|
||
|
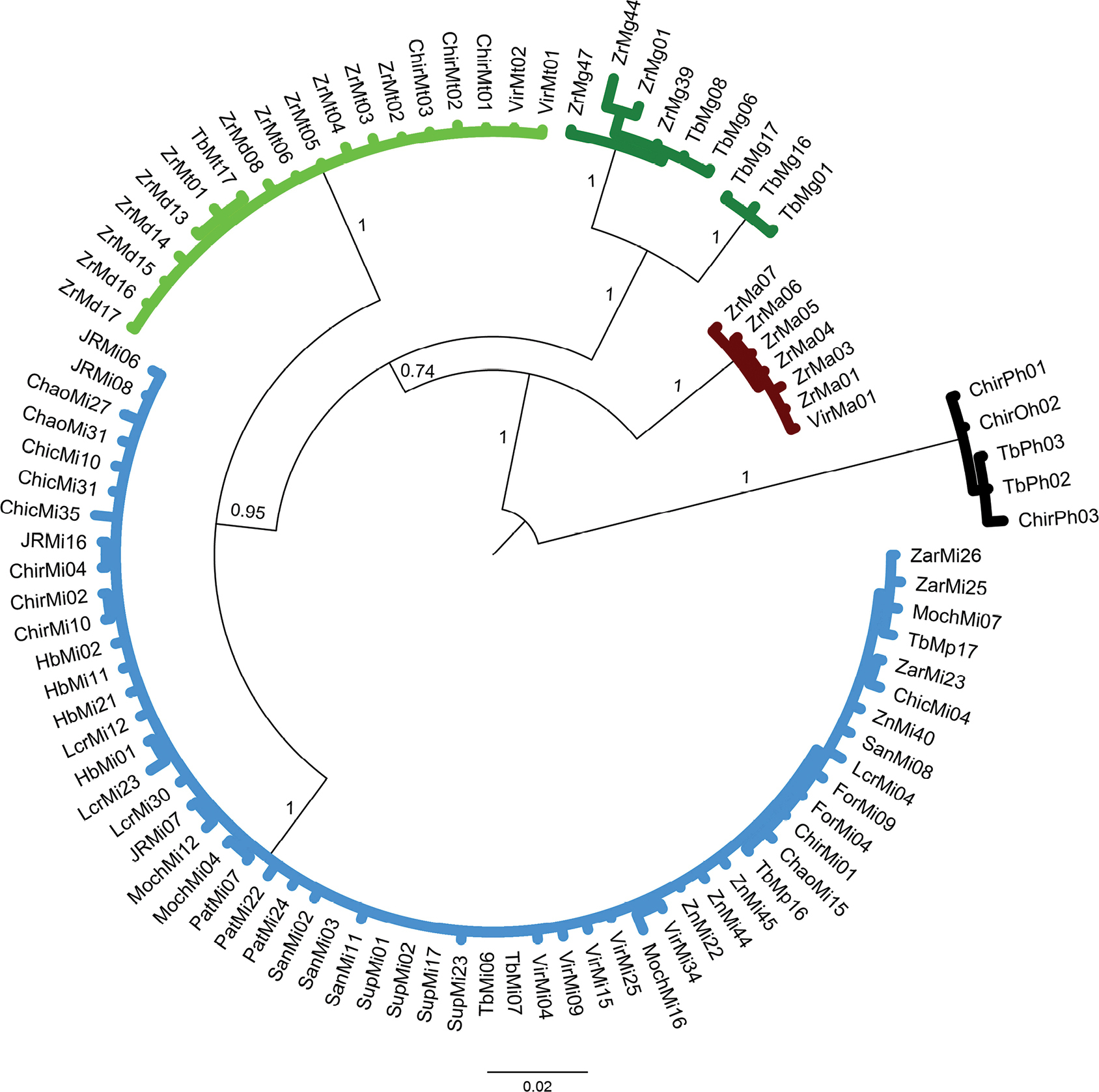
Phylogenetic tree based on Bayesian Inference approach generated under the K80+I+G substitution model using 93 partial sequences of the mitochondrial 16S rRNA gene from six Macrobrachium species collected in Peruvian rivers of the Pacific slope. Bootstrap values and posterior probabilities ≥ 50% are shown. P. hancocki was used as outgroup. GenBank accession numbers OR941603–OR941697. Abbreviations: Mt: M. transandicum; Mp: M. panamense; Mi: M. inca; Mg: M. gallus; Ma: M. americanum; Md: M. digueti; Ph: P. hancocki; Chir: Chira River; Tb: Tumbes River; Zr, Zar: Zarumilla River; Vir: Virú River; Pat: Pativilca River; JR: Juana Ríos River; Chao: Chao River; Lcr: Lacramarca River; Hb: Nepeña River; For: Fortaleza River; San: Santa River; Moch: Moche River; Chic: Chicama River; Sup: Supe River; Zn: Zaña River. |