|
||
|
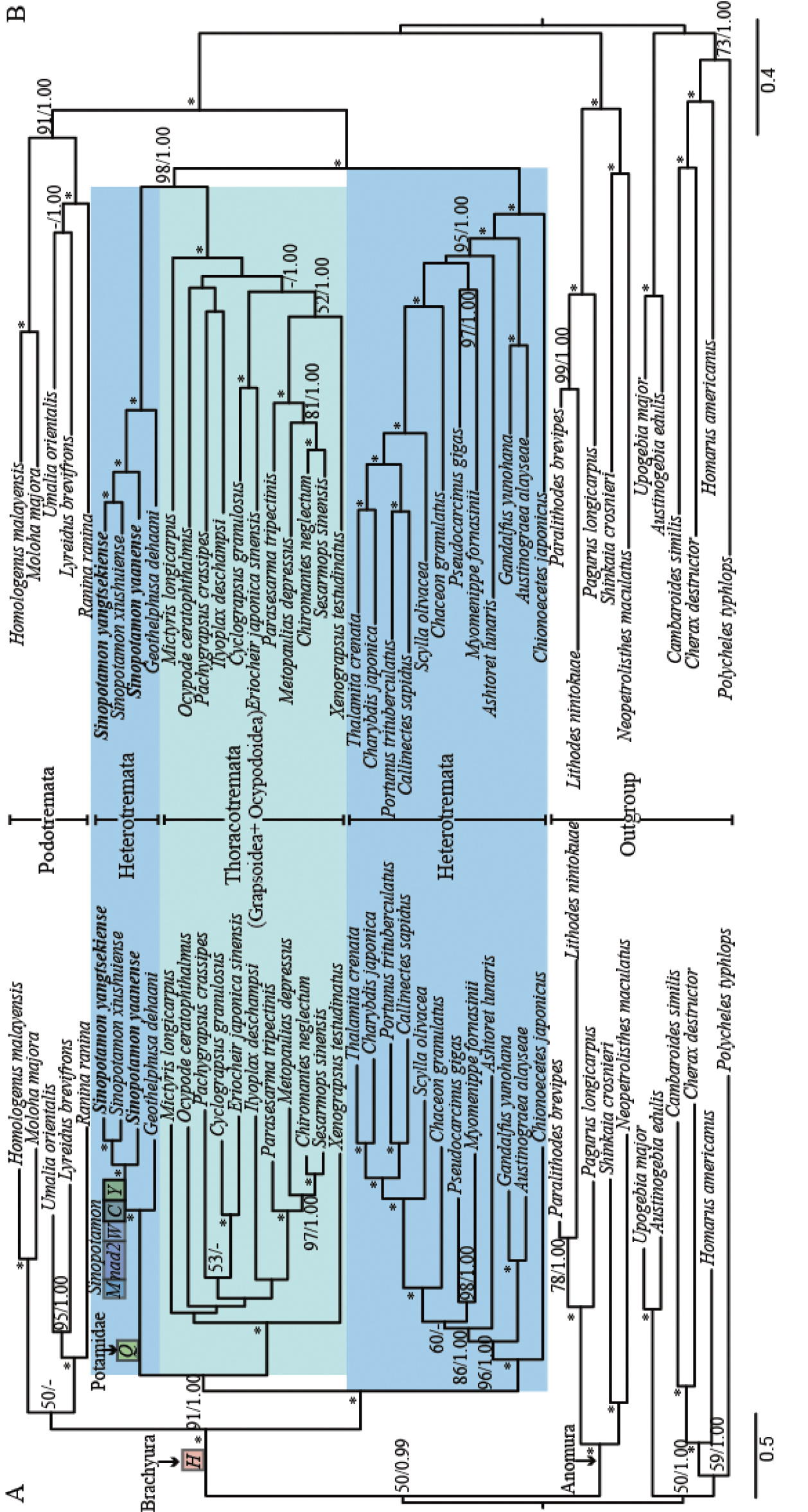
Phylogenetic analyses derived for brachyurans using the maximum likelihood (ML) analyses and Bayesian inferences (BI) using dataset A (13 PCGs) and dataset B (13 PCGs + two rRNAs). Branch lengths and topologies came from ML analysis. Values at the branches represent BP (Bootstrap value)/BPP (Bayesian posterior probability). 100/1.00 is denoted by an asterisk. The horizontal line stands for BP under 50 or BPP under 0.9 ML analyses. The gene rearrangement is denoted by the block on (A): (I) the translocation of trnH shared by the Brachyura taxa sampled; (II) the transposition of trnQ shared by potamid species; (III) the five-gene block, (trnM-nad2-trnW-trnC-trnY), translocation shared by three Sinopotamon crabs sampled. |