(C) 2012 Andre Skale. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Carabdytes upin tindige ssp. n. is described from the Arfak Mountains Bird’s Head Indonesian Papua. It is morphologically very similar to Carabdytes upin upin Balke et al. 1992 known from eastern Indonesian Papua eastward to the western limits of the Papuan Peninsula of Papua New Guinea. For 726 bp at the 3’ end of the mitochondrial cox1 gene the subspecies differ by 8.1–9.2% uncorrected p-distance. However we also document considerable cox1 divergence within Carabdytes upin upin. We find few diagnostic positions in the nuclear genes argenine kinase as well as elongation factor 1 alpha that suggest there are indeed two isolated groups of Carabdytes but evidence in elongation factor 1 alpha is not unambiguous. We decided to highlight this phenomenon of ambiguous evidence for ongoing/just attained speciation by describing a subspecies. We argue that such cases are actually common once mitochondrial sequence data are routinely added to the taxonomist’s toolkit and sometimes simply adding data from few nuclear genes will not suffice the solve taxonomic riddles. Here detailed population genetic investigations would be required – for which sufficient numbers of specimens from a sufficiently wide geographical sampling might be nearly impossible to acquire.
Coleoptera, integrative taxonomy, cryptic species, DNA sequencing, DNA barcoding
Carabdytes Balke et al., 1992, is a genus of New Guinea Colymbetinae diving beetles which to date only contains Carabdytes upin (
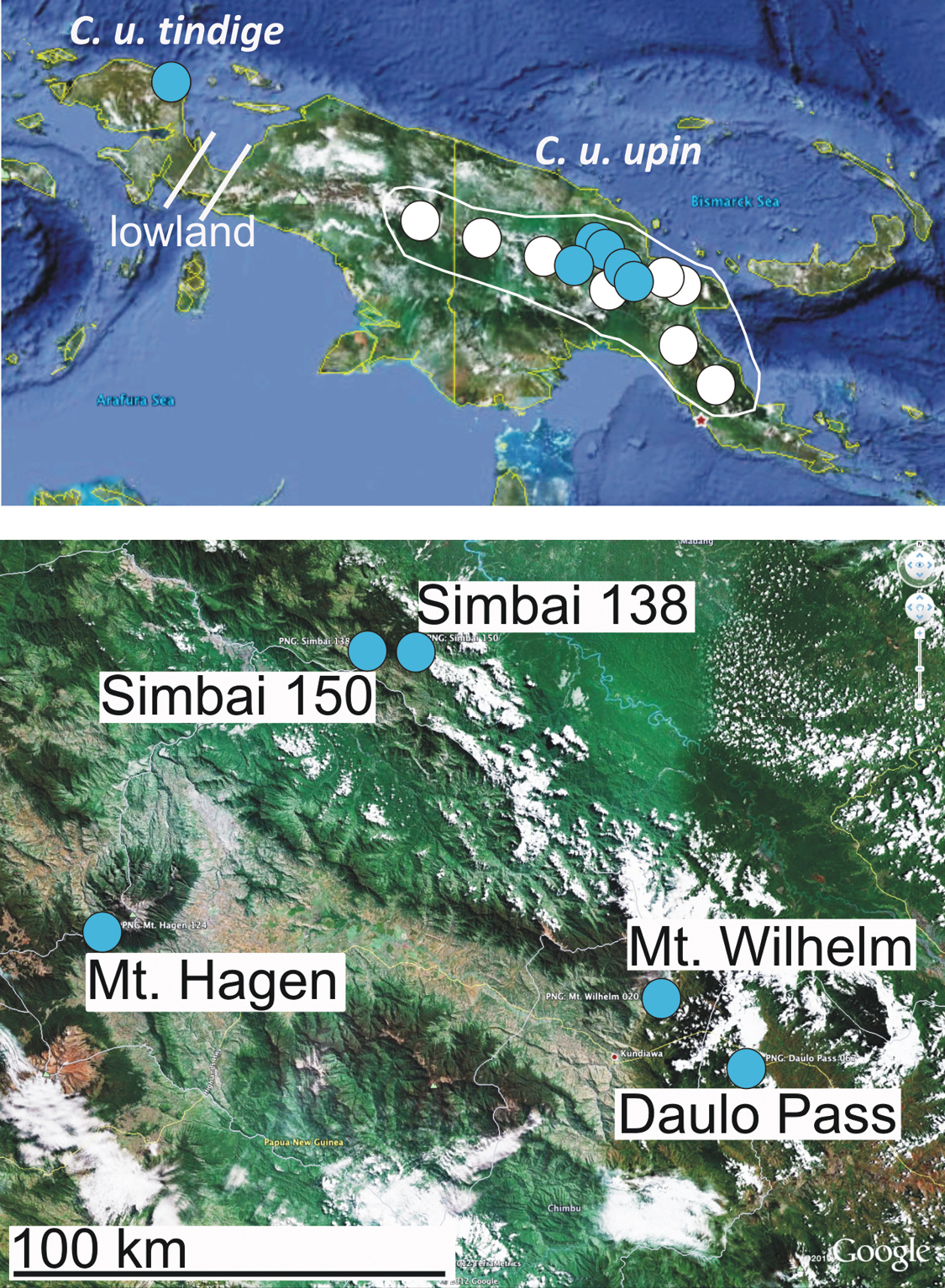
Distribution of Carabdytes upin, blue dots = sequenced specimens, white dots = other records. Lowland gap indicated by bars in the Bird’s neck. Below, detailed map of central Papua New Guinea with localities for sequenced specimens.
Recent molecular phylogenetic analyses revealed that Carabdytes upin belongs to an isolated clade of the Colymbetinae (
During an extensive survey across the island of New Guinea, we obtained Carabdytes samples from several new localities, pushing its range boundary approximately 700 kilometers westward to the Bird’s Head peninsula (Fig. 1). The fresh tissue was used for DNA purification and sequencing to study intraspecific variation. Here, we report surprisingly high mitochondrial DNA divergence in Carabdytes upin. We present evidence from nuclear protein coding genes and morphology that the Bird’s Head beetles might belong to a different species. We also describe the taxonomist’s dilemma when there is some evidence for the presence of cryptic species but perhaps not enough and there is no straightforward solution as the required additional localities are very remote and extremely difficult to visit. We argue this scenario might be not so rare, and new technology eagerly awaited by the traditional taxonomist does not always provide a fast and complete solution of the “old problems”.
Beetles were preserved in 95% ethanol and flight muscle tissue was used for DNA purification. The laboratory methods employed are detailed on our DNA laboratory wiki: http://zsm-entomology.de/wiki/The_Beetle_D_N_A_Lab . PCR conditions with Mango Taq (Bioline) were for cox1: (primers: Jerry/Pat,
We use GARLI V.0.951 (
Digital images were taken with a Nikon D3X with a Voigtländer Apo Lanthar 90 mm attached to a bellows; fitted to a custom built macro rail (image steps used: 0.4 mm). Image stacks were aligned and assembled with the computer software Helicon Focus 4.77TM.
Institutional abbreviations:
CSH Coll. Andre Skale, Hof/Saale, Germany
MZB Museum Zoologicum Bogoriense, LIPI, Cibinong, Indonesia
NMW Naturhistorisches Museum Wien, Austria
ZSM Zoologische Staatssammlung München, Germany
Other abbreviation:
PNG Papua New Guinea
Locality data for specimens of Carabdytes upin studied for this paper (* sequence data available, see Table 1):
Papua New Guinea
* Papua New Guinea: Simbu, Mt Wilhelm, lower lake from Keglsugl, 3500–3700 m, 23.ix.2002, ca. 05.53.733S, 145.02.742E, Balke & Sagata leg. (PNG 020) (ZSM)
* Papua New Guinea: Eastern Highlands, Goroka, Daulo Pass, 2500m, 19.v.2006, 06.02.432S, 145.13.333E, John & Balke leg. (PNG 67) (ZSM)
Papua New Guinea: Southern Highlands, Sopulkul, 30–35Km NE Mendi, from swamp that drains into stream, 2679m, 16.vi.2006, 06.02.944S, 143.46.485E, John leg. (PNG 79) (no DNA sequence data) (ZSM)
* Papua New Guinea: Enga, Kumul Lodge @ foot of Mt Hagen, 2700m, 5.xii.2006, 05.47.548S, 143.58.761E, Balke & Kinibel leg. (PNG 124) (ZSM)
* Papua New Guinea: Western Highlands, Simbai, 1800–2000m, 1.iii.2007, 05.14.276S, 144.28.741E, Kinibel leg. (PNG 138) (ZSM)
* Papua New Guinea: Western Highlands, Simbai area, 2500m, 8.iii.2007, 05.14.202S, 144.33.651E, Kinibel leg. (PNG 150) (ZSM)
Indonesia: Papua
Jayawijaya Mts., Aipomek-Diruemna, 2600m, 3.ix.1992, 04.26S, 139.57E, Balke leg. (no DNA sequence data) (NMW)
Indonesia: West Papua
* Bird’s Head, Manokwari, Mokwam (Siyoubrig), 1400–1800m, 24.–28.II.2007, 01.06.26S, 133.54.41E, Skale leg. (CSH, MZB, ZSM)
Sequenced Carabdytes specimens and EMBL accession numbers.
| cox1 | EF1α | ARK | ||
|---|---|---|---|---|
| Carabdytes upin tindige MB 3084 | Papua: Arfak | HF558675 | HF558686 | HF558698 |
| Carabdytes upin MB 3328 | PNG 138: Simbai | HF558676 | HF558687 | HF558699 |
| Carabdytes upin MB 3354 | PNG 067: Daulo Pass | HF558677 | HF558688 | HF558700 |
| Carabdytes upin MB3452 | PNG 150: Simbai | HF558678 | HF558689 | |
| Carabdytes upin MB3453 | PNG 150: Simbai | HF558679 | HF558690 | |
| Carabdytes upin MB3454 | PNG 150: Simbai | HF558680 | HF558691 | |
| Carabdytes upin MB3455 | PNG 150: Simbai | HF558681 | HF558692 | |
| Carabdytes upin MB3045 | PNG 124: Mt. Hagen | HF558682 | HF558693 | HF558701 |
| Carabdytes upin MB4316 | PNG 067: Daulo Pass | HF558683 | HF558694 | |
| Carabdytes upin MB4317 | PNG 138: Simbai | HF558684 | HF558695 | HF558702 |
| Carabdytes upin MB4318 | PNG 138: Simbai | HF558685 | HF558696 | HF558703 |
| Carabdytes upin MB0306 | PNG 020: Mt. Wilhelm | FN263070.1 | HF558697 |
Elongation factor 1 alpha. We obtained a 555 bp fragment in the 5’ region of EF1α for all specimens shown in Figure 2. There are 3 diagnostic nucleotide substitutions for a specimen from the Bird’s Head (MB3084, see Fig. 1) (positions 36, 75, and 357, all 3rd codon positions) delineating this specimen from the other Carabdytes.
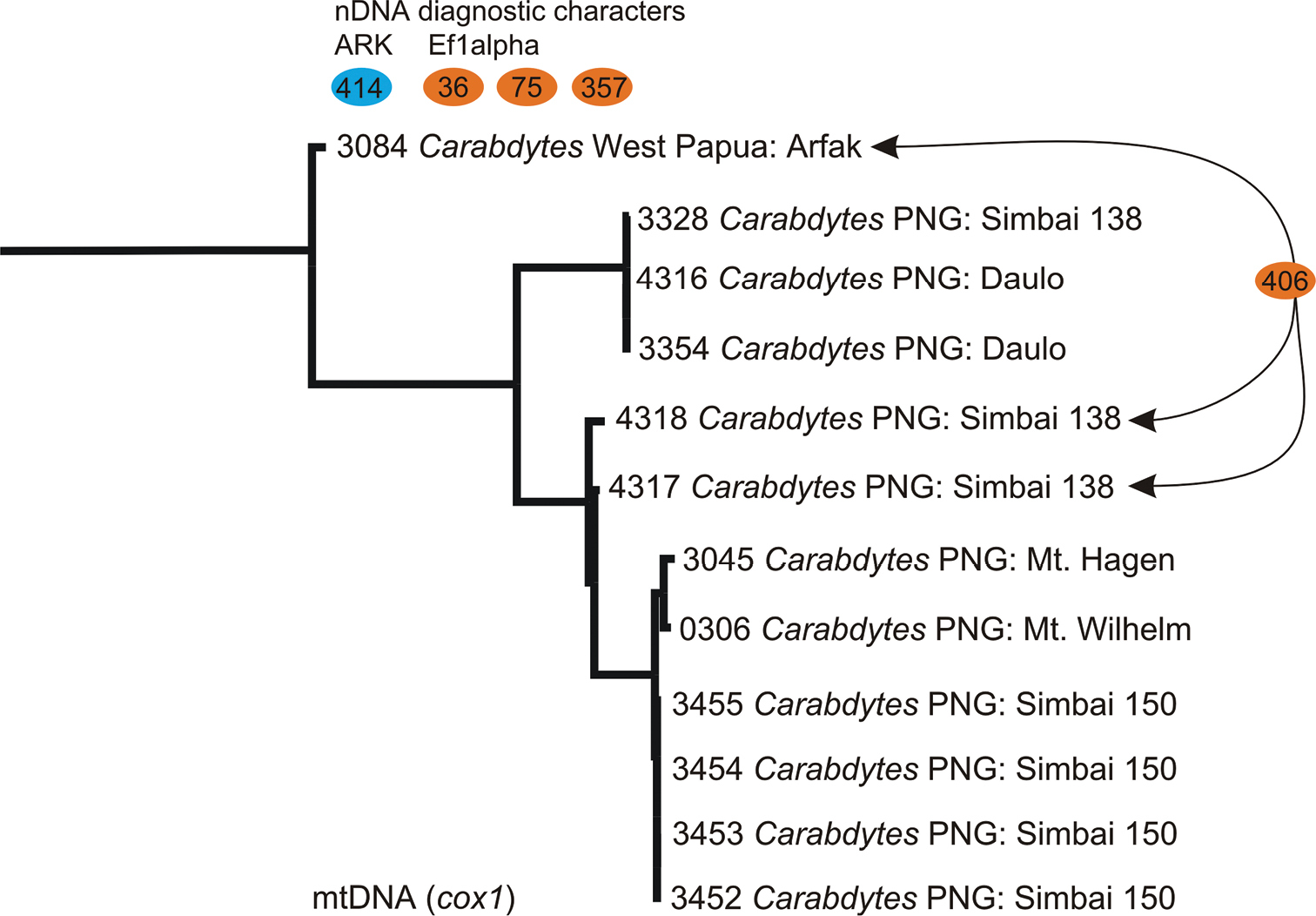
Maximum likelihood tree based on cox1 data, the tree was rooted with Rhantus guadalcanalensis Balke (pruned here), colored buttons represent diagnostic nuclear DNA characters and their positions in our sequence alignment (there are found characters supporting the node between Carabdytes 3084 and all others, and one that is shared between 3084 as well as the two specimens 4317 & 4318 in Simbai 138) .
Specimens MB3084 (Bird’s Head) and MB4317, 4318 (PNG: Simbai) share a non synonymous substitution in position 406 (1st codon position).
Arginine Kinase. We obtained 656 bp of sequence data for 6 individuals of Carabdytes (Table 1). There was one diagnostic character, a 3rd codon substitution in position 414 of our alignment. This character delineates the Bird’s Head specimen from the other Carabdytes. The sequences are otherwise identical.
Cytochrome c oxidase 1. We obtained a 726 bp fragment at the 3’ of cox1 for 12 individuals of Carabdytes (Table 1). Sequence data were surprisingly divergent, although most of the samples all originate from one major region in eastern New Guinea (Figs 1, 2). Uncorrected p-distances were 0–9.23%. There are 29 unambiguous diagnostic characters delineating the Bird’s Head specimen from the other Carabdytes, all of them in 3rd codon positions.
The clade MB3328 / MB3354 / MB4316 has 20 diagnostic characters (and two 1st codon substitutions resulting in amino acid change, pos. 316 and 415 in our alignment) and MB0306 / MB 3045 / MB3452–55 has 5 diagnostic characters (Fig. 2).
Cluster analysis. At 3% preset threshold, SpeciesIdentifier finds four cox1 clusters, which agree with the three main lineages of the tree in Fig. 2 (MB4317 and MB4318 form one cluster). For the nuclear markers, all data only form a single cluster at 3%.
Morphology.Four specimens were available from the Bird’s Head for morphological study. The distinguishing feature between these specimens and Carabdytes upin from eastern localities in New Guinea is: Pronotum and elytra conspicuously shining with very indistinct punctuation (the elytra have a conspicuous coarse punctuation, especially on the apical half, in eastern Carabdytes upin) (Fig. 3). Specimens of Carabdytes upin studied for this comparison come from the localities mentioned above, covering most of its range (no specimens studied from Huon Peninsula and Wau). The specimens from Simbai (locality PNG138) have an intermediate elytral punctation, with only few coarse punctures on the apical part, while specimens from Simbai (loc. PNG150) which is less than 20 kilometers apart (Fig. 1) are coarsely punctate. The Simbai localities both have specimens with attached sequence data.
Morphological characters, above habitus, left Carabdytes upin tindige from the Bird’s Head (12.0 mm long), right Carabdytes upin upin from Papua: Aipomek area (12.9 mm long); below left median lobe of aedeagus in lateral view of Carabdytes upin tindige (Carabdytes upin upin is identical), its paramere, right, paramere of Carabdytes upin upin from Papua: Aipomek area. Scale for genitalia is 0.1 mm.
For Carabdytes upin, we do suggest to flag the Bird’s Head beetles with a subspecies name, assuming the combined, congruent observations described above are evidence for longer periods of interrupted gene flow. We suggest the use of a subspecies name to stimulate further investigation to verify or falsify this hypothesis.
West Papua, Arfak Mountains, Siyoubrig , 01°06.26'S, 133°54.41'E.
Holotype: ♂ (MZB): Indonesia, West Papua, Arfak Mountains, stream near Siyoubrig 01°06.26'S, 133°54.41'E, 1400–1800 m, 24. –28.II.2007, leg. A. Weigel. Paratypes: 2♂♂ 1 ♀ (CSH, ZSM): same locality data as holotype (the female was sequenced).
Habitus as in Fig. 3; total length: 11.6–12.0 mm; total width: 5.3–5.5 mm. Dark brown to almost black; labrum, lateral margin of pronotum and all body appendages paler reddish brown; elongate.
Head black, labrum reddish brown, clypeus with indistinctly reddish colour almost reaching eyes; head with indistinctly reddish patch on frons. Pronotum black, with indistinctly median reddish patch, posterior angles reddish. Ventral surface blackish. Venter dark brown.
Structures. Head with fine, sparse punctation interspersed with coarser punctures between eyes and behind anterior clypeal margin. Pronotum shining, posterior angles with irregular, coarse punctures, lateral margin very strongly. Elytra shining with very indistinctly punctures; each elytra with four rows of coarser, moderately arranged punctures. Lateral wing of metaventrite broad and tongue-shaped; outer margin slightly sinuate; last abdominal ventrite medially emarginate. Legs long and slender.
Male. Pro- and mesotarsal claws of similar structure; anterior and posterior claws moderately long and evenly curved; Median lobe of aedeagus relatively slender (Fig. 3, shape in Carabdytes upin upin is identical); paramere slender, with distinctly longitudinal striation; setation more or less long (Fig. 3), the setation might be basally shorter in some specimens of Carabdytes upin upin (Fig. 3), but this difference does not appear constant.
Distinguished from Carabdytes upin upin through the molecular and morphological characters mentioned in the results section under “morphology”. With Carabdytes upin upin there is no overall size difference (10.1–12.2 mm).
Two individuals collected with an aquatic net from the rough gravel at the edge of a stream bed, the stream was rather dry at the time of collection (Fig. 4). The species co-occured with Platynectes spp., Exocelina (=Papuadytes) spp. and Hydraena cristatigena Jäch & Diaz. Two exemplars were collected with the help of a light trap, approx. 50 m away from the stream.
Type locality of Carabdytes upin tindige ssp.n.
So far known only from the type locality (Figs 1, 4).
In loving memory of Samkris “Kris” Tindige, relentless conservationist in Papua, who left us too early. The beetles were collected in the stream bed very close to a birdwatching guesthouse set up by Kris and Shita Prativi above Siyoubrig village.
Here, we document mitochondrial DNA divergence of up to 9% within Carabdytes upin. Our samples mainly originate from the core range of this species, from eastern New Guinea. One specimen from the Bird’s Head Peninsula in the west of New Guinea, about 700 km west of the next known locality for Carabdytes upin, is well separated geographically from other populations. It is also most divergent genetically. The mountain regions between the known localities are understudied, but some (wider Wamena area eastwards to Diruemna; Nabire area up to Enarotalia; Cyclops Mountains near Jayapura) have specifically been screened for diving beetles. Carabdytes upin was not yet collected there. The vast expanse of karst as well as tropical lowland in the Bird’s neck region, roughly from Lake Yamur westwards to Arfak Mountains, offers few obvious habitats for Carabdytes upin, with Wandammen Peninsula as a potential stepping stone (though the species was not yet detected there) (Fig. 1, “lowland”).
Intraspecific mitochondrial cox1 divergences >3 % are considered high in Dytiscidae. For the Australian fauna, largest intraspecific distances reported by
Cryptic species are apparently more common and phylogenetically more widespread than assumed previously (
In the morphologically highly similar Carabdytes upin, we find geographical separation and high cox1 divergence. In the nDNA marker Arginine Kinase, we find one diagnostic character for the Bird’s Head beetle, in elongation factor 1 alpha(EF1α) there are three, but all of these are synonymous substitutions not altering the amino acid sequence and thus protein derived from the nucleotide sequence. For EF1α, there is another substitution, but this one is shared between the Bird’s Head specimen and two specimens from eastern New Guinea (Simbai, PNG138, MB4317 & 4318) (Figs 1, 2). A third specimen from the Simbai PNG138 locality has the same EF1α genotype as all other Carabdytes upin. Within Carabdytes upin from eastern New Guinea, we also observe considerable mtDNA variation, up to 7.7% (Table 2). Importantly, this also concerns close localities such as Simbai PNG138 and PNG150, less than 10 km apart. Moreover, haplotypes from locality Simbai PNG138 also differ around 7% from each other.
Uncorrected cox1 p-distances for Carabdytes specimens (* the new subspecies from the Bird’s Head).
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0306 Carabdytes PNG Mt Wilhelm | 1 | __ | |||||||||||
| 3045 Carabdytes PNG Mt Hagen | 2 | 0.010 | __ | ||||||||||
| 3452 Carabdytes PNG150 Simbai | 3 | 0.010 | 0.011 | __ | |||||||||
| 3453 Carabdytes PNG150 Simbai | 4 | 0.010 | 0.011 | 0.000 | __ | ||||||||
| 3454 Carabdytes PNG150 Simbai | 5 | 0.010 | 0.011 | 0.000 | 0.000 | __ | |||||||
| 3455 Carabdytes PNG150 Simbai | 6 | 0.010 | 0.011 | 0.000 | 0.000 | 0.000 | __ | ||||||
| 3328 Carabdytes PNG138 Simbai | 7 | 0.077 | 0.077 | 0.073 | 0.073 | 0.073 | 0.073 | __ | |||||
| 4316 Carabdytes PNG067 Daulo | 8 | 0.076 | 0.076 | 0.072 | 0.072 | 0.072 | 0.072 | 0.001 | __ | ||||
| 3354 Carabdytes PNG067 Daulo | 9 | 0.076 | 0.076 | 0.072 | 0.072 | 0.072 | 0.072 | 0.001 | 0.000 | __ | |||
| 4318 Carabdytes PNG138 Simbai | 10 | 0.040 | 0.047 | 0.039 | 0.039 | 0.039 | 0.039 | 0.068 | 0.066 | 0.066 | __ | ||
| * 3084 Carabdytes West Papua Arfak | 11 | 0.087 | 0.080 | 0.086 | 0.086 | 0.086 | 0.086 | 0.093 | 0.091 | 0.091 | 0.090 | __ | |
| 4317 Carabdytes PNG138 Simbai | 12 | 0.032 | 0.039 | 0.030 | 0.030 | 0.030 | 0.030 | 0.070 | 0.069 | 0.069 | 0.014 | 0.084 | __ |
Thus, there is considerable cox1 variation, as expected in running water organisms, or species in highly fragmented habitats in general (
What are the practical implications from the beetle taxonomist’s point? Mitochondrial DNA variation alone does not provide sufficient evidence. While divergence between eastern and western localities is high, such is divergence even within one of the eastern localities, as well. Thus, we tried to find other congruent evidence that might indicate presence of a cryptic species. In the nuclear genes Arginine Kinase and elongation factor 1 alpha, we count a total of 4 diagnostic characters (Fig. 2). This is additional evidence, combined with the high mtDNA divergence, of interrupted gene flow over longer periods. It is interesting to note that two specimens from Simbai (locality PNG138) which also diverge highly from other Carabdytes upin share 1 diagnostic EF1α position with the Bird’s Head specimen. As described above, the Simbai (PNG138) specimens are morphologically intermediate between the Bird’s Head specimens and other studied Carabdytes upin. Overall molecular evidence suggests they belong to the eastern clade, presence of a shared substitution in EF1α between PNG138 and EF1α can not be explained based on the available data. The generally high mitochondrial divergence indicates complex mechanisms are at work, and the mtDNA data are not necessarily the answer but rather a starting point for a population genetic study in its own right.
This project was supported by Deutsch Forschungsgemeinschaft through various grants to Michael Balke (e.g. DFG BA2152/7-1). This work was considerebly improved through comments from two reviewers, as well as from journal editor Dr. Martin Fikacek (Prague).