(C) 2013 Rimantas Rakauskas. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Rakauskas R, Havelka J, Zaremba A (2013) Mitochondrial COI and morphological specificity of the mealy aphids (Hyalopterus ssp.) collected from different hosts in Europe (Hemiptera, Aphididae). In: Popov A, Grozeva S, Simov N, Tasheva E (Eds) Advances in Hemipterology. ZooKeys 319: 255–267. doi: 10.3897/zookeys.319.4251
Forty three European population samples of mealy aphids from various winter and summer host plants were attributed to respective species of Hyalopterus by means of their partial sequences of mitochondrial COI gene. Used Hyalopterus samples emerged as monophyletic relative to outgroup and formed three major clades representing three host specific mealy aphid species in the Neighbor joining, Maximum parsimony, Maximum likelihood and Bayesian inference trees. Hyalopterus pruni and Hyalopterus persikonus emerged as a sister species, whilst Hyalopterus amygdali was located basally. Samples representing different clades in the molecular trees were used for canonical discrimination analysis based on twenty two morphological characters. Length of the median dorsal head hair enabled a 97.3 % separation of Hyalopterus amygdali from the remaining two species. No single character enabled satisfactory discrimination between apterous viviparous females of Hyalopterus pruni and Hyalopterus persikonus. A modified key for the morphological identification of Hyalopterus species is suggested and their taxonomic status discussed.
Europe, Hyalopterus amygdali, Hyalopterus pruni, Hyalopterus persikonus, molecular phylogeny, mitochondrial COI, morphological key to species
Mealy aphids of the genus Hyalopterus Koch are reported to be serious pests of stone fruits all over the World (
The aim of this study was to elaborate morphological identification key of the genus Hyalopterus based on the material from Europe that was identified by means of partial CO-I sequences.
Forty three population samples of mealy aphids from five European countries were collected from various winter and summer host plants (Table 1). The entire data set has been subdivided: 21 samples (bolded in Table 1) were used for canonical discrimination procedures and subsequent evaluation of the received discrimination functions was performed on remaining 22 samples.
Aphid material used in the present study. Samples used for the morphological discrimination analysis with a priori specified group membership are given in bold.
| Place, date, collection No | GenBank Accession No |
|---|---|
| Prunus domestica (plum) | |
| Galata, Bulgaria, 2012.06.18, z12-101 | JX943533 |
| Costinesti, Romania, 2012.06.13, z12-67 | JX943536 |
| Gilau, Romania, 2012.06.19, z12-114 | JX943537 |
| Toplita, Romania, 2012.06.10, z12-46b | JX943538 |
| Constanta, Romania, 2012.06.14, z12-78 | JX943539 |
| Valu lui Traian, Romania, 2012.06.14, z12-77 | JX943540 |
| Michalovce, Slovakia, 2012.06.08, z12-43a | JX943545 |
| Mezopeterd, Hungary, 2012.06.20, z12-121 | JX943541 |
| Derecske, Hungary, 2012.06.20, z12-123 | JX943542 |
| Gemzse, Hungary, 2012.06.08, z12-44 | JX943543 |
| Jieznas, Prienai distr., Lithuania, 2012.05.30, 12-24 | JX943544 |
| Daugai, Alytus distr., Lithuania, 2012.05.30, 12-31 | JX943547 |
| Ignalina, Ignalina distr., Lithuania, 2012.06.19, 12-65 | JX943549 |
| Prunus cerasifera (cherry plum) | |
| Ditrau, Romania, 2012.06.11, z12-52 | JX943534 |
| Gheorheni, Romania, 2012.06.11, z12-53 | JX943535 |
| Blagojevgrad, Bulgaria, 2012.06.25, 12-81 | JX943550 |
| Alytus, Alytus distr., Lithuania, 2012.05.30, 12-28 | JX943546 |
| Eišiškės, Šalčininkai distr., Lithuania, 2012.06.13, 12-41 | JX943548 |
| Prunus cerasifera var. Pissardii (red plum) | |
| Costinesti, Romania, 2012.06.13, z12-65 | JX943553 |
| Prunus armeniaca (apricot) | |
| Costinesti, Romania, 2012.06.15, z12-88 | JX943551 |
| Murfatlar, Romania, 2012.06.13, z12-64 | JX943531 |
| Vama Veche, Romania, 2012.06.16, z12-93 | JX943552 |
| Mezopeterd, Hungary, 2012.06.20, z12-120 | JX943555 |
| Kairėnai, Vilnius distr., Lithuania, 2010.07.01, z10-5 | JX943558 |
| Prunus persica (peach) | |
| Goron, Bulgaria, 2012.06.09, z12-111 | JX943519 |
| Bucuresti, Romania, 2012.06.13, z12-58 | JX943521 |
| Constanta, Romania, 2012.06.14, z12-79 | JX943522 |
| Costinesti, Romania, 2012.06.15, z12-86 | JX943523 |
| Murfatlar, Romania, 2012.06.13, z12-63 | JX943524 |
| Pieta Porta Alba, Romania, 2012.06.14, z12-70 | JX943525 |
| Valu lui Traian. Romania, 2012.06.14, z12-75 | JX943526 |
| Mezopeterd, Hungary, 2012.06.20, z12-119 | JX943527 |
| Szikso, Hungary, 2012.06.20, z12-124 | JX943528 |
| Csobad, Hungary, 2012.06.20, z12-126 | JX943529 |
| Foro, Hungary, 2012.06.20, z12-127 | JX943530 |
| Prunus persica var. nectarina (nectarine) | |
| Pieta Porta Alba, Romania, 2012.06.14, z12-73 | JX943520 |
| Prunus dulcis (almond) | |
| Varna, Bulgaria, 2012.06.18, z12-104 | JX943517 |
| Varna, Bulgaria, 2012.06.18, z12-108 | JX943518 |
| Prunus maritima (beach plum) | |
| Kairėnai, Vilnius distr., Lithuania, 2010.07.01, z10-4 | JX943557 |
| Phragmites australis (common reed) | |
| Vama Veche, Romania, 2012.06.16, z12-91 | JX943532 |
| Biharkeresztes, Hungary, 2012.06.20, z12-118 | JX943554 |
| Baltupiai, Vilnius, Lithuania, 2010.06.30, z10-1 | JX943556 |
| Palanga, Klaipėda distr., Lithuania, 2010.07.15, z10-24 | JX943559 |
For molecular analysis, a single aphid individual from one sampled plant was considered as a unique sample. Total genomic DNA was extracted from a single aphid using the DNeasy Blood & Tissue kit (Qiagen), which involved at least a 2 h digestion of tissue with proteinase K. Partial sequences of mitochondrial COIwere PCR-amplified using previously published primers (
Forty three sequences of three Hyalopterus species were analyzed. Sequences of Aphis gossypii Glover, 1877 (Aphidini) and Nasonovia ribisnigri (Mosley, 1841) (Macrosiphini) were selected as outgroups for the phylogenetic analyses, which included Neighbor joining (NJ), Maximum parsimony (MP), Maximum likelihood (ML) and Bayesian inference in phylogeny (BI). NJ, MP and ML analyses were performed using MEGA 5 (
Samples representing different clades in the molecular trees were used for canonical discrimination analysis: 2 samples from almond (Hyalopterus amygdali clade), 10 samples from cultivated plums (Hyalopterus pruni clade), and 9 samples from peaches (Hyalopterus persikonus clade) (Table 1).
Based on the earlier references (
A2L – length of antennal segment 2; A2W – width of antennal segment 2; A3BW – basal width of antennal segment 3; A3L – length of antennal segment 3; A3SL – length of the longest hair on antennal segment 3; A4L – length of antennal segment 4; A5L – length of antennal segment 5; A6BL – length of basal part of antennal segment 6; A6TPL – length of terminal process of antennal segment 6; AT8SL – length of submedian hair on abdominal tergite 8; BL – body length (excluding cauda); CL – length of cauda; DT3L – length of the second segment of hind tarsus; F3L – length of hind femur; FSL – length of the frons hair; HW – width of the head across eyes; MDHSL – length of median dorsal head hair; MDHSW – distance between the bases of median dorsal head hairs. SL – length of siphunculus; T3L – length of hind tibia; URL – length of ultimate rostral segment; URW – basal width of ultimate rostral segment.
Measurements of the slide-mounted apterous viviparous females were performed by means of interactive measurement system Micro-Image (Olympus Optical Co. GmbH). STATISTICA 8 version software (Statsoft 2007) was exploited for data analysis. Pearson’s correlation coefficients were calculated to evaluate the correlation of morphometric characters with body length. Characters with strong (| r | ≥ 0.50) statistically significant (p<0.05) correlation with body length were removed from the further analysis: BL (r=1.00), F3L (r=0.58), T3L (r=0.59), A2L (r=0.57), HW (r=0.51). Remaining seventeen characters were used for forward stepwise discriminant analysis with host plant species as grouping variable followed by canonical analysis. Discriminant analysis was conducted in three steps. The first step was performed to discriminate between the all three mealy aphid species emerged in the COI dendrogram (Hyalopterus amygdali, Hyalopterus persikonus and Hyalopterus pruni). The second step was carried out to discriminate between Hyalopterus persikonus and non- Hyalopterus persikonus (Hyalopterus amygdali and Hyalopterus pruni) samples. The third step of the discriminant analysis was performed on Hyalopterus amygdali - Hyalopterus pruni data set (Hyalopterus persikonus samples excluded) to separate almond and plum mealy aphid species. Canonical scores were visualized as scatter plots. The morphological interrelationships among different samples were examined using hierarchical cluster analysis based on squared Mahalanobis distances (linkage method – UPGA).
Characters that contributed most in canonical discrimination functions were evaluated as having potential for species separation. The eventual species identification key based on these morphological characters and host plant information was constructed. Afterwards, it was applied on mealy aphid samples that were not used for the construction of the identification key (Table 1).
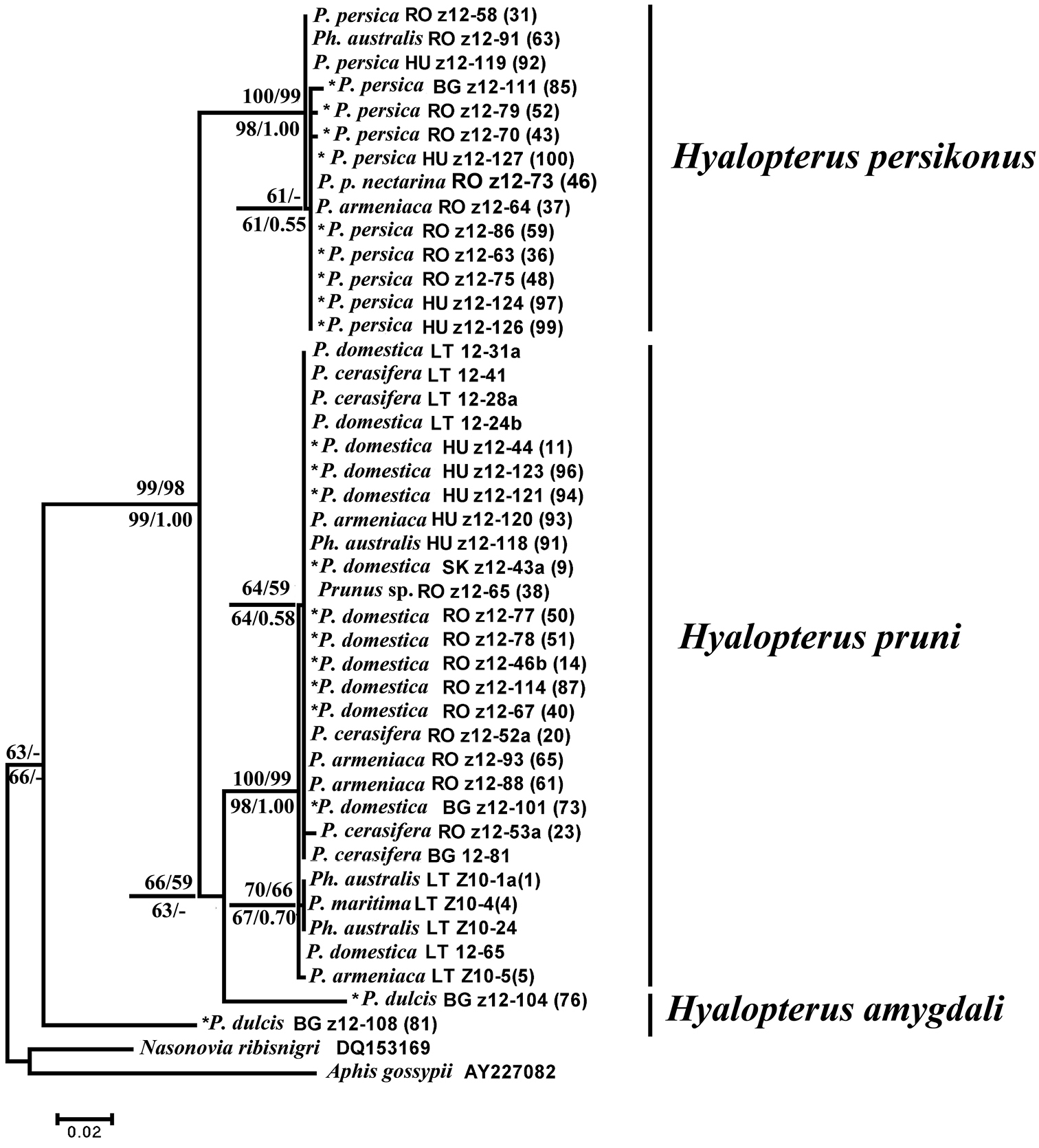
The maximum parsimony (MP) analysis of partial COI sequences resulted in 425 equally parsimonious trees (length = 152, CI=0.76, RI=0.95). ML tree (T92+G model) showed similar topology, the same as NJ analysis (Kimura 2-parameter distances) and BI (GTR+G model) analyses. NJ, MP and ML bootstrap values over 50 % together with BI posterior probabilities over 0.50 are given at respective nodes of the same tree in Fig. 1. One can ensure that used Hyalopterus samples emerge as monophyletic relative to outgroup and form three major clades representing three host specific mealy aphid species. Hyalopterus pruni and Hyalopterus persikonus are placed as a sister species, whilst Hyalopterus amygdali is located basally.
Maximum likelihood (ML) tree showing phylogenetic relationships among three Hyalopterus species based on partial sequences of mitochondrial COI (564 positions in final set). Numbers above branches indicate support of NJ (left) and MP (right) bootstrap test with 1000 replicates, and numbers below branches indicate support of ML (left) bootstrap test with 1000 replicates and posterior probabilities of BI analysis (right). Samples used for the discriminant analysis with a priori specified group membership followed by the construction of identification key are asterisked (*). The remaining samples were used for the post hoc classification. Sample numbers are the same as given in Table 1, together with the abbreviated symbol of respective country: BG Bulgaria, HU Hungary, LT Lithuania, RO Romania, SK Slovakia.
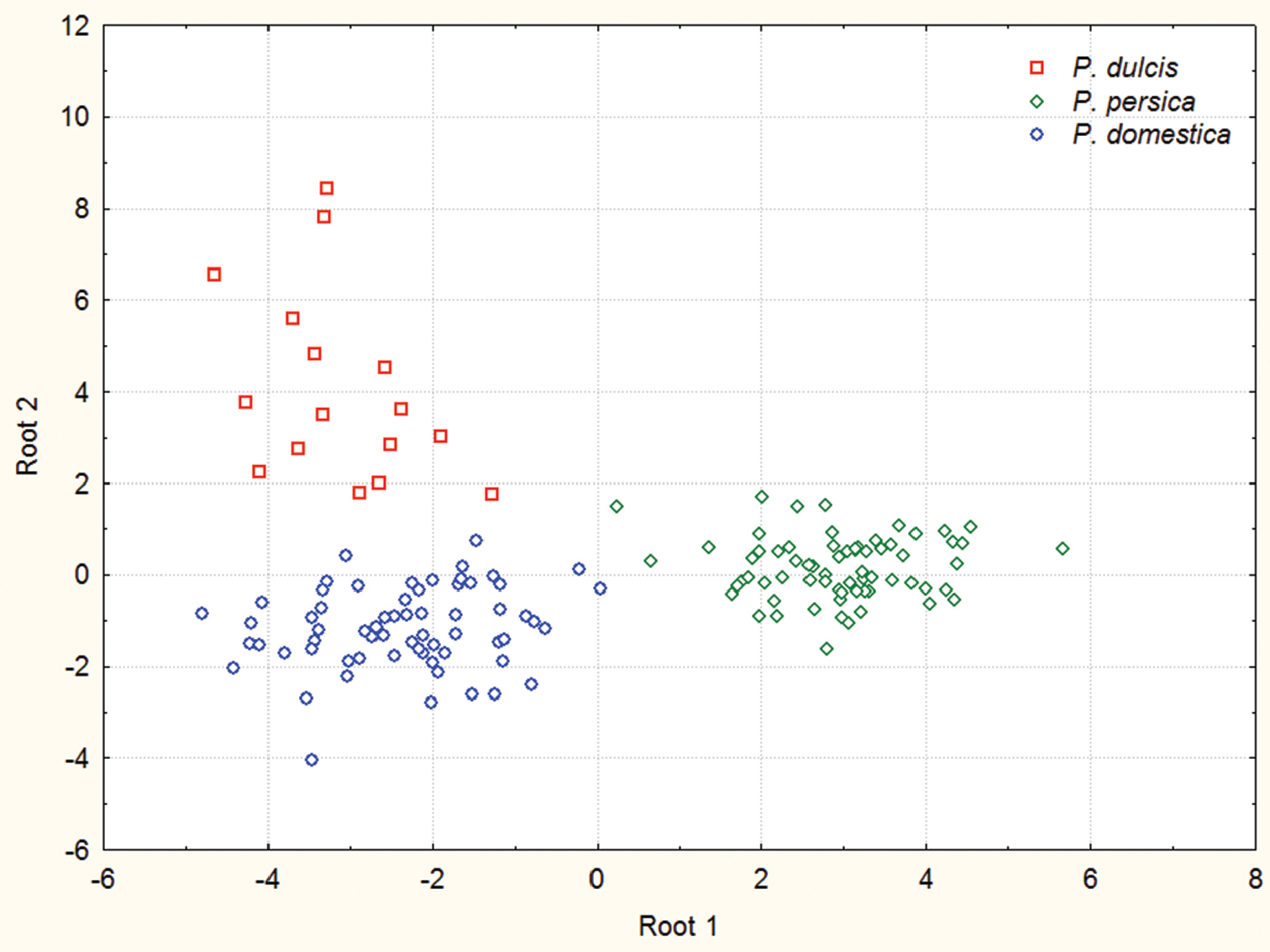
The scatter plot of the first two canonical variates for samples from 18 different geographical localities representing three mealy aphid species (apterous viviparous females) is shown in Fig. 2. All individuals were reclassified correctly into their a priori specified groups. The following characters proved to be important predictors when separating between three Hyalopterus species: MDHSL, URW, T3L/CL (Table 2). The post hoc classification of samples gave 96.7 % correct identification of Hyalopterus persikonus (n=46), 100 % of Hyalopterus amygdali (n=10) and 99% of Hyalopterus pruni (n=94) specimens.
Scatter-plot of the individual canonical scores of the first two canonical variates discriminating 21 samples of Hyalopterus collected from different host plants in five European countries (Bulgaria, Hungary, Lithuania, Romania, Slovakia).
Contribution of eleven morphological characters to the canonical functions discriminating 23 European samples of Hyalopterus. Character abbreviations the same as in the text (Material and methods).
| Wilks’ Lambda | Partial Wilks’ Lambda | F-remove (2, 135) | p-level | Toler. | 1-Toler. (R-Sqr.) | |
| T3L/CL | 0, 05 | 0, 66 | 34, 70 | 0, 00 | 0, 71 | 0, 29 |
| MDHSL | 0, 04 | 0, 81 | 15, 40 | 0, 00 | 0, 14 | 0, 86 |
| URW | 0, 04 | 0, 82 | 14, 33 | 0, 00 | 0, 86 | 0, 14 |
| URL | 0, 04 | 0, 89 | 8, 37 | 0, 00 | 0, 81 | 0, 19 |
| DT3L | 0, 04 | 0, 97 | 1, 98 | 0, 14 | 0, 69 | 0, 31 |
| A6TPL | 0, 04 | 0, 86 | 11, 14 | 0, 00 | 0, 60 | 0, 40 |
| MDHSW | 0, 06 | 0, 58 | 48, 13 | 0, 00 | 0, 12 | 0, 88 |
| MDHSW/MDHSL | 0, 06 | 0, 58 | 49, 50 | 0, 00 | 0, 07 | 0, 93 |
| A5L | 0, 04 | 0, 90 | 7, 57 | 0, 00 | 0, 40 | 0, 61 |
| SL | 0, 04 | 0, 92 | 6, 30 | 0, 00 | 0, 75 | 0, 25 |
| A6BL | 0, 04 | 0, 96 | 3, 04 | 0, 05 | 0, 60 | 0, 40 |
To discriminate between apterous viviparous females of Hyalopterus persikonus and non- Hyalopterus persikonus (Hyalopterus amygdali and Hyalopterus pruni) samples the following canonical function (for character acronyms see above) was obtained: 74.6150*URW-1.2696*T3L/CL+1. The values of canonical scores were >0 for Hyalopterus persikonus and <0 for Hyalopterus amygdali + Hyalopterus pruni. This combination of canonical variables separated 100 % of Hyalopterus persikonus (n=71) specimens involved in the analysis with a priori specified group membership. The post hoc classification gave 94.4 % correct identification of Hyalopterus persikonus (n=46) specimens.
To discriminate between apterous viviparous females of Hyalopterus amygdali and Hyalopterus pruni samples the following canonical function (for character acronyms see above) was obtained: -2.2645*SL-18.6609*MDHSL+1. The values of canonical scores were >0 for Hyalopterus amygdali and <0 for Hyalopterus pruni. This combination of canonical variables separated 94.5 % of Hyalopterus amygdali (n=18) and 100% of Hyalopterus pruni (n=67) specimens involved in the analysis with a priori specified group membership. The post hoc classification gave 100 % correct identification of Hyalopterus amygdali (n=10) and 94.7% of Hyalopterus pruni (n=94) specimens.
Out of eleven morphological characters included in the canonical function discriminating between sampled apterous viviparous females of mealy aphidspecies complex, the length of median dorsal head hair (MDHSL) enabled separation of 97.3 % Hyalopterus amygdali specimens. Namely, the lengths of median dorsal head hair from 0.026 to 0.039 mm were characteristic of Hyalopterus amygdali, whilst 0.036 – 0.067 mm – for other two species. Yet we failed to find any single character or ratio enabling satisfactory discrimination between apterous viviparous females of Hyalopterus pruni and Hyalopterus persikonus. For the present, the following morphological identification key might be suggested to identify apterous viviparous females of the mealy aphid species complex.
| 1 | Canonical discrimination function 74, 6150*URW - 1, 2696*T3L/CL + 1 value exceeding 0. Setae on frons stout. On peaches, nectarines, apricots or reeds | Hyalopterus persikonus |
| – | Canonical discrimination function value less than 0. Setae on frons filiform. On almonds, plums, apricots or reed | 2 |
| 2 | Length of the median dorsal head hair (MDHSL) 0.026 – 0.039 (average 0.031) mm. Canonical discrimination function -2.2645*SL - 18.6609* MDHSL + 1value exceeds 0. On almond or reeds | Hyalopterus amygdali |
| – | MDHSL 0.036 – 0.067 (0.05) mm. Canonical discrimination function value less than 0. On plums, apricots or reeds | Hyalopterus pruni |
Our analysis shows the morphological separation of mealy aphid species complex being a really difficult task which is in accordance with the earlier references (
When performing discriminant analyses, the body length should be eliminated from the data set together with characters that have strong and statistically significant correlation with the body length. In our case, when the entire data set of morphological characters was used for discriminant analysis, samples from reeds appeared the most different (not shown). Contrary, after the body length and correlated characters were removed from analysis, samples from reeds scattered amongst samples from plum and peach.
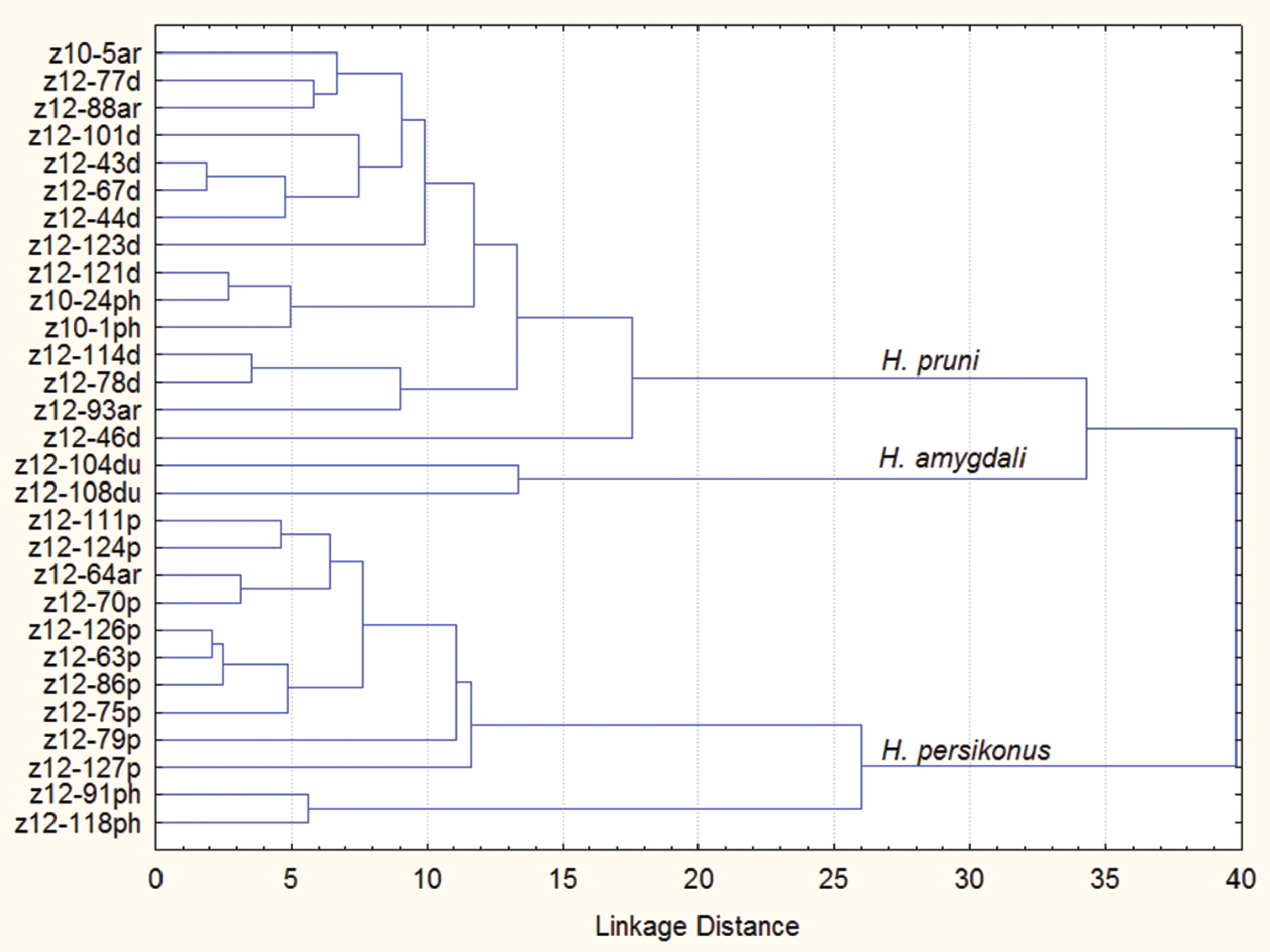
The results of cluster analysis based on morphological data (Figure 3) show Hyalopterus persikonus being more distantly related with Hyalopterus pruni and Hyalopterus amygdali. This contradicts the results of morphological analysis by
Dendrogram of hierarchical cluster analysis based on 17 morphological characters (squared Mahalanobis distances) using unweighted pair-group average linkage among 29 samples of Hyalopterus. Sample numbers the same as in Table 1. ar samples from Prunus armeniaca, d Prunus domestica, du Prunus dulcis, p Prunus persica, ph Phragmites communis.
This research was funded by a grant (No LEK-04/2012) from the Research Council of Lithuania.