|
||
|
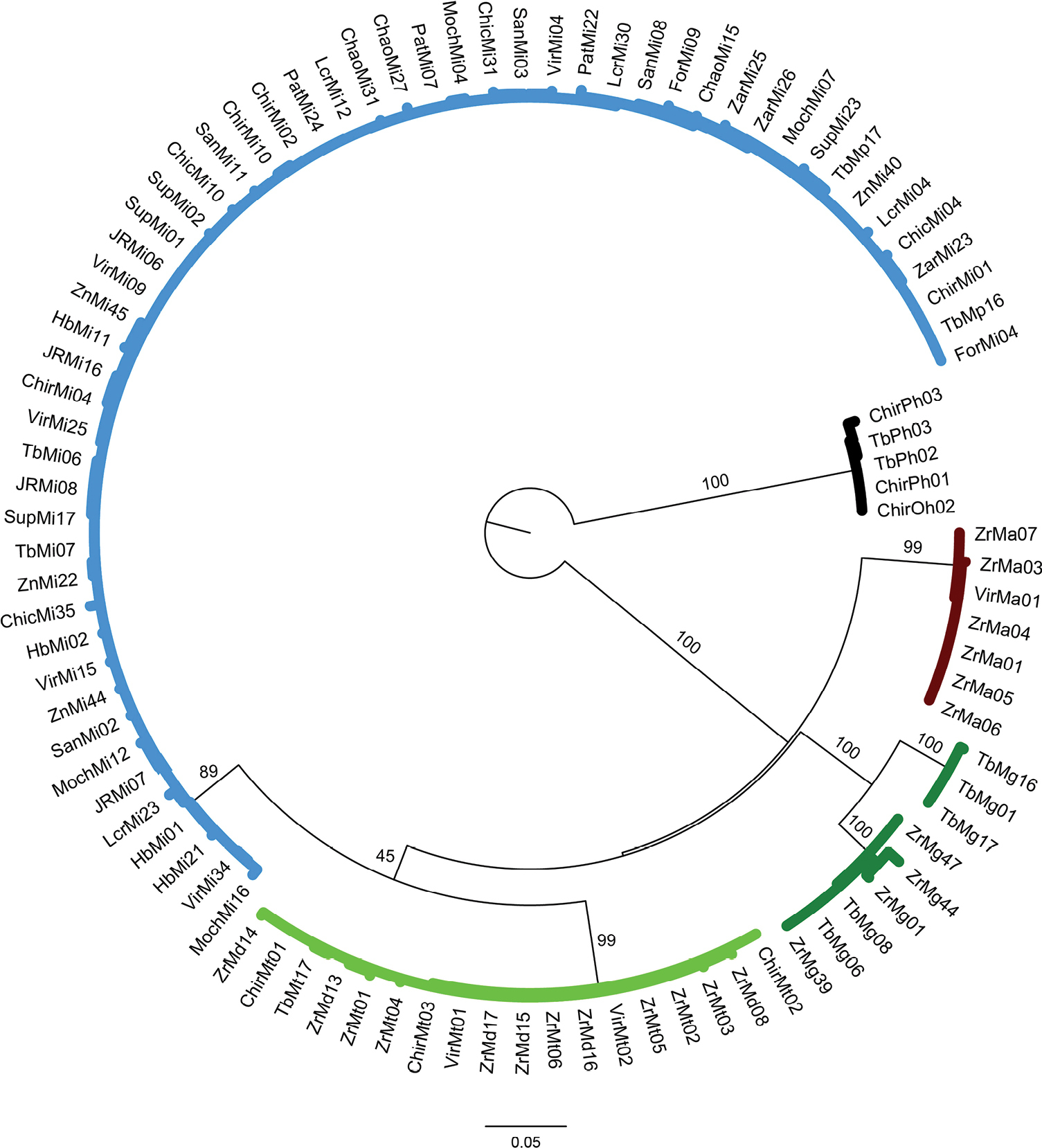
Phylogenetic tree based on maximum likelihood inference generated under the GTRGAMMA substitution model using 93 partial sequences of the mitochondrial 16S rRNA gene from six Macrobrachium species collected in Peruvian rivers of the Pacific slope. Bootstrap values ≥ 50% are shown. P. hancocki was used as outgroup. GenBank accession numbers OR941603–OR941697. Abbreviations: Mt: M. transandicum; Mp: M. panamense; Mi: M. inca; Mg: M. gallus; Ma: M. americanum; Md: M. digueti; Ph: P. hancocki; Chir: Chira River; Tb: Tumbes River; Zr, Zar: Zarumilla River; Vir: Virú River; Pat: Pativilca River; JR: Juana Ríos River; Chao: Chao River; Lcr: Lacramarca River; Hb: Nepeña River; For: Fortaleza River; San: Santa River; Moch: Moche River; Chic: Chicama River; Sup: Supe River; Zn: Zaña River. |