sequences of spionids obtained from Japan in the present and previous studies (provided in Table 1). The gene sequences of adult and larval spionid polychaetes are indicated by solid squares and circles in front of each species name, respectively. SH-aLRT/approximate Bayes support/ultrafast bootstrap support values of ≥ 80%, ≥ 0.95, ≥ 95%, respectively are given beside the respective nodes. Nodes with red circles indicate triple high support values of SH-aLRT ≥ 80, approximate Bayes support ≥ 0.95, and ultrafast bootstrap support ≥ 95. The scale bar represents the number of substitutions per site. Sequences of Laonome sp. and Sabella pavonina Savigny, 1822 obtained from DDBJ/EMBL/GenBank were used for outgroup rooting. |