|
||
|
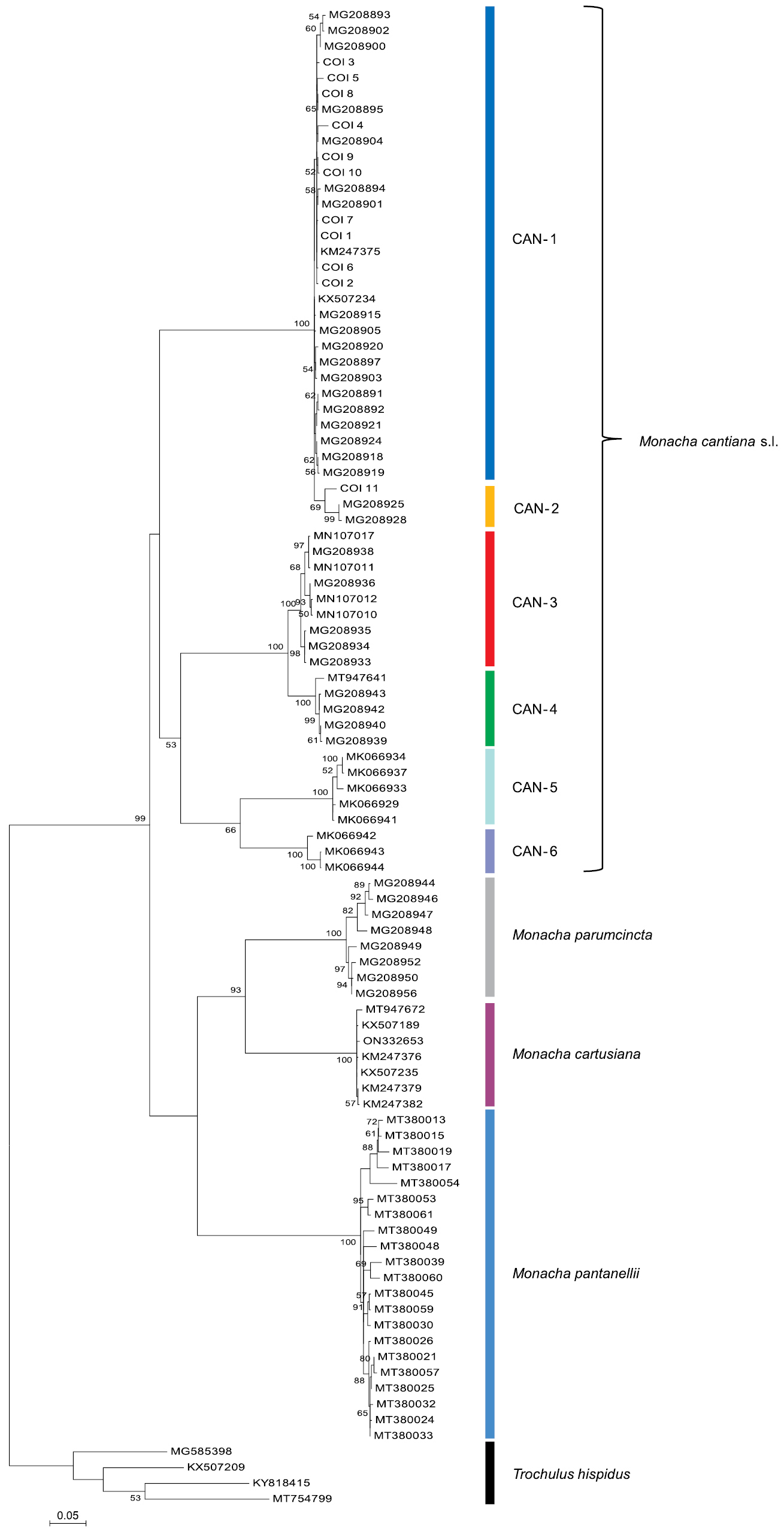
Maximum Likelihood (ML) tree of COI haplotypes of Monacha cantiana. New COI sequences of M. cantiana (Table 1) were compared with COI sequences of M. cantiana s.l., M. parumcincta, M. pantanellii and M. cartusiana obtained from GenBank (Suppl. material 1). Sequences were cut to 591 bp. HKY+G+I was the best nucleotide substitution model according to the Bayesian Information Criterion (BIC). The tree was rooted with Trochulus hispidus sequences obtained from GenBank (Suppl. material 1). |