|
||
|
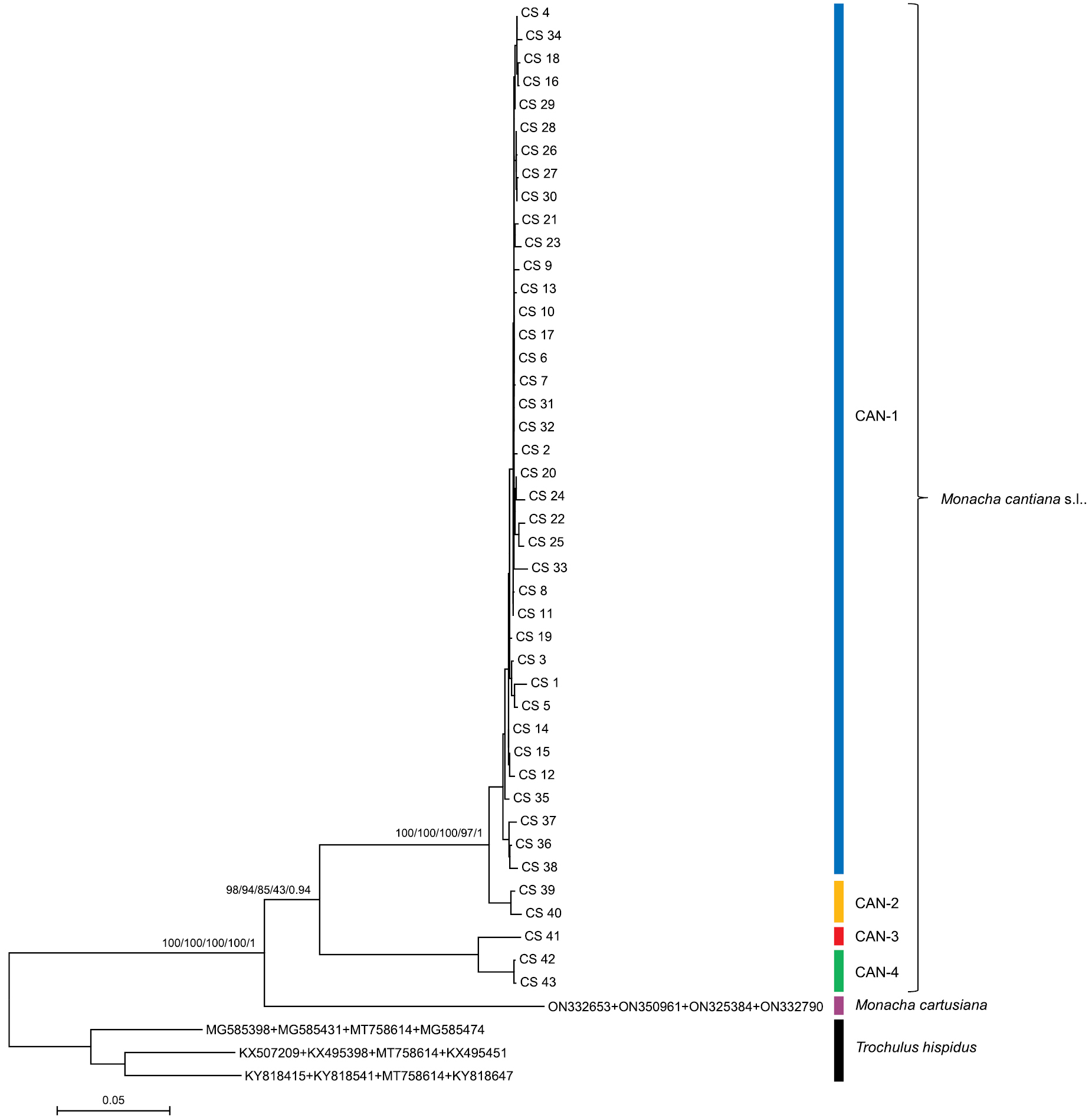
Maximum Likelihood (ML) tree of concatenated COI, 16SrDNA, H3, and ITS2 (flanked with 5.8S and 28SrDNA) haplotypes of Monacha cantiana. COI, 16SrDNA, H3, and ITS2 sequences of M. cantiana were compared with sequences of M. cantiana s.l. and M. cartusiana obtained from GenBank (Suppl. materials 1–4, 7). Length of sequences was 2498 positions (615 of COI, 829 of 16SrDNA, 279 of H3, and 775 of ITS2). Bayesian Information Criterion (BIC) specified GTR+G+I the best nucleotide substitution model in MEGA7, or HKY+F+G4 for COI, TIM2+F+I for 16SrDNA, TIM3e+I+G4 for H3, and K3P+I+G4 for ITS2 partition in IQ-Tree, RAxML, and MrBayes. Numbers next to main branches indicate (left to right): bootstrap support above 50% calculated by NJ-MEGA7 (Saitou and Nei 1987), ML-MEGA7 (Kumar et al. 2016), IQ-Tree (Trifinopoulos et al. 2016), RAxML (Stamatakis 2014), and posterior probabilities by BI (Ronquist et al. 2012). The tree was rooted with Trochulus hispidus concatenated sequences obtained from GenBank (Suppl. material 7). |