(C) 2011 Baoping Li. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The grasshopper family Catantopidae is a well-known group, whose members include some of the most notorious agricultural pests. The existing classifications of the family are mostly utilitarian rather than being based on phylogenetic analysis and therefore unable to provide the stability desired for such an economically important group. In the present study, we present the first comprehensive phylogenetic analysis of the family based on morphology. By extensively sampling from the Chinese fauna, we included in the present analysis multiple representatives of each of the previously recognized tribes in the family. In total, we examined 94 genera represented by 240 species and evaluated 116 characters, including 84 for external morphology and 32 for male genitalia. The final matrix consists of 86 ingroup taxa and 88 characters. Our phylogenetic analyses resulted in a high resolution of the basal relationships of the family while showed considerable uncertainty about the relationships among some crown taxa. We further evaluated the usefulness of morphological characters in phylogeny reconstruction of the catantopids by examining character fit to the shortest trees found, and contrary to previous suggestions, our results suggest that genitalia characters are not as informative as external morphology in inferring higher-level relationship. We further suggest that earlier classification systems of grasshoppers in general and Catantopidae in particular most probably consist of many groups that are not natural due the heavy reliance on genitalia features and need to be revised in the light of future phylogenetic studies. Finally, we outlined a tentative classification scheme based on the results of our phylogenetic analysis.
Orthoptera, Acridoidea, Catantopidae, China, Phylogeny, Morphology, Systematics, Male Genitalia
Zhiwei Liu would like to dedicate this paper to the honor of Professor Kumar Krishna for his friendship, kindness, professional encouragement, and the good times at the AMNH.
IntroductionCatantopidae (Acridoidea, Orthoptera) is a well-known grasshopper family; its members include some of the most notorious pests in agriculture, including Schistocerca gregaria (Forsköl), Oxya spp, and Melanoplus spp (Hill 1987). The family is by far the largest and the most diverse acridoid family, consisting of over 3000 species in about 640 genera mainly distributed in the tropical and subtropical areas of the world (
The previous classifications of Acridoidea (Orthoptera) have been predominantly utilitarian; existing classifications of the superfamily almost entirely ignored phylogenetic relationships among taxa. Among the various classification systems or schemes of acridoids (
Classification systems of the Catantopid fauna from China
|
|
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|
| Cyrtacanthacrinae | Catantopinae | Acrididae | Catantopidae | Hemicarididae | Oedipodidae | Catantopidae | Acrididae |
| Hemiacridinae | |||||||
| Conophymatini | Hemiacaridinae | Conophyminae | Conophyminae | Conophyminae | Conophyminae | ||
| Spathosternini | Spathosterninae | Spathosterninae | Spathosterninae | Spathosterninae | |||
| Leptacri | Leptacrinae | Leptacrinae | |||||
| Caryandae | Catantopidae | ||||||
| Dericorythini | Dericorythinae | Dericorythinae | Dericorythinae | Dericorythinae | Dericorythinae | (Dericorythidae) | |
| Oxyae | Oxyini | Oxyinae | Oxyinae | Oxyinae | Oxyinae | Oxyinae | Oxyinae |
| Catantopini | Catantopini | Catantopinae | Catantopinae | Catantopinae | Catantopinae | Catantopinae | Catantopinae |
| Calliptamini | Calliptamini | Calliptaminae | Calliptaminae | Calliptaminae | Calliptaminae | Calliptaminae | |
| Eyprepocnemini | Eyprepocnemidini | Eyprepocneminae | Eyprepocneminae | Eyprepocneminae | Eyprepocneminae | Eyprepocnemidinae | |
| Cyrtacanthacridini | Cyrtacanthacridini | Cyrtacanthacridinae | Cyrtacanthacridinae | Cyrtacanthacridinae | Cyrtacanthacridinae | Cyrtacanthacridinae | |
| Coptacrae | Coptacrini | Coptacrinae | Coptacrinae | Coptacrinae | Coptacridinae | ||
| Podisminae | Podismini | Podisminae | Podisminae | Podisminae | Podismini | ||
| Tropidopolini | Tropidopolinae | Tropidopolinae | Tropidopolinae | Tropidopolinae | |||
| Tristrini | Tristriini | Tristrinae | (Tristiridae) | ||||
| Hieroglyphini | Hieroglyphinae | Hieroglyphinae | |||||
| Trauliae | Trauliini | ||||||
| Oxyrrhepini | Habrocneminae | Habrocneminae | Habrocneminae | ||||
| Xenacanthippi | Melanoplinae | Melanoplodinae | |||||
| Tauchirae | Acrididae | Acridinae | |||||
| Incolacri | Leptacrinae | ||||||
| Egnatiinae | Egnatiidae | Egnatiinae | Egnatiinae | Egnatiinae | Egnatiinae | Egnatinae | |
| Acrididae | Gomphoceridae | Gomphoceridae | Gomphocerinae |
The need for a classification of the grasshoppers and locusts based on phylogeny, rather than based on overall similarity, is obvious.
There has been an increased interest in recent years in the phylogenetic relationship of orthopteroid insects in general (
This lack of higher-level phylogenetic study of Catantopidae leads to a lack of stability in the classification within the family (Table 1), which is unusual for such a well-known and economically important group. In this paper, we present the first comprehensive phylogenetic analysis of the family Catantopidae based on morphology by sampling extensively from the Chinese fauna. Our purpose is to (1) conduct an exploratory phylogenetic analysis of the phylogenetic relationship within the family represented by the Chinese members, (2) provide an objective evaluation of the usefulness of morphological characters in phylogeny reconstruction of the acridoids in general and the catantopids in particular, and (3) provide a general framework for taxon sampling in future studies of acridoid phylogeny on a global basis.
Materials and methods I. MonophylyThe name Catantopidae, or its original form Catantopinae as subfamily, has had a long history of divergent usages (Key and Colles 1993). The modern definition of Catantopidae took after the name of Cyrtacanthacrinae (
Catantopidae in our view is readily defined by the unmistakable synapomorphy of having a distinct prosternal process between the forecoxae. Although some species of Pamphagidae and Pyrgomorphidae have a lamellate process on the prosternum, the process in these species is on the anterior margin of the prosternum and is obviously an independently evolved feature not homologous to the prosternal process between the forecoxae observed in Catantopidae. Nonetheless, as shown in Table 1, there was considerable disagreement among earlier authors about the definition of Catantopidae, which obviously arose from the fact that earlier acridologists defined higher-level taxa on basis of overall similarities, instead of on synapomorphies. Our interpretation of Catantopidae in the present paper, as defined by the presence of prosternal process between the forecoxae, is in accordance with Catantopinae of
About 327 species in 96 genera of Catantopidae (sensu
The majority of the study materials of the present project were provided by the following institutions (curators in parentheses):
Entomological Museum, Shaanxi Normal University, Xi’an, Shaanxi Province (Shengquan Xu)
Entomological Museum, Zhongshan University, Guangzhou, Guangdong (Geqiao Liang)
Entomological Museum, Research Institute of Entomology, Chinese Academy of Sciences, Shanghai (Kailing Xia)
Entomological Museum, Beijing Institute of Zoology, Chinese Academy of Sciences, Beijing (Chunmei Huang)
Zoological Museum, Northwest Plateau Institute of Biology, Chinese Academy of Sciences, Xining, Qinghai (Xiangchu Yin)
III. Selection of outgroupsBecause of the lack of phylogenetic analysis of Acridoidea at levels above subfamily, we had to rely on previous systematic studies on Acridoidea for outgroup selection. All existing classifications of Acridoidea treated Catantopinae, Egnatiinae, Acridinae, and Oedipodinae as being closer to each other than they are to Pyrmorphinae and Pamphaginae (
Terms and abbreviations used in the present study followed
Specimens for the study were selected in the following order of priority: 1) type specimens, 2) specimens determined by the author of the taxon, and 3) specimens determined by experts of the taxon. All characters were coded from direct observation of specimens, except in a few instances where characters of a species were coded based on illustrations and descriptions from monographs or reviews (
External morphology was surveyed before specimens were dissected for examination of genitalia characters. When available, multiple individuals were examined for each species and multiple species for each genus. For polymorphism at species level, we took an approach similar to, but much more restricted than, the “majority state rule” proposed by
The final matrix contained 87 terminals, including outgroup and 86 catantopid genera in the ingroup, and 88 characters, of which 79 were phylogeny-informative and the other nine were autapomorphies (Appendix 2-3). The autapomorphic characters were excluded from the final cladistic analyses and not counted when calculating tree length, CI, or RI. Nonetheless, they were kept in the matrix for their taxonomic values and potential use in future phylogenetic studies involving the included taxa.
V. Phylogenetic AnalysisPAUP version 4.0 beta10 (
We examined a total of 116 characters, including 84 characters of external morphology and 32 characters of male genitalia morphology. Twenty-eight characters were excluded from out analysis either because they were too variable across examined species of a genus to reach a generic consensus or because they were continuous and discrete coding of character states was impossible. In addition, characters of body color patterns, although important for identification of some species of the family, were found to be too variable, both among individuals of species and among species of genera, to be of much use in resolving phylogenetic relationships within Catantopidae and were therefore excluded from the present study. The eighty-eight characters included in the final character matrix consist of 71 external morphological characters and 17 genitalia characters (Appendix 2). Character fit on the shortest trees, as expressed by the consistency index (CI) and retention index (RI), was lower for characters of male genitalia morphology in comparison to characters of external morphology (Table 2).
Fit on shortest trees of different categories of characters, as expressed by the consistency index (CI) and retention index (RI) (n = number of characters; autapomorphis excluded).
| Character Category | n | CI | RI |
| External Morphology | 63 | 0.19 | 0.58 |
| Body shape | 1 | 0.25 | 0.63 |
| Head | 10 | 0.17 | 0.54 |
| Mesosoma | 29 | 0.20 | 0.66 |
| Metasoma | 23 | 0.20 | 0.45 |
| Male Genitalia | 16 | 0.12 | 0.49 |
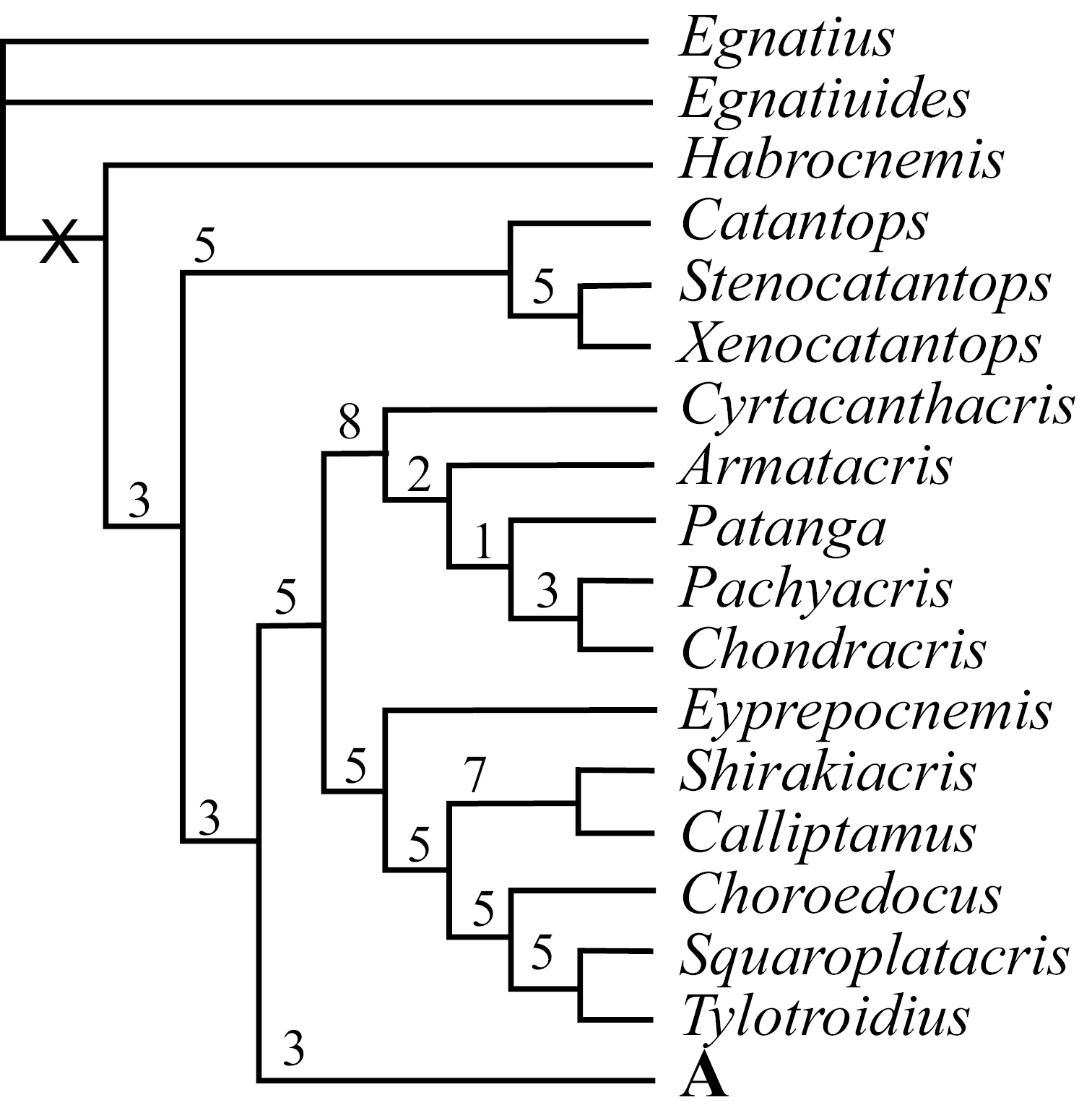
Using maximum parsimony analysis with Nixon’s ratchet method, we found in thirteen of our 30 replications and 218 of the 6, 000 iterations a tree with the shortest length of 688 steps (L=688, CI = 0.17, RI = 0.55). With duplicate trees deleted, the final number of the shortest trees was 204;subsequent swapping of these optimal trees using TBR did not find shorter trees, but found a total of 22, 354 equally most parsimonious trees. Figs. 1–2 and Fig. 3 show the strict consensus tree with Bremer Support for completely resolved branches and the 50% majority consensus tree with percentage of branches appearing in all shortest trees summarized, respectively.
Strict consensus tree of the 22, 355 found shortest trees using Parsimony Ratchet method in PAUP 4.0 beta10 (30 repetitions and 200 iterations per run, followed by TBR swapping). Above each resolved branch is the Bremer Support value (/decay index) for the branch estimated using NONA2.0. Only the completely resolved basal part is shown.
Searching with NONA 2.0 (Hold=10, 000–30, 000, Mult*50, and Max*) did not find trees shorter than those found with PAUP 4.0 using parsimony ratchet method. Although we were always able to find trees of the shortest length in a few minutes with NONA, our searches invariably resulted in only about 50 trees with MAX*, even when we increased the number of trees to be held in memory to 30, 000. Further swapping using SSWAP*2 and MSWAP*2 apparently would take a long time (3.2GHZ CPU frequency and 1G RAM) and were terminated after a few hours. Comparison of the NONA trees with PAUP trees showed that they were a (small) subset of the trees we found using ratchet method in PAUP. Searching with TNT, either ratchet method or other new technology methods, did not resulted in shorter trees.
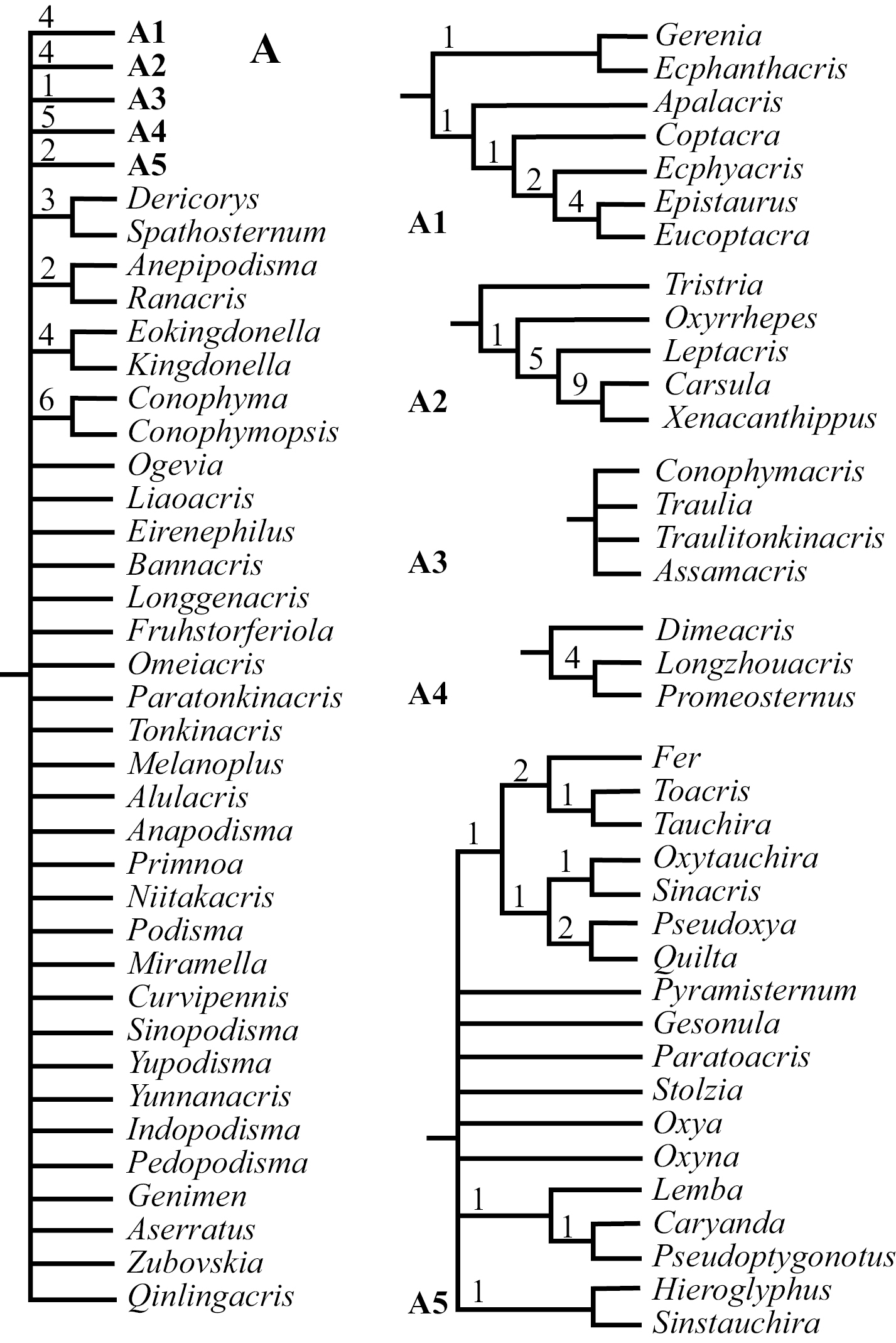
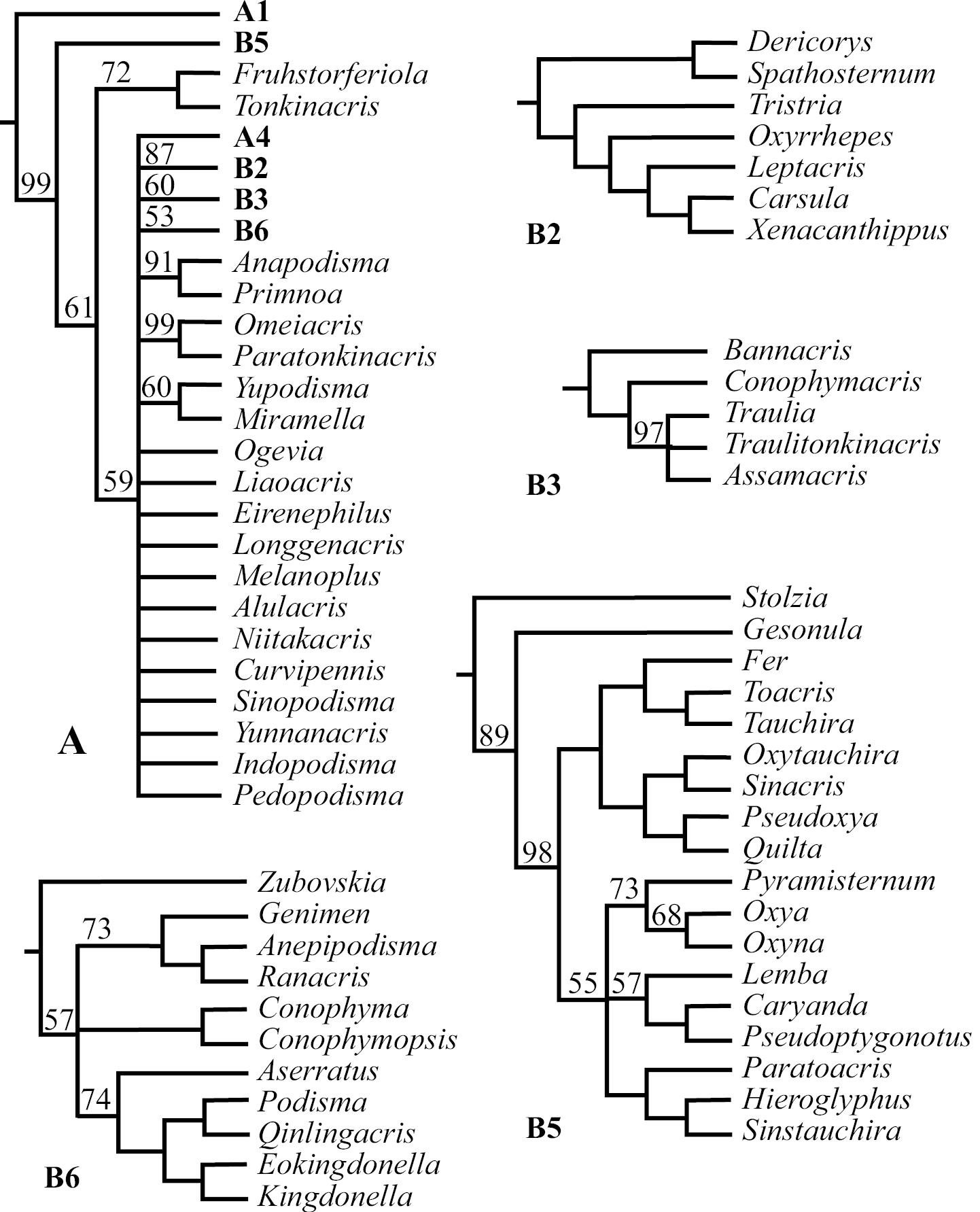
III. Phylogenetic RelationshipAlthough the number of shortest trees found by our cladistic analyses is huge, the phylogenetic relationship among genera at the base of the cladogram was well resolved, and all basal clades were also relatively well supported with Bremer Support values ranging mostly from 3 to 8 (Fig. 1). The majority of genera, 71 out of 88, fell into the monophyletic Clade A (Fig. 1), which is a polytomy consisting of several relatively well-supported monophyletic clades (Fig. 2: A, A1–5; clade A3 is only supported by a Bremer Support value of 1) as well as a number of unresolved genera / pairs of genera (Fig. 2: A). When a 50% majority consensus tree was calculated, better resolution within Clade A is achieved (Fig. 3, A, B2–B6). In comparison to the strict consensus tree, a sister relationship between A1 and the rest of the clade is supported by 99% of all shortest trees (Fig. 3: A), and A5 (Fig. 2: A5) is supported as the sister clade of the clade consisting of the rest of the genera with improved within-clade resolution (Fig. 3: B5), and (Fruhstorferiola + Tonkinacris) becomes the sister clade to the clade including all members of Clade A except clade A1 and B5 (Fig. 3: A). This terminal clade, while supported by 59% of all shortest trees, form a polytomy consisting of several monophyletic, relatively well resolved clades, 12 distinct genera, and three genera pairs. In addition, there is also an increased resolution at the base of Clade A -- B2 consists of A2 and (Dericorys + Spathosternum) (Fig. 3: B2), B3 includes A3 and Bannacris, and an additional clade is resolved (Fig. 3: B6).
Strict consensus tree of the 22, 355 found shortest trees using Parsimony Ratchet method in PAUP 4.0 beta10 (30 repetitions and 200 iterations per run, followed by TBR swapping). Shown in the figure is the expansion of Clade A of Figure 1. Several completely resolved clades are further expanded as A1, A2, A3, A4 and A5 respectively.
Majority (50% and above) consensus tree of the 22, 355 found shortest trees using Parsimony Ratchet method in PAUP 4.0 beta10. The basal part of the majority consensus tree is completely resolved and is the same as in Figure 1, and the figure shows only the phylogenetic relationship within Clade A as resolved by MJ consensus tree. The clades better resolved in comparison with strict consensus tree are further expanded as B2, B3, and B5. B5 is the same as A5 of Figure 2, but with better internal resolution. B2 is A2 plus (Dericorys, Spathosternum) at the base, and B3 is A3 plus Bannacris added at base and has higher internal resolution. B6 consists of several pairs of genera unresolved in the strict consensus tree. A1 and A4 are each completely resolved and remain the same as in Figure 1, and are thus not expanded here in. More differences between strict and MJ consensus trees are found in the basal part of Clade A (cf. Figure 2: A). Number above each branch is frequency of occurrence of a particular branch among all 22, 354 found shortest trees, and branches not indicated with a number have 100% occurrence.
Male genital morphology received special attention from
An earlier attempt to study the phylogenetic relation within Catantopidae from Australia found almost no resolution, especially at the base (
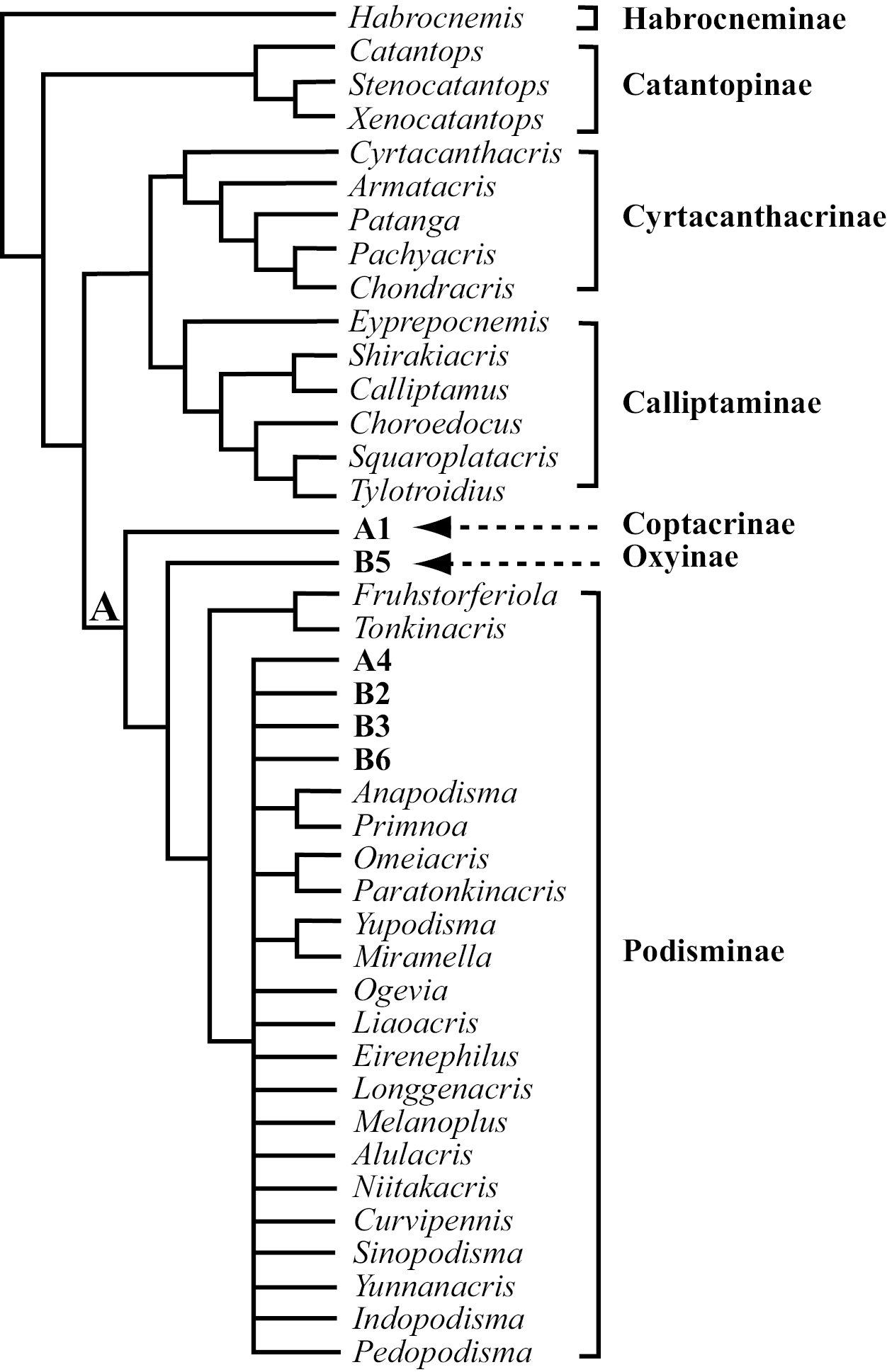
A possible scheme classification of Catantopidae from China based on parsimony phylogenetic analysis of 86 genera and 79 phylogeny–informative morphological characters. Details of Coptacridae and Oxynae are found in Figure 2 (Clade A1 Coptacridae and A5 Oxynae) and Figure 3 (B5 Oxynae). Podisminae is further divided into six tribes, of which five are supported as monophyletic by the 50% majority consensus tree of all shortest trees found while the other ‘tribe’ Melanoplini is suggested as a ‘sink’ to temporarily keep the genera that do not belong to any of the supported clades. The Fruhstorferiolini is the most basal tribe consisting of Fruhstorferiola and Tonkinacris, while details of Melanoplini are found in Figure 2 (A4: Promeosternini) and Figure 3 (B2 Dericorythini, B3 Traulini, B6 Podismini).
To our knowledge, the present study is the most comprehensive of its kind to study the higher level phylogeny of orthopteran insects in terms of the number of taxa sampled and characters examined and coded. Through this study we were able to demonstrate that the external morphology of orthopteran insects can be a very useful source for assessing higher-level phylogeny. For example, the study provided complete resolution for the basal relationships of the Catantopidae (Fig. 1), Nonetheless, our dataset were unable to provide an unambiguous solution for the relationships within the largest terminal clade that comprise 80% of all sampled genera in this study (Figs. 2, 3).It is generally accepted that phylogenetic hypotheses basing on as many independent lines of evidence as possible have the highest explanation value (
Based on the strict consensus tree and the 50% Majority-rule consensus of the 22, 355 shortest trees, we hereby outline a scheme for the classification for the family Catantopidae from China. As we discussed above, a comprehensive phylogenetic study based on a more inclusive taxon sampling from all regions of the world and including both morphology and molecular sequences is needed for highly resolving the phylogenetic relationship within the family, especially with regard to the relationship between and within the subfamilies Coptacridinae, Oxyinae, and especially Podisminae (see below). Therefore, the purpose of our outline is to serve as a basis for further studies, rather than as formal classification.
According to this scheme, the Chinese Catantopidae can be classified into seven subfamilies: Habrocneminae, Catantopinae, Cyrtacanthacrinae, Calliptaminae, Coptacridinae, Oxyinae, and Podisminae (Fig. 4). Among the seven recongnized subfamilies, Habrocneminae, Catantopinae, Cyrtacanthacrinae, and Calliptaminae are unambiguously supported as monophyletic clades, and the relationship of each to the rest of the family are completely resolved (Fig. 1, Fig. 4). Coptacridinae and Oxyinae, although each relatively well supported as monophyletic clade, are part of a crown clade that is highly unresolved in terms of within clade relationship (Clade A, Fig. 2). The monophyly of Podisminae, and the resolution of its relationship with Coptacridinae and Oxyinae are only supported by the 50% Majority-rule consensus, which is considered as a compromised solution in phylogenetic systematics (
We wish to thank Geqiao Liang, Chunmei Huang, Kailing Xia, and Xiangchu Yin for their generous support in arranging the specimen loans for the project. We also wish to thank David Hollis, Xinbao Jing, Zhibing Liu, Tieqiao Zhao for discusions and/or comments on earlier versions of the manuscript. Jim Carpenter, Gordon Tucker, and several anonymous reviewers have kindly provided valuable comments / criticisms on the final drafts of the manuscript. BL wishes to thank Shengquan Xu, Xiaohong Ou, Zhenghui Xu, Jun Chen, Wei Zhang, Fuming Shi, and other former colleagues at the Institute of Zoology, Shaanxi Normal University for their generous help and friendship.Part of the work was completed when ZL was a research associate under Richard Brusca of Arizona-Sonora Desert Museum and Terry Markow of the Center for Insect Science, University of Arizona. The work was funded in part by a National Science Foundation of China Grant to BL (# 30370237), NIH Training Grant (# 1 K12 Gm00708) in the form of fellowship to ZL through the Center for Insect Science, University of Arizona, and a Summer Research and Creative Activity Award to ZL from the EIU Council on Faculty Research.
List of sampled taxa († outgroups. *indicates genus not included in the final analysis. All ingroup genera are listed alphabatically).
| Genus | Species | Examined specimens | |
|---|---|---|---|
| ♂ | ♀ | ||
| Egnatius Voss.† | |||
| apicalis Stål | 10 | 5 | |
| Egnatioides Liu† | |||
| xinjiangensis Liu | 6 | 4 | |
| Arcyptera Serv. †* | |||
| coreona Shiraki | 4 | 4 | |
| fusca fusca (Pall.) | 4 | 4 | |
| Epacromius Uv. †* | |||
| tergestinus (Charp.) | |||
| Alulacris Zheng | |||
| shilingensis (Cheng) | 11 | 8 | |
| Anapodisma Dov.-Zap. | |||
| miramae Dov.-Zap. | 10 | 8 | |
| rufipenna Zheng | 2 | ||
| Anepipodisma Huang | |||
| punctata Huang | 1 | 1 | |
| Apalacris Walker | |||
| hyaline Walker | 6 | 5 | |
| nigrogeniculata Bi | 5 | 5 | |
| tonkinensis Ramme | 1 | ||
| varicornis Walker | 5 | 5 | |
| viridis Huang et Xia | 1 | ||
| xizangensis Bi | 14 | 11 | |
| Armatacris Yin | |||
| xishanensis Yin | 1 | 5 | |
| Assamacris Uv. | |||
| curticerca ( Huang ) | 1 | ||
| longicerca ( Huang ) | 6 | 12 | |
| Bannacris Zheng | |||
| punctonotus Zheng | 2 | 2 | |
| Calliptamus Serv. | |||
| abbreviatus Ikonn. | 15 | 10 | |
| barbarus (Costa.) | 15 | 10 | |
| coelesyriensis (G.-T.) | 7 | 2 | |
| italicus (L.) | 15 | 10 | |
| turranicus Tarb. | 7 | 15 | |
| Carsula Stal | |||
| brachycerca Huang et Xia | 1 | ||
| brachyptera Huang et Xia | 2 | 1 | |
| yunnana Zheng | 1 | ||
| Caryanda Stal | |||
| bambusa Liu et Yin | 3 | 3 | |
| bidentata Zheng et Liang | 1 | ||
| elegans Bol. | 15 | 15 | |
| glauca You | 6 | 5 | |
| gracilis Liu et Yin | 2 | 10 | |
| hunana Liu et Yin | 2 | 3 | |
| methiola Chang | 1 | ||
| nigrovittata Lian et Zheng | 4 | 3 | |
| omeiensis Cheng | 1 | ||
| pieli Chang | 4 | 5 | |
| quadrata Bi et Jin | 1 | 1 | |
| vittata Li et Jin | 4 | 5 | |
| Catantops Schaum | |||
| pinguis (Stal) | 10 | 7 | |
| simlae Dirsh | 2 | 2 | |
| Chondracris Uv. | |||
| rosea brunneri Uv. | 6 | 8 | |
| rosea (De Geer) | 10 | 10 | |
| Choroedocus I. Bol. | |||
| capensis (Thunb.) | 11 | 10 | |
| robusta (Serv.) | 13 | 10 | |
| violaceipes Miller | 15 | 10 | |
| Conophyma Zub. | |||
| almasyi almasyi Kuthy | 10 | 10 | |
| zhaosuensis Uv. | 2 | 1 | |
| Conophymacris Will. | |||
| chinensis Chang | 10 | 10 | |
| szechwanensis Chang | 10 | 10 | |
| viridis Zheng | 10 | 10 | |
| yunnanensis Zheng | 2 | 2 | |
| Conophymopsis Huang | |||
| labrispinus Huang | 10 | 10 | |
| linguspinus Huang | 6 | 8 | |
| Coptacra Stal | |||
| hainanensisTink. | 1 | ||
| tonkinensisWill. | 2 | 3 | |
| Cuvipennis Huang | |||
| wixiensis Huang | 10 | 10 | |
| Cyrtacanthacris Walk | |||
| tatarica L. | 10 | 7 | |
| Dericorys Serv. | |||
| annulata roseipennis (Redt.) | 1 | ||
| tibialis (Pall.) | |||
| Dimeacris Niu et Zheng | |||
| prasina Niu et Zheng | 2 | 2 | |
| Ecphanthacris Tink. | |||
| mirabilis Tink. | 4 | 3 | |
| Ecphymacris Bi | |||
| lofaoshana (Tink.) | 2 | 5 | |
| Eirenephilus Ikonn. | |||
| longipennis (Shir.) | 10 | 7 | |
| Epistaurus I. Bol. | |||
| aberrans r.-W. | 10 | 10 | |
| meridionalis Bi | 15 | 12 | |
| Eucoptacra I. Bol. | |||
| binghami Uv. | 4 | 2 | |
| kwangtungensis Tink. | 10 | 11 | |
| motuoensis Yin | 5 | 6 | |
| praemorsa Stal | 5 | 5 | |
| Eyprepocnemis Fieb. | |||
| hoktuensis Shiraki | 2 | 6 | |
| perbrevipennis Bi et Xia | 2 | ||
| Fer I. Bol. | |||
| bimaculatus You et Li | 4 | 4 | |
| nonmaculatus Zheng | 1 | ||
| yunnensis Huang et Xia | 2 | 2 | |
| Fruhstorferiola Will. | |||
| huangshanensis Bi et Xia | 6 | 11 | |
| huayinensis Bi et Xia | 3 | 3 | |
| kulinga (Chang) | 10 | 10 | |
| omei (Rehn et Rehn) | 1 | 5 | |
| tonkinensis Will. | 10 | 10 | |
| viridifemorata (Caud.) | 12 | 8 | |
| Genimen I.-Bol. | |||
| burmanum Ramme | 1 | ||
| yunnanensis Zheng | 7 | 4 | |
| Gerenia Stal | |||
| intermedia Br.-W. | 1 | 1 | |
| Gesonula Uv. | |||
| mundataszemaoensis Cheng | 3 | 3 | |
| punctifrons Stal | 8 | 6 | |
| Habrocnemis Uv. | |||
| sinensis Uv. | 1 | 4 | |
| Hieroglyphus Krauss. | |||
| annuliconis (Shir.) | 10 | 5 | |
| banian (Fabr.) | 13 | 7 | |
| concolor (Walk.) | 1 | ||
| tonkinensis I.-Bol. | 10 | 3 | |
| Indopodisma Dov.-Zap. | |||
| kingdoni (Uv.) | 7 | 10 | |
| Kingdonella Uv. | |||
| hanburyi Uv. | 15 | 3 | |
| kozlovi Mistsh. | 14 | 13 | |
| nigrofemora Yin | 2 | 2 | |
| nigrotibia Zheng | 1 | ||
| parvula Yin | 5 | 8 | |
| pienbaensis zheng | 1 | 1 | |
| qinghaiensis Zheng | 2 | ||
| rivuna Huang | 3 | 1 | |
| Lemba Huang | |||
| bituberculata Yin et Liu | 2 | 7 | |
| daguanensis Huang | 1 | ||
| viridatibia Niu et Zheng | 2 | 2 | |
| yunnana Ma et Zheng | 1 | ||
| zhengi Li | 2 | ||
| Leptacris Walk. | |||
| taeniata (Stal) | 3 | 4 | |
| vittata (Fabr.) | 8 | 7 | |
| Liaoacris Zheng | |||
| ochropteris Zheng | 2 | 4 | |
| Longgenacris You et Li | |||
| maculacorina You et Li | 2 | 2 | |
| Longzhouacris You et Bi | |||
| hainanensis Zheng et Liang | 4 | 5 | |
| jinxiuensis Li et Jin | 14 | 8 | |
| rufipenns You et Bi | 9 | 8 | |
| Melanoplus Stal | |||
| frigidus (Boh.) | 4 | 7 | |
| Miramella Dov.-Zap. | |||
| sinensis Chang | 2 | 1 | |
| solitaria (Ikonn.) | 5 | 3 | |
| Niitakacris Tinkham | |||
| goganzanensisTink. | 4 | 5 | |
| rosaeceanum (Shir) | 8 | 1 | |
| Emeiacris Zheng | |||
| maculata Zheng | 2 | 2 | |
| Ognevia Ikonn. | |||
| sergii Ikonn. | 2 | 1 | |
| Oxya Saerv. | |||
| adentata Will. | 10 | 10 | |
| agavisa Tsai | 14 | 10 | |
| anagavisa Bi | 11 | 9 | |
| chinensis (Thunb.) | 12 | 10 | |
| hainanensis Bi | 11 | 10 | |
| intricata (Stål) | 10 | 10 | |
| ningpoensis Chang | 13 | 13 | |
| tinkhami Uv. | 13 | 12 | |
| velox(Fabr.) | 6 | 3 | |
| yunnana Bi | 8 | 10 | |
| Oxyina Hollis | |||
| sinobidentata (Hollis) | 13 | 14 | |
| Oxyrrhepes Srtal | |||
| cantonensis Tink. | 5 | 11 | |
| obtuse (De Haan) | |||
| quadripunctata Will. | |||
| Oxytauchira Ramme | |||
| brachyptera zheng | 1 | 1 | |
| elegans Zheng et Liang | 2 | ||
| Pachyacris Uv. | |||
| vinosa (Walk.) | 3 | 3 | |
| Paratoacris Li et Jin | |||
| reticulipennis Li et Jin | 4 | 3 | |
| Patanga Uv. | |||
| apicerca Huang | 1 | 1 | |
| humilis Bi | 12 | 10 | |
| japonica (I.-Bol.) | 10 | 7 | |
| succincta(Johan.) | 6 | 5 | |
| Pedopodisma Zheng | |||
| emeiiensis (Yin) | 3 | 3 | |
| huangshana Huang | 1 | 1 | |
| protrucula Zheng | 4 | 4 | |
| shennongjiana Huang | 1 | 1 | |
| tsinlingensis (Chang) | 2 | 2 | |
| Podisma Berthold | |||
| aberrans Ikonn. | 4 | 3 | |
| pedestris (L.) | 3 | 5 | |
| Prumna Motschulsky | |||
| arctica Zhang et Jin | 10 | 12 | |
| cavicerca Zhang | 3 | 3 | |
| jingpohu Huang | 1 | 3 | |
| primnoa F.-W. | 10 | 10 | |
| primnoides (Ikonn.) | 3 | ||
| wuchangensis Huang | 1 | 1 | |
| Promesosternus Yin | |||
| himalayicus Yin | 1 | ||
| vittatus Yin | 1 | ||
| Pseudoptygonotus Zheng | |||
| gunshensis Zheng etal | 1 | ||
| kunmingensis Cheng | 7 | 6 | |
| Pseudoxya Yin et Liu | |||
| diminuta (Walk.) | 15 | 15 | |
| Pyramisternum Huang | |||
| herbaceum Huang | 1 | 1 | |
| Qinlingacris Yin et Chou | |||
| elaeodes Yin et Chou | 3 | 4 | |
| taibaiensis Yin et Chou | 3 | 4 | |
| Quilta Stal | |||
| oryzae Uv. | 7 | 8 | |
| Shirakiacris Dirsh | |||
| brachyptera Zheng | 13 | 10 | |
| shiraki (I.-Bol.) | 15 | 8 | |
| yunkweiensis (Chang) | 9 | 6 | |
| Sinacris Tinkham | |||
| longipennis Liang | 1 | 1 | |
| oreophilus Tink. | 1 | 1 | |
| Sinopodisma Chang | |||
| bidenta Liang | 1 | 4 | |
| formosana (Shir.) | 5 | 4 | |
| houshana Huang | 2 | 2 | |
| huangshana Huang | 1 | ||
| jiulianshana Huang | 2 | 2 | |
| kawakamii (Shir.) | 1 | 2 | |
| kelloggii (Chang) | 10 | 10 | |
| kodamae (Shir.) | 1 | 2 | |
| lofaoshana (Tink.) | 11 | 19 | |
| pieli (Chang) | 10 | 8 | |
| quizhouensis Zheng | 10 | 10 | |
| rostellcerca Zheng et Liang | 8 | 10 | |
| shiraki (Tink.) | 3 | 2 | |
| spinocerca Zheng et Liang | 1 | 2 | |
| splendida (Tink.) | 2 | 3 | |
| tsai (Chang) | 13 | 15 | |
| yingdensis Liang | 7 | 4 | |
| Sinstauchira Zheng | |||
| gressitti (Tink.) | 1 | 1 | |
| pui Liang et Zheng | 11 | 11 | |
| ruficornis Huang | 10 | 10 | |
| yunnansis Zheng | 1 | 1 | |
| Spathosternum Krauss | |||
| prasiniferum (Walk.) | 15 | 13 | |
| Squaroplatacris Liang et Zheng | |||
| elegans Zheng et Cao | 4 | 3 | |
| violatibialis Liang | 1 | ||
| Stenocatantops Dirsh | |||
| splendens (Thunb.) | 15 | 10 | |
| Stolzia Will. | |||
| hainanensis (Tink.) | 1 | 1 | |
| jianfengensis Zheng et Liang | 1 | 1 | |
| Tauchira Stal | |||
| damingshana Zheng | 1 | 1 | |
| Toacris Tink. | |||
| shaloshanensisTink. | 1 | 1 | |
| yaoshanensisTink. | 1 | 1 | |
| Tonkinacris Carl. | |||
| decoratus Carl. | 1 | 1 | |
| meridionalis Li | 4 | 4 | |
| sinensis Chang | 10 | 8 | |
| Traulia Stal | |||
| lofaoshana Tink. | 4 | 2 | |
| minuta Huang et Xia | 5 | 5 | |
| nigrotibialis Bi | 3 | 3 | |
| orientalis Ramme | 4 | 3 | |
| szetshuanensis Ramme. | 7 | 4 | |
| orchotibialis Liang et Zheng | 1 | 1 | |
| ornate Shir. | 4 | 4 | |
| tonknensis C. Bol. | 3 | 3 | |
| Tristria Stal | |||
| palvinata Uv. | 1 | 1 | |
| pisciform (Serv.) | 1 | ||
| Tylotropidius Stal | |||
| sp. | 2 | 3 | |
| yunnanensis Zheng et Liang Ge-qiu | 2 | 5 | |
| Xenacanthippus Mill. | |||
| hainanensis Tink. | 4 | 1 | |
| Xenocatantops Dirsh | |||
| brachycerus (Will.) | 10 | 8 | |
| humilis (Serv.) | 15 | 10 | |
| Yunnanacris Chang | |||
| yunnaneus (Ramme) | 10 | 10 | |
| Yupodisma Zhang et Xia | |||
| rufipennis Zhang et Xia | 2 | 2 | |
| Zubovskia Dov.-Zap. | |||
| koeppeni (Zub.) | 4 | 3 | |
| parvula (Ikonn.) | 8 | 10 | |
| planicaudata Zhang et Jin | 5 | 3 | |
| striata Huang | 10 | 10 | |
Character list
1. Shape of body: (0) stout, ratio of body length to width equal at most 4; (1) moderate; ratio of body length to width is between 4–8; (2) elongated and cylindrical, ratio of body length to width is at least 8. (ordered)
I. Head
2. Obliquity of frons in profile: (0) not oblique, forming with vertex an right angle; (1) oblique, forming with vertex an acute angle of over 40º; (2) strongly oblique, forming with vertex an very acute angle less than 40º. (ordered)
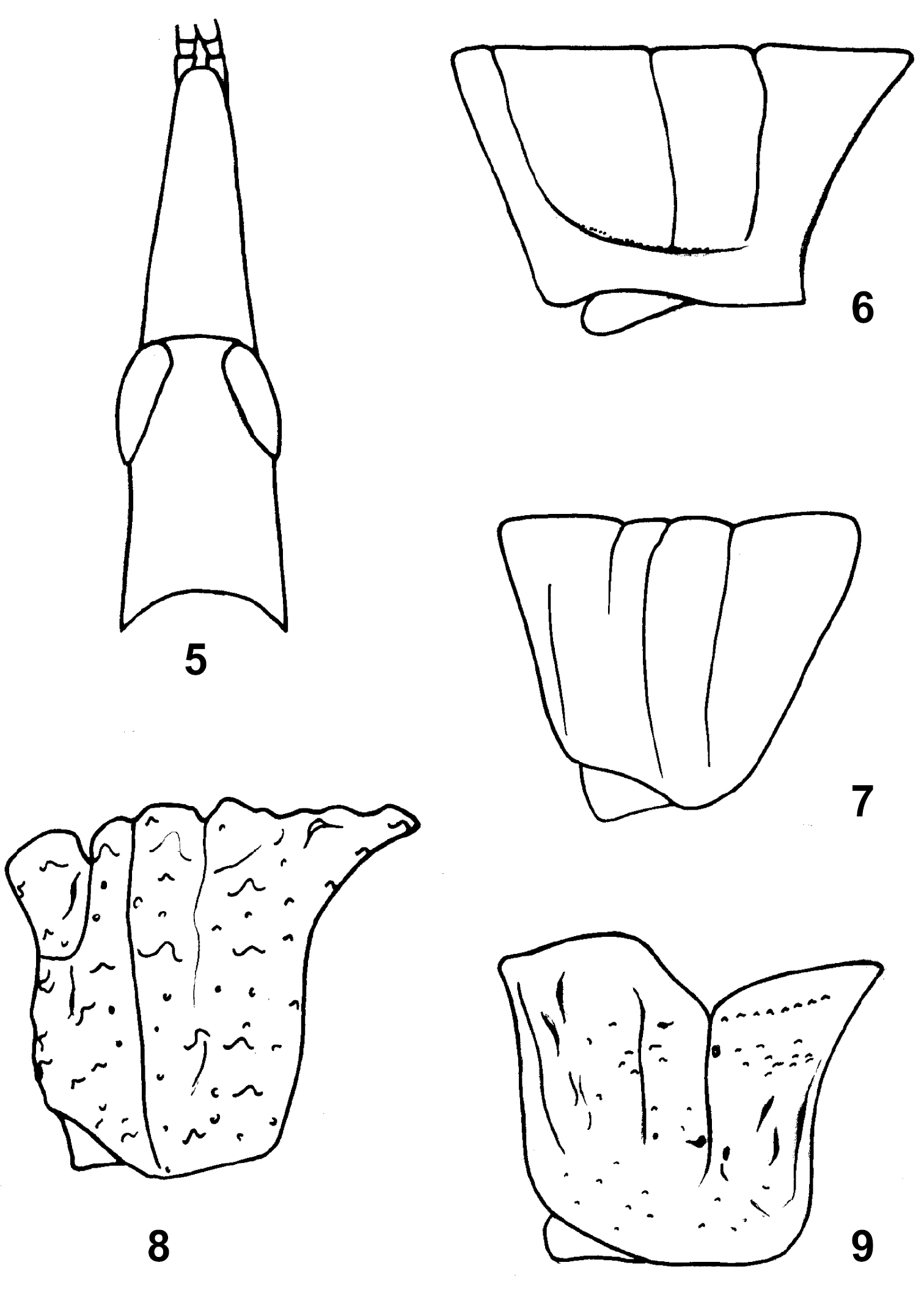
3. Shape of fastigium in dorsal view: (0) normal, not strongly projected anteriorly, the distance from anterior margin of eyes to the apex of fastigium equal or less than the horizontal diameter of eye; (1) strongly projected anteriorly, the distance from anterior margin of eyes to the apex of fastigium obviously greater than the horizontal diameter of eye (Fig. 5).
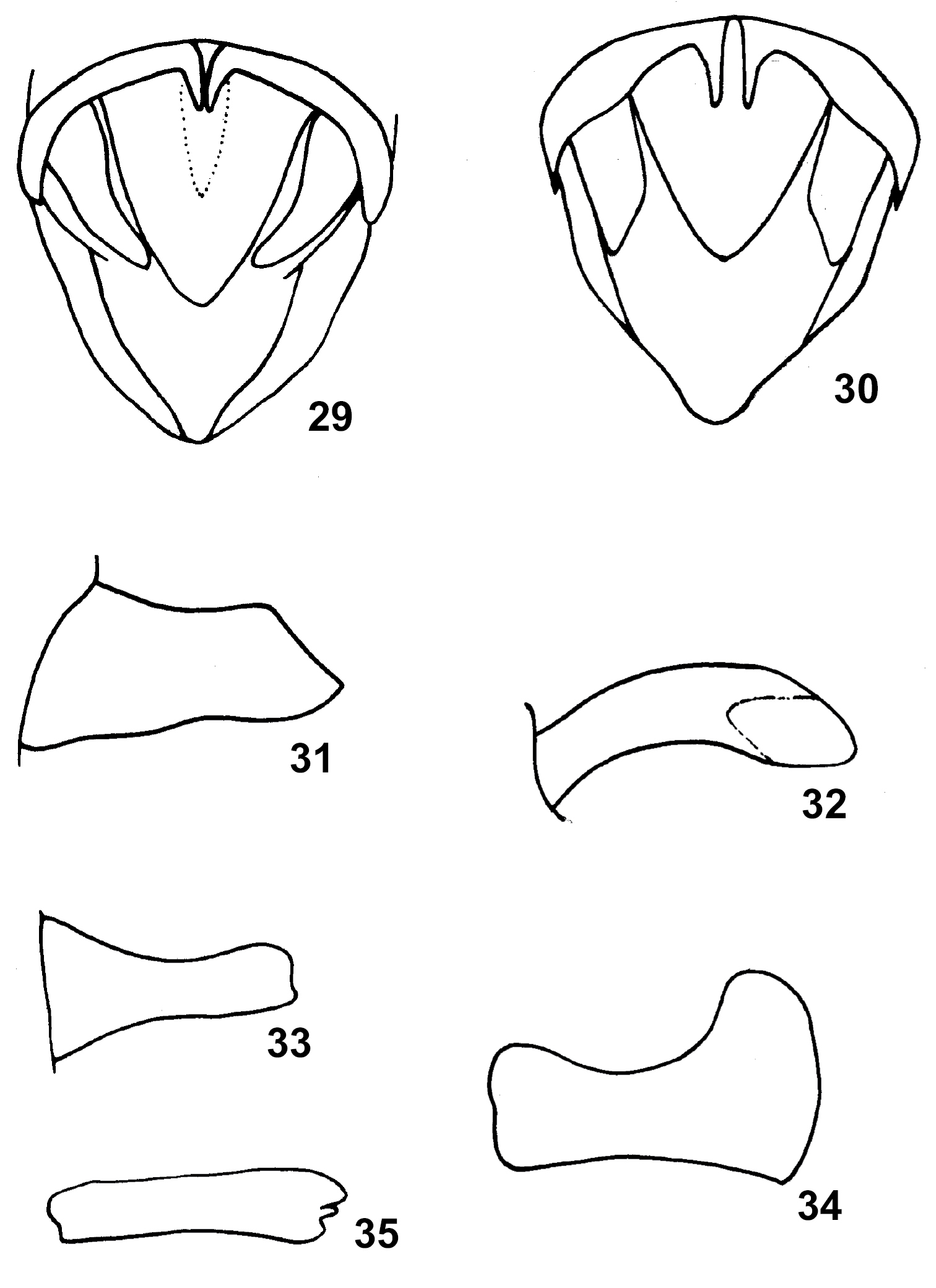
Head and pronotum: 5–6 Carsula brachyptera Huang et Xia, female: head and pronotum, dorsal view and lateral view, respectively; 7 Yunnanacris yunaeus (Ramme), pronotum, lateral view; 8 Ecphanthacris mirabiis Tinkham, male, pronotum, lateral view; 9 Dericorys roseipennis (Redt.), male, pronotum, lateral view.
4. Transverse groove at base of fastigium: (0) absent; (1) present and fine, not interrupting lateral carinae of vertex. (2) present and distinct, cutting through lateral carinae of vertex (Fig. 6). (ordered)
5. Interorbital distance of vertex: (0) obviously wider than the width of the frontal costa between antennae; (1) almost as broad as the frontal costa between antennae; (2) obviously narrower than the frontal costa between antennae. (ordered)
6. Foveola: (0) distinct; (1) absent or not perceptible.
7. Frontal costa between antennae: (0) not obviously projected; (1) obviously projected forward.
8. Shape of eye: (0) long oval, vertical diameter of eye greater than 1.3 times its horizontal diameter; (1) oval, vertical diameter of eye equal to or less than its horizontal diameter.
9. Size of eye: (0) large, vertical height greater than 1.3 times length of subocular groove; (1) small, vertical height less than 1.2 times length of subocular groove.
10. Shape of antennae: (0) filiform; (1) sward-shaped, width of basal segments greater than length.
11. Length of male antennae: (0) short, tip distinctly not reaching to base of hind femur; (1) long, tip distinctly reaching to or beyond base of hind femur.
II. Mesosoma
12. Convexity of median posterior margin of pronotum: (0) smoothly round or broadly angular; (1) projected into a right or acute angle.
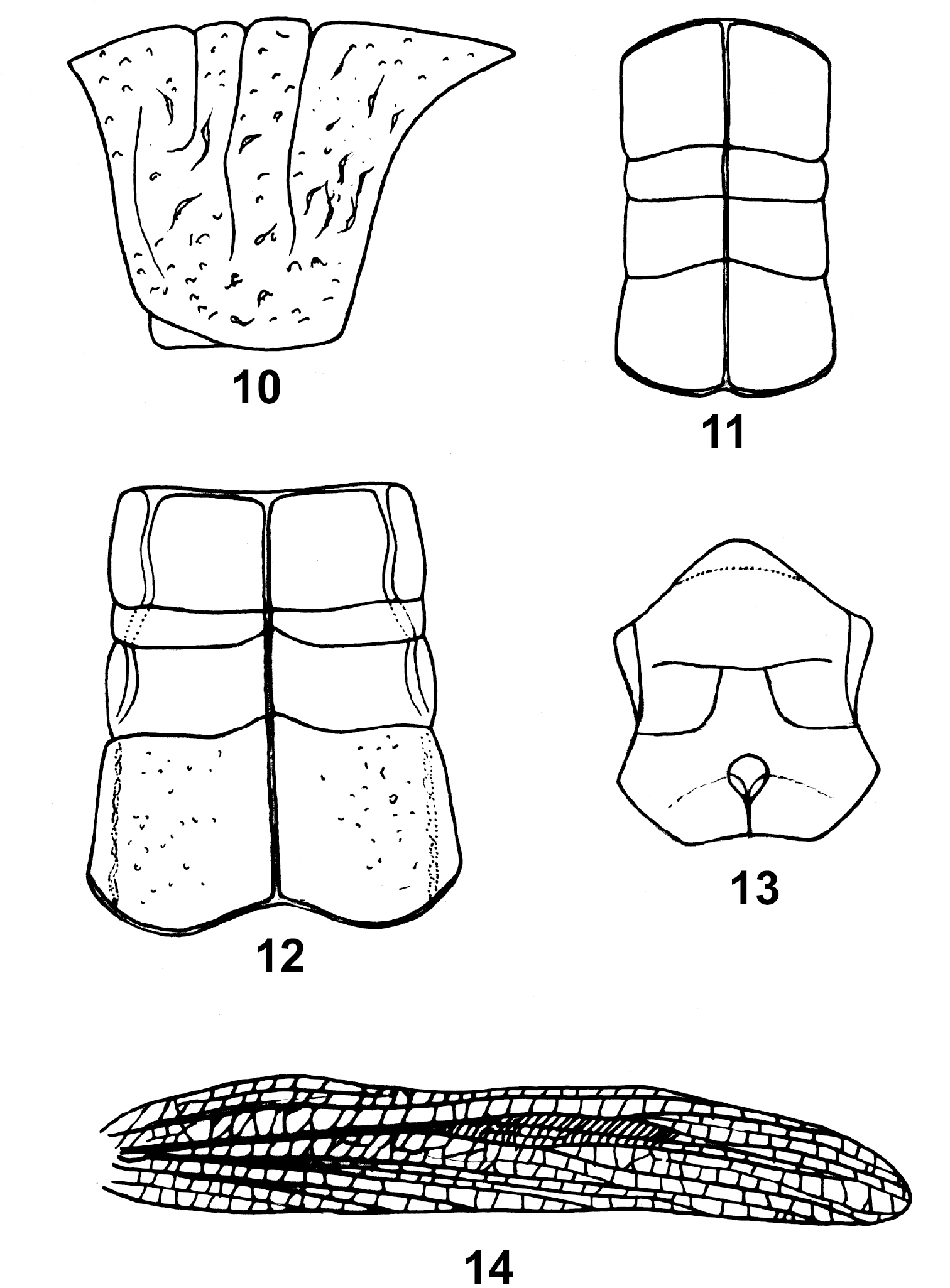
13. Concavity of median posterior margin of pronotum: (0) not concave; (1) broadly concave (Fig. 12); (2) distinctly concave, forming a triangle (Fig. 11). (ordered)
Thorax: 10 Chondracris rosae rosae (De Geer), male, pronotum, lateral view; 11 Caryanda elegans I –Bol., male, pronotum, dorsal view; 12 Niitakacris rosaceanum (Shiraki), male, pronotum, dorsal view; 13 Longzhouacris hainanensis Zheng et Liang, male, mesosternum and metasternum, ventral view; 14 Tauchira damingshana Zheng, female, elytron.
14. Longitudinal margins of dorsal surface of pronotum: (0) constricted in the middle; (1) parallel.
15. Surface of pronotum: (0) smooth to finely sculptured; (1) coarsely granuate, irregularly carinulate, or tuberculate (Fig. 8).
16. Median carina on prozona of pronotum: (0) flat; (1) distinctly elevated and roundly pectinate (Fig. 9).
17. Distinctness of median carina on pronotum: (0) distinct, from almost complete to complete; (1) barely discernible to absent.
18. Median carina on pronotum in lateral view: (0) straight; (1) strongly elevated medially, forming a distinct round ridge (Fig. 10).
19. Incision on median carina of pronotum by principal sulcus: (0) shallow to indistinct; (1) very deep (Fig. 8).
20. Presence of additional incisions of median carina of pronotum by minor transverse carina(e): (0) absent; (1) present.
21. Ratio of length of prozona to length of metazona of pronotum measured along median carina: (0) 1.0–1.2; (1) 1.5–2.0; (2) more than 2.3. (ordered)
22. Lateral carinae on pronotum: (0) absent or slightly elevated, distinctly not reaching to posterior margin of pronotum; (1) distinctly elevated, complete or nearly so.
23. Ventral posterior angle of lateral lob of pronotum: (0) broadly round (Fig. 7); (1) roundly angular to anglular (Fig. 6, 10).
24. Posterior margin of lateral lob of pronotum: (0) not concave to slightly arched; (1) strongly concave.
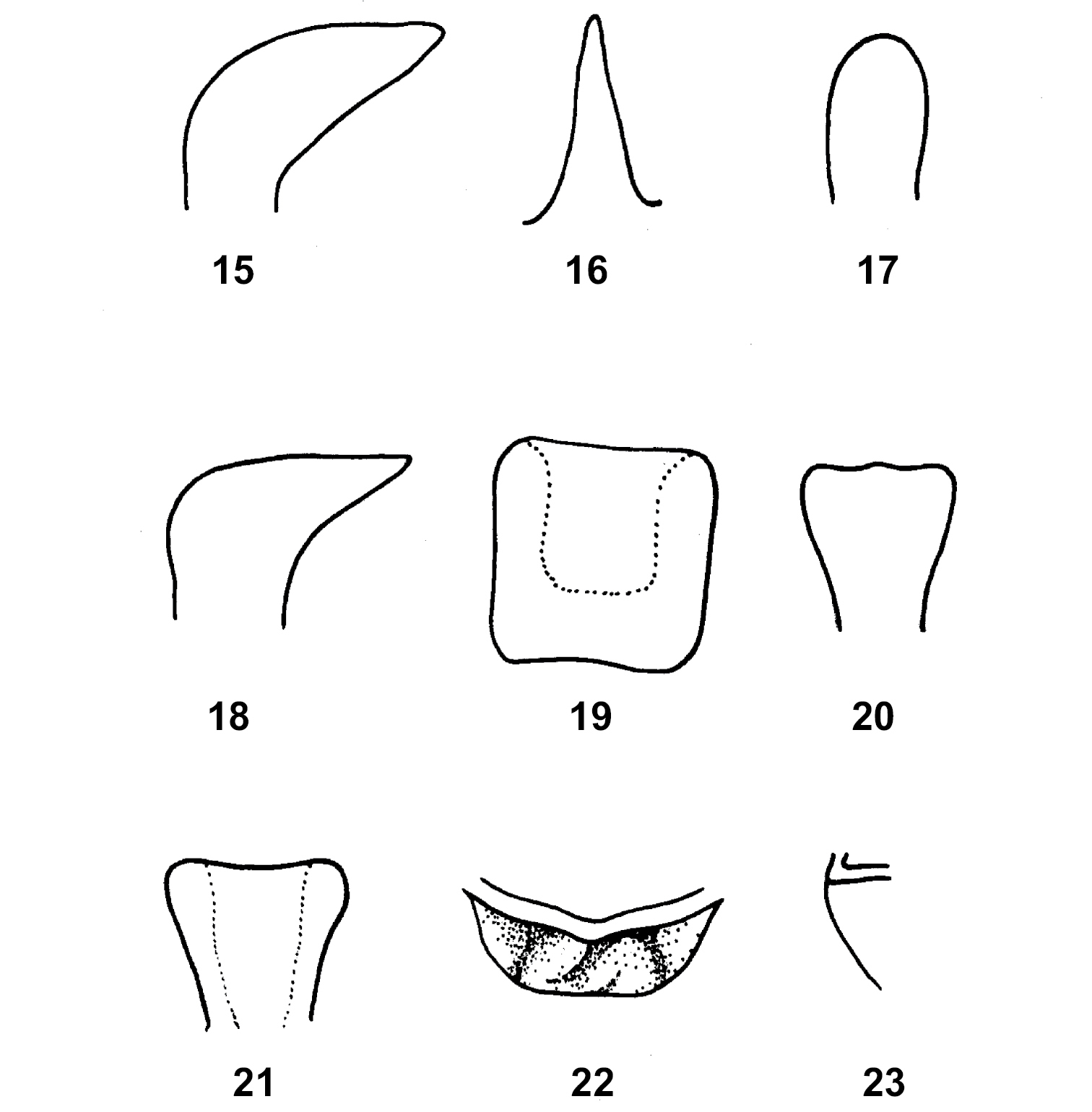
25. Shape of prosternal process: (0) conical (Fig. 16); (1) cylindrical (Fig. 17); (2) transverse and lobular (Fig. 20, 21); (3) mushroom-shaped (Fig. 18, 19).
Prosternal process: 15 Cyrtacanthacris tatarica L., male lateral view; 16 Caryanda elegans I –Bol., male lateral view; 17 Calliptamus barbarus (Costa), male lateral view; 18–19 Tristria pulvinata Uv., male lateral view and ventral view respectively; 20 Sinstauchira yunnana Zheng, male rear view; 21 Spathosternum prasiniferum (Walk), male front view; 22–23 Conophymopsis labrispinus Huang, male ventral view, and lateral view respectively (from Huang 1983).
26. (25:0) Apical part of cone-shaped prosternal process: (0) straight; (1) strongly bent posteriorly.
27. (25:1) Apical part of cylindrical prosternal process: 0 straight or slightly bent posteriorly; (1) strongly bent posteriorly, almost reaching anterior margin of mesosternum, (2) compressed laterally and flat apically.
28. (25:2) Ventral margin of lobular prosternal process: (0) truncate or slightly serrated (Fig. 21); (1) with 2–3 apically rounded, triangular processes (Fig. 20); (2) medially projected into a large triangle; (3) triangular as state 2 and turned anteriorly (Fig. 22, 23).
29. Anterior border of mesosternum: (0) straight or slightly arched; (1) broadly projected in the middle (Fig. 13).
30. Shape of mesosternal interspace: (0) wide, as long as or less than width; (1) elongate, length at least 1.3 times its narrowest width; (2) very reduced, lateral margins partly or completely contiguous. (ordered)
31. Contact of lateral lobes of metasternum medially: (0) separated; (1) contiguous.
32. Inner posterior corners of lateral lobes of mesosternum: (0) obtusely round or angularly round; (1) right angular to acutely angular.
33. Relative length of dorsal and ventral basal lobe of hind femur: (0) dorsal lobe as long as ventral lobe; (1) dorsal lobe longer than ventral lobe.
34. Shape of ventral genicular lobe of hind femur: (0) round or roundly angular distally; (1) spined distally.
35. Shape of dorsal genicular lobe of hind femur: (0) round distally; (1) spined distally.
36. Serration of dorsal carina of hind femur: (0) absent, smooth; (1) present, finely serrated.
37. Shape of distal end of dorsal carina of hind femur: (0) round or slightly broadly angular: (1) spined, acutely pointed, or narrowly obtuse-angular.
38. Outer apical spine on hind tibia: (0) absent; (1) present.
39. Number of spines on outer margin of hind tibia: (0) 5–6; (1) 8–10; (2) over 12. (ordered)
40. Distance between 1st and 2nd spines of inner spine series on hind tibia: (0) as long as any other inter-spine distance; (1) longer than any other inter-spine distance.
41. Distal half of hind tibia: (0) not obviously broaden toward apex, without obvious edges running through the spines; (1) broadened toward apex, with distinct outer and inner edges running through the spines; (2) strongly broadened toward apex, with sharp outer and inner edges running through the spines. (ordered)
42. Size of male tegmina: (0) developed, at least in contact with each other above abdomen; (1) abbreviated, lobate, and lateral, not in contact above abdomen, but reaching to posterior margin of metanotum; (2) rudimentary, not reaching to posterior margin of metanotum; (3) absent. (ordered)
43. Distal margin of tegmina: (0) round; (1) obliquely truncated.
44. Cells of distal part of tegmina: (0) rectangular or irregular; (1) oblique.
45. Radial cells in the middle of tegmina: (0) with irregular cross-veins; (1) with parallel cross-veins (Fig. 14).
III. Metasoma
46. Development of tympanal organ: (0) developed, distinct; (1) vestigial with just discernible opening, or absent.
47. Presence of tubercle on sides of apical field of male supra-anal plate: (0) absent, (1) present.
8. Presence of transverse groove on apical field of male supra-anal plate: (0) absent, (1) present.
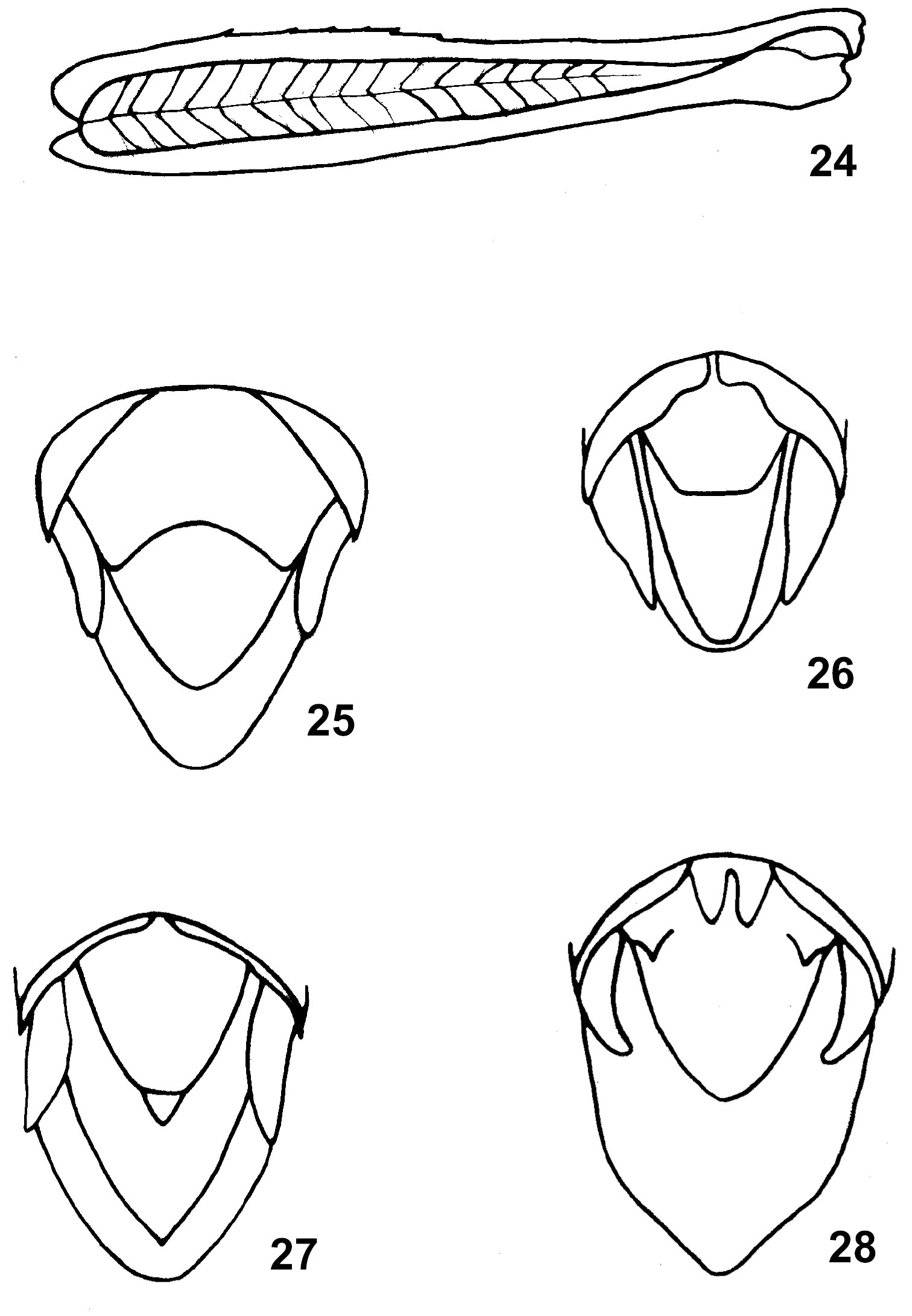
49. Presence of transverse ridge on middle field of male supra-anal plate: (0) absent, (1) present (Fig. 25).
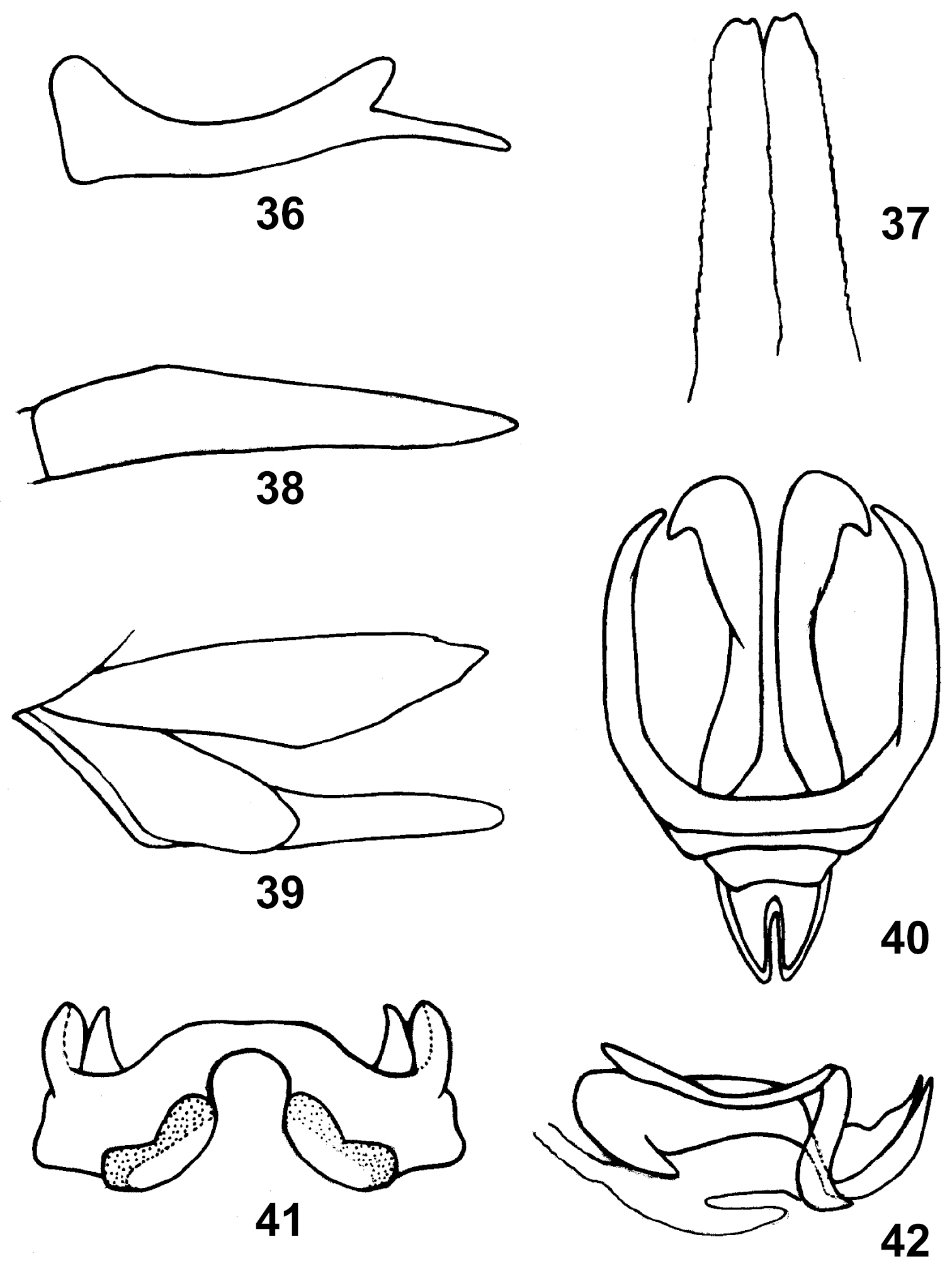
24 Dericorys roseipennis (Redt.), male, hind femur, lateral view; 25–28 End of male abdomen in dorsal view: 25 Dericorys roseipennis (Redt.); 26.Ecphanthacris mirabiis Tinkham; 27 Spathosternum prasiniferum (Walk); 28 Anapodisma miramae Dov.–Zap.
50. Presence of tubercle on sides of middle field of male supra-anal plate: (0) absent, (1) present.
51. Presence of transverse groove on middle field of male supra-anal plate in male: (0) absent, (1) present.
52. Presence of hair tufts on last sternum of abdomen: (0) absent; (1) present.
53. Presence and size of furcula: (0) absent; (1) present and small (Fig. 26); (2) present, and large and long (Fig. 28–30). (ordered)
54. Basal field of male supra-anal plate: (0) smooth without tubercles near lateral margins; (1) with two digitiform tubercles near lateral margins (Fig. 28).
55. Shape of male supra-anal plate: (0) triangular; (1) rectangular or trapezoid; (2) scutate.
56. Shape of male cerci: (0) conical; (1) compressed laterally.
57. (55:0) Length of conical cerci in male: (0) short; (1) long.
58. Curvature of male cerci: (0) straight; (1) curved inward posteriorly (Fig. 28); (2) curved upward posteriorly; (3) curved downward posteriorly.
59. Apex of cerci in males: (0) pointed; (1) round (Fig. 34); (2) truncated (Fig. 31); (3) bifurcated (Fig. 29); (4) dentate (Fig. 35). (ordered)
29–30 End of male abdomen in dorsal view: 29 Niitakacris rosaceanum (Shiraki); 30 Indopodisma kingdoni (Uv.); 31–34 Cercus in lateral view: 31 Indopodisma kingdoni (Uv.); 32 Squaroplatacris elegans Zheng et Cao; 33 Sinopodisma tsaii (Chang); 34 Fruhstorferiola omei (Rehn et Rehn); 35 Calliptamus barbarus (Costa).
60. Shape of male cerci in lateral view: (0) strongly tapering toward apex, width at apical part less than at middle; (1) broadened toward apex, width at apical part slightly greater than at middle (Fig. 32, 33); (2) strongly broadened toward apex, width at apical part much greater than at middle (Fig. 35). (ordered)
61. Shape of male subgenital plate in ventral view: (0) very short, length equal to or less than basal width; (1) long, length greater than basal width, but not more than 1.5 times; (2) strongly elongated, more than twice basal width (Fig. 38). (ordered)
36. Assamacris longicerca (Huang), cercus, male, lateral view; 37 Anapodisma miramae Dov.–Zap., upper ovipositor valve of female, dorsal view; 38 Leptacris vittata (Fabr.), subgenital plate of male, lateral view; 39 Longzhouacris hainanensis Zheng et Liang, ovipositor, lateral view. 40–42 Egnatius apicalis Stål 40: phallic organ of male, dorsal view 41 epiphallus dorsal view and 42 phallic organ, lateral view.
62. Shape of posterior part of male subgenital plate in ventral view: (0) conical; (1) trapezoid.
63. (61:0) Compression of posteriorly conical subgenital plate in males in ventral view: (0) not compressed; (1) compressed laterally.
64. Shape of male subgenital plate in dorsal view: (0) strongly tapering toward apex, end pointed or blunt; (1) gradually tapering toward apex, end round or concave; (2) not tapering, sometimes even slightly broaden, toward the apex, end truncated. (ordered)
65. Presence of tubercle at apex of male subgenital plate: (0) absent; (1) present and short (Fig. 29, 30); (2) present and much prolonged, forming prominent pointed projection. (ordered)
66. Posterior margin of female subgenital plate in ventral view: (0) triangularly projected posteriorly in the middle; (1) straight or broadly rounded.
67. Presence of lateral teeth on posterior margin of female subgenital plate in ventral view: (0) absent; (1) present.
68. Shape of dorsal valves of ovipositors in profile: (0) stout, less than 3 times as long as broad when in a position coalesced with ventral valves; (1) slender, more than 3.5 times as long as broad when in a position coalesced with ventral valves.
69. Serration of dorsal external margin of dorsal ovipositor valves: (0) smooth or weakly serrated; (1) distinctly serrated.
70. Presence of a notch on apex of dorsal external margin of dorsal valves of ovipositor: (0) absent; (1) distinctly present.
71. Apex of dorsal valves of ovipositor: (0) not bidentate; (1) bidentate.
IV. Male genitalia
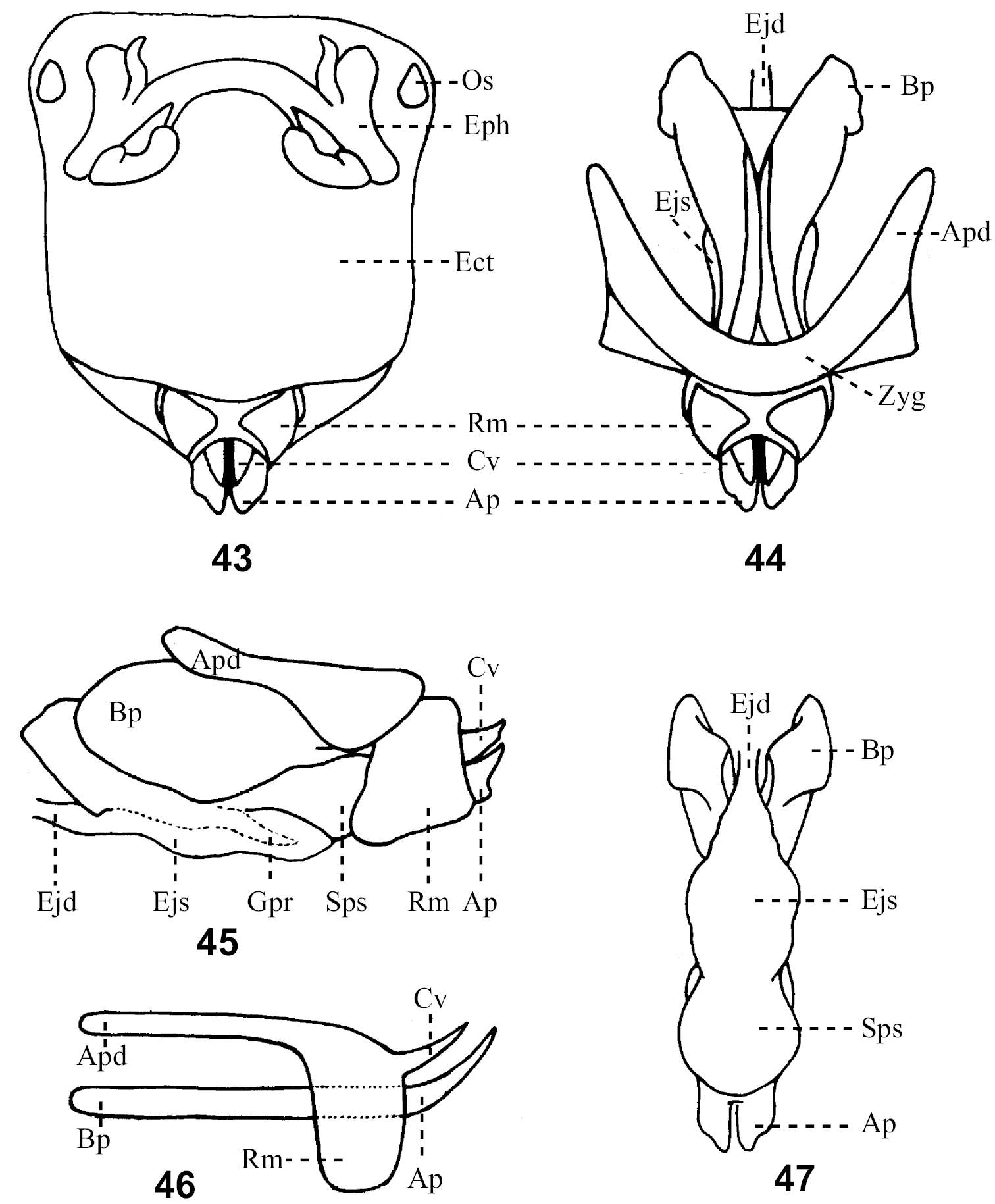
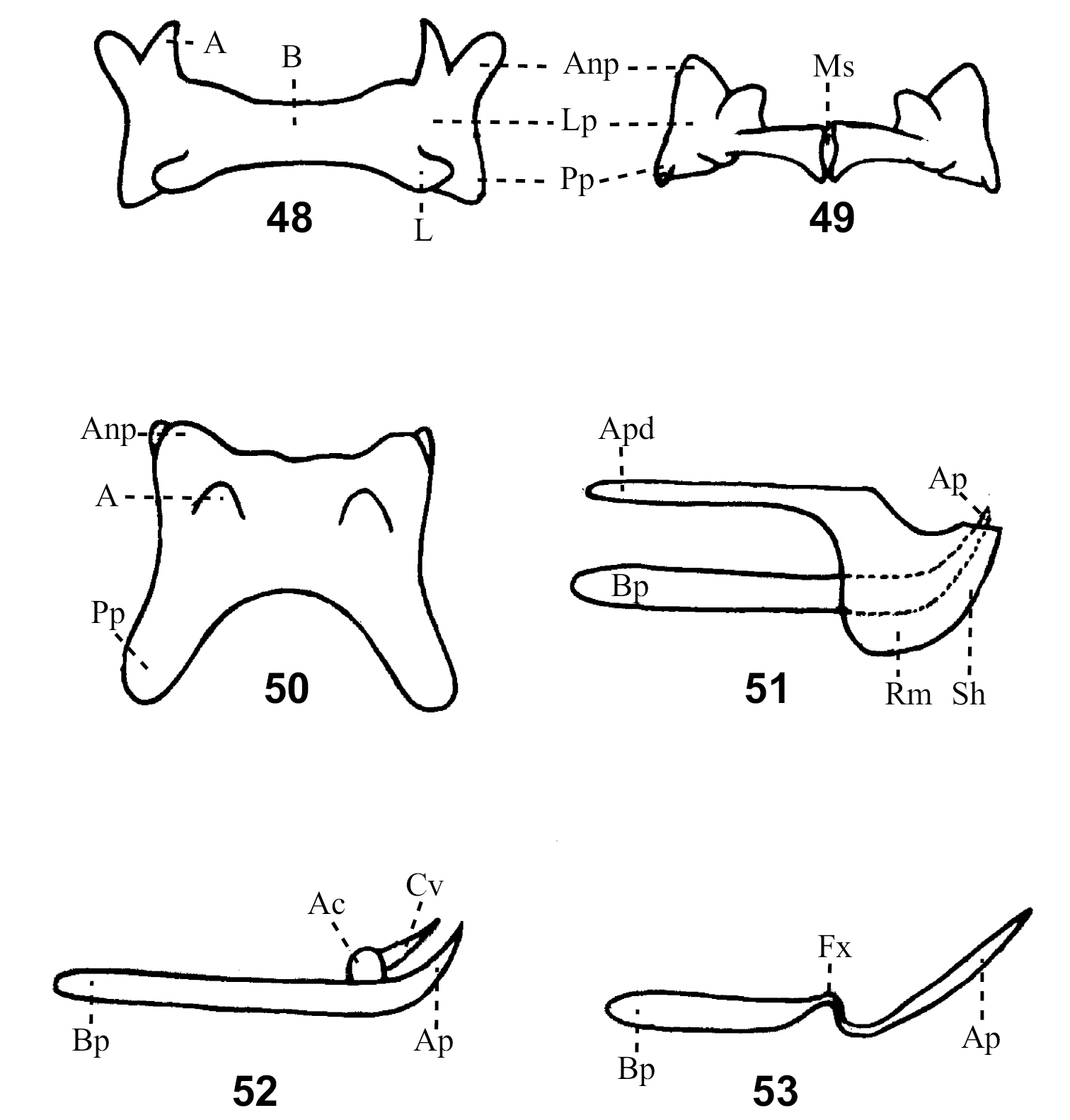
Details of the male genitalia morphology are explained in Figures 43-53. Terminology for genital structures followDirsh (1956). Acronyms used in description of the listed genital characters are:
General morphology of phallic complex. Terminology and abbreviations used in the figures follow Dirsh (1956). 43 Whole phallic complex, dorsal view; 44 Phallic organ (phallic complex with epiphallus removed) dorsal view; 45. Phallic organ lateral view; 46 Penis and cingulum of simple form lateral view; 47 Phallic organ with zygoma apodems and rami removed ventral view. Abbreviations: Ap – apical valves of penis, Apd – apodemes, Bp – basal valves of penis, Cv – valves of cinglum and Rm – rami of cinglum Zyg – zygoma. Additional abbreviations for characters not coded: Ect – ectophalus Ejd – ejaculatory duct, Ejs – ejaculatory sac, Eph – epiphallus, Gpr – gonopore process, Os – oval sclerite of epihallus, Sps – spermatophore sac.
Variations of phallic complex: 48–50 Variation of epiphallus in dorsal view; 51. Penis with sheath formed from cingulum and exposed apex of penis; 52 Penis with arch of cingulum and valve derived from it; 53 Valve of penis with flexture. Abbreviations: A – ancora of epiphallus, Ac – arc of cinglum of phallic complex, Anp – anterior projections of epiphallus, Ap – apical valves of penis of phallic complex, Apd – apodeme of phallic complex, B – bridge of epiphallus, Bp – basal valves of penis of phallic complex, Cv – valves of cinglum of phallic complex, Fx – flexture L – lophi of epiphallus, Ms – median slit of epiphallus, Pp – posterior projections of epiphallus, Rm – rami of cinglum and Sh – sheath
Ac: arc of cinglum (of phallic complex)
A: ancora (of epiphallus)
Ap: apical valves of penis (of phallic complex)
Anp: anterior projection (of epiphallus)
Apd: apodeme (phallic complex)
B: bridge (of epiphallus)
Bp: basal valves of penis (of phallic complex)
Cv: valves of cinglum (of phallic complex)
L: lophus (of epiphallus)
Rm: rami of cinglum (of phallic complex).
72. Rami of cinglum (Rm) of phallic complex: (0) undeveloped, narrowly sclerotized; (1) developed, broadly sclerotized (Fig. 45).
73. Length of the apodemes (Apd): (0) far from reaching to apex of the basal valves of penis (Bp); (1) reaching to apex of basal valves of penis; (2) reaching beyond apex of the basal valves of penis. (ordered)
74. Shape of apodemes (Apd) (dorsal view): (0) slender, more than 7 times as long as broad; (1) stout, less than 6 times as long as broad.
75. Prominence of arc of cingulum (Ac): (0) well developed and large; (1) weak, but perceptible; (2) absent. (ordered)
76. Bp and apical valves of penis (Ap): (0) connected by strongly scleorotized flexure (Fx) (Fig. 53); (1) separated, being connected by membrane.
77. Apex of Ap (in profile): (0) distinctly bent upward (Figs. 42); (1) straight; (2) distinctly bent sideward.
78. Length of the valves of cingulum (Cv): (0) very long, apex distinctly reaching beyond apex of Ap (Fig. 42) ; (1) long, apex reaching to or almost to apex of Ap; (2) reduced, apex reaching at most to middle of Ap; (3) completely absent. (ordered)
79. Shape of epiphallus: (0) bridge-shaped (Fig. 41); (1) shield-shaped (Fig. 49).
80. Integrity of epiphalus: (0) complete, not divided (Fig. 41); (1) longitudinally divided into two parts along midline, connected by membrane (Figs. 49).
81. Bridge of epiphalus in dorsal view in relation to width of lateral plate (width of plate refers to its width at ancora without including the latter): (0) broad; width in the middle broader than 1/2 of, but narrower than width of lateral plate (Fig. 49); (1) narrow; width in the middle narrower than half of the width of lateral plate (Fig. 41); (2) absent (Fig. 50). (ordered)
82. Presence of ancorae (A) and its size in relation to width of bridge of epiphalus: (0) developed, distinctly projected, longer than 1/2 of width of bridge (Fig. 41); (1) small, obviously less than 1/2 of width of bridge; (2) absent. (Ordered)
83. Development of lophi (L): (0) well developed, large; (1) undeveloped, small but perceptible; (2) absent. (ordered)
84. Shape of lophi: (0) lobiform with 2 or 3 lobes (Fig. 41); (1) lobiform with only one lobe.
85. Shape of anterior projections of epiphalus (Anp): (0) distinctly projected (Fig. 41); (1) slightly projected.
86. Posterior projections of epiphallus (Pp): (0) not or slightly projected; (1) distinctly projected.
87. Apex of ancorae: (0) pointed, (1) bluntly round; (2) truncated.
88. Length of Bp relative to Ap: (0) Bp more than 1.5 times length of Ap; (1) Bp as long as Ap (Fig. 42); (2) Bp less than 0.8 times length of Ap. (ordered).
Character Matrix (Taxa in bold are outgroups).
| CHARACTERS | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TAXA | 1 | 11 | 21 | 31 | 41 | 51 | 61 | 71 | 81 |
| Egnatius | 1100000002 | 0000000001 | 0000----00 | 0000000010 | 0000000000 | 0000000000 | 0000000000 | 0010000100 | 10000000 |
| Egnatiuides | 1100000002 | 0000000001 | 0000----00 | 0000000010 | 0000000000 | 0000000000 | 0000000000 | 0010000100 | 10000000 |
| Alulacris | 0000110100 | 0101001000 | -01000--11 | 0000001010 | 0100000000 | 101001-010 | 0000100010 | 1000001100 | 00000100 |
| Anapodisma | 0000110100 | 1011001000 | 200000--11 | 0000001010 | 0100001000 | 1021000000 | 0000100110 | 1000011100 | 11000002 |
| Anepipodisma | 0000000000 | 0011101000 | 200000--11 | 0000000000 | 03---10000 | 1010001000 | 0000000000 | 0111000100 | 01001100 |
| Apalacris | 0001-00000 | ---1000000 | 001000--11 | 0000011010 | 0010000000 | 10000010-0 | 0000000010 | 011-000001 | 00010100 |
| Armatacris | 0000110000 | 0100000000 | 001001--10 | 1100010010 | 0000001000 | 101101-100 | 2000000000 | 0101001100 | 11011101 |
| Aserratus | 0000010000 | 0001001000 | 101000--10 | 0000001010 | 03---00000 | 1010000100 | 0000000010 | 0020101100 | 00000100 |
| Assamacris | 0000011100 | 1101100000 | 101000--11 | 0000011010 | 0000000000 | 1010200031 | 0000000110 | 0111000000 | 00000000 |
| Bannacris | 0000210000 | 1101000000 | 101000--11 | 0000001010 | 0000000000 | 1010000200 | 0000100010 | 0001000100 | 10000000 |
| Calliptamus | 1100110000 | 010100000- | 01001-0-11 | 0000011010 | 0000000000 | 100001-141 | 0000000000 | 011100-110 | 112--110 |
| Carsula | 2212000011 | 0101000000 | 10103---12 | 1000000120 | 1000100000 | 111001-000 | 1000000110 | 0111010101 | 020101-1 |
| Caryanda | 0001010000 | ---100-000 | -01000---0 | 1001001110 | 1100000000 | 1110000000 | 00000--010 | 00-1000101 | 01000-1- |
| Catantops | 0000210000 | 0101000000 | 00101-0-10 | 1000011010 | 0000000000 | 10-0000111 | 0000000000 | 00--000000 | 10000000 |
| Chondracris | 0000010000 | 0021100100 | 001001--10 | 0100010010 | 0000001000 | 100001-100 | 1000000000 | 0111000100 | 11010011 |
| Choroedocus | 0000010000 | 0101000000 | 1010111-10 | 0000011020 | 0000000000 | 101001-112 | 0000000000 | 0-01000000 | -000110- |
| Conophyma | 0000010000 | 0001001000 | 211000--11 | 0000000110 | 03---100-- | -0-011-0-0 | 000-000000 | 0000211300 | 00010010 |
| Conophymacris | 0001010000 | 0101000000 | 111000--11 | 0000001110 | 0100000000 | 1000200311 | 00000-1010 | 0000000-00 | 00000000 |
| Conophymopsis | 0000010110 | 0001000000 | 20102--311 | 0000000110 | 03---10000 | 1020100000 | 0000000000 | 001021-300 | 10010110 |
| Coptacra | 0002100000 | 0021100000 | 001000--11 | 0000011010 | 0010000000 | 1020000100 | 0000000000 | 0110000101 | 00000100 |
| Curvipennis | 0000110000 | 0001101000 | 100000--11 | 0000001010 | 0100000000 | 1010000010 | 0000000010 | 0110001000 | 10000101 |
| Cyrtacanthacris | 0000110000 | 0100000000 | 0010111-10 | 0100010000 | 0000001000 | 1010000000 | 1000000000 | 0101000200 | 120101-1 |
| Dimeacris | 0002010000 | 1001000000 | 200000--11 | 1000000000 | 0100000000 | 1110101000 | 0000000000 | 0001010100 | 00010100 |
| Dericorys | 0100000000 | 0101110001 | 000000--10 | 0010010120 | 0000000010 | 0000001010 | 0000000001 | 0101011100 | 00010010 |
| Ecphanthacris | 0102000010 | 1021100110 | 001000--11 | 0000011010 | 0010000000 | 1010000000 | 0000000000 | 0100100100 | 00001100 |
| Ecphyacris | 0102100000 | 0021100000 | 001000--11 | 0000011010 | 0011000000 | 1010001000 | 00000--010 | 0110000101 | 00000100 |
| Eirenephilus | 0000010110 | 0101101000 | 000000--11 | 0000001010 | 0000000000 | 1010000000 | 0000100010 | 0011010100 | 00000100 |
| Emeiacris | 0000010000 | 0101001000 | 101000--11 | 0000000010 | 0000000001 | 0010001010 | 0000100010 | 0101011100 | 00010100 |
| Eokingdonella | 0000010110 | 0001001000 | 111000--11 | 0000000010 | 03---00001 | 0020001000 | 0000000010 | 0--------- | -------- |
| Epistaurus | 0102200000 | 0101000001 | 001000--11 | 0000011010 | 0011000000 | 1020100-00 | 0000000000 | 0010001001 | 00000000 |
| Eucoptacra | 0102200000 | 0021000000 | 000000--11 | 0000011010 | 0011000000 | 1020000--0 | 0000000010 | 00-1000001 | 0000--00 |
| Eyprepocnemis | 0000010000 | 0101000000 | -0101-0-10 | 0000011010 | 0000000000 | 1010000000 | 0000000000 | 011-000-00 | 10010100 |
| Fer | 0001010000 | 0101001000 | 10101-0-10 | 0001001110 | 1000100000 | 1110000000 | 0000000010 | 0100000301 | 01000110 |
| Fruhstorferiola | 0000110000 | 0101001000 | 001000--10 | 0000001010 | 0000000000 | 1010000012 | 00001-1010 | 01--00--00 | 00000100 |
| Genimen | 0000210100 | 1001001001 | 200000--11 | 1000000010 | 03---00000 | 10-0000000 | 0000000000 | 0110000300 | 00010100 |
| Gerenia | 1000000000 | 0021000000 | 001000--11 | 0000011010 | 0011000000 | 1010000000 | 0000000010 | 0100100200 | 11101100 |
| Gesonula | 0000-10000 | 0101001000 | 001001--11 | 1001001111 | 2000100000 | 11-0000-00 | 0000000010 | 01000-0101 | 00000100 |
| Habrocnemis | 1000000000 | 0101100000 | 10001-0-10 | 0000011010 | 0100000010 | 002001-000 | 0000000000 | 0101000101 | 20000000 |
| Hieroglyphus | 0001010000 | 1101001000 | 101000--10 | 1000001110 | -000101000 | 11-0-001-- | 00000000-0 | 01010-1001 | 0000-000 |
| Indopodisma | 0000210000 | 0001001000 | 100000--11 | 0000001010 | 0200000000 | 102001-020 | 0000100010 | 0000111100 | 00000020 |
| Kingdonella | 1100010010 | 0--100-00- | 110000--11 | 0000000010 | 03---10001 | 00-00010-0 | 0000000010 | 012--11100 | 11001100 |
| Lemba | 0001010000 | 0101000000 | 101000--10 | 0011001110 | 1000000000 | 1110000000 | 1001010010 | 0120100101 | 00000001 |
| Leptacris | 2202000001 | 0101000000 | 101000--12 | 1000000120 | 0000100000 | 1010001000 | 2010000000 | 0000011100 | 10100-02 |
| Liaoacris | 0000010100 | 0101001000 | 000000--11 | 0000000010 | 0000000000 | 1010001000 | 0000000010 | 0001011100 | 01000100 |
| Longgenacris | 0000110000 | 0101001000 | 101000--11 | 0000001010 | 0000000000 | 101001-010 | 0000100000 | 0000101100 | 10010101 |
| Longzhouacris | 0001110000 | 1101101000 | 10-000--01 | 1000011010 | 0100000000 | 1110100000 | 00000-1110 | 0001010100 | 10011101 |
| Melanoplus | 0000110100 | 0101001000 | -00000--11 | 0000000010 | 0000000000 | 102001-010 | 0000100000 | 0100011100 | 00000100 |
| Miramella | 0000010110 | 1101001000 | 100000--11 | 0000000010 | 0-000000-- | -020000000 | 0000200010 | 10-0101100 | 0100000- |
| Niitakacris | 0000110000 | 0011001000 | 100000--11 | 0000001010 | 0100000000 | 1020000010 | 0000000010 | 0000111100 | 00000110 |
| Ognevia | 0000010110 | 0101000000 | 001000--11 | 0000000010 | 0000000000 | 1010000000 | 0000100010 | 0--------- | -------- |
| Oxya | 0001-10000 | 0101001000 | -01000--10 | 1001001110 | 20000000-- | -1000000-0 | 00000--010 | 0100000-01 | 02000--- |
| Oxyna | 0001110000 | 0101001000 | 101000--10 | 1001000110 | 2000000000 | 11000000-0 | 0000000010 | 0---00--01 | 0000000- |
| Oxyrrhepes | 2000000000 | 0101000000 | 001000--12 | 0000010120 | 0000000001 | 00100--100 | 1000000000 | 0---011100 | 00-10-01 |
| Oxytauchira | 0000010000 | 010100-000 | 10102--110 | 0001001110 | 1000000000 | 1110000000 | 0000010010 | 0100001101 | 00000100 |
| Pachyacris | 0000-00000 | 0101100100 | 001000--10 | 1100011010 | 0001001000 | 1000000100 | 1000000000 | 0111000100 | 11010011 |
| Paratoacris | 0001010000 | 0101001000 | 101000--10 | 1001001010 | 1000000000 | 1110000000 | 0000000010 | 0100000001 | 00000000 |
| Paratonkinacris | 0000010010 | 110100100- | 101000--11 | 0000000010 | 0000000001 | 0010000310 | 0000100010 | 0--------- | -------- |
| Patanga | 0000110000 | 0100000000 | 0010111-10 | 1100010010 | 0000001000 | 100001-0-0 | 1000000010 | 01--000100 | 01010011 |
| Pedopodisma | 0000110000 | 000100100- | -01000--10 | 000000-010 | 0200000000 | 1010000--0 | 0000100010 | 0100-1-100 | 01000100 |
| Podisma | 0000210110 | 0101000000 | 001000--1- | 0000000010 | 0-00000000 | 1020201000 | 0000000010 | 0121110100 | 01000100 |
| Primnoa | 0000110110 | -011001000 | 201000--11 | 000000-010 | 010000--00 | 1010---0-0 | 01-20000-0 | 0021014200 | 11010102 |
| Promeosternus | 0001010000 | 1101101000 | 101000--00 | 1000010000 | 010-000001 | 0020100111 | 0000000100 | 0000010100 | 00011110 |
| Pseudoxya | 0000210000 | 0101001000 | 101000--10 | 0011001110 | 2000000000 | 1110000000 | 0000000010 | 0100001101 | 01000010 |
| Pseudoptygonotus | 0000010000 | 0001000000 | 201000--10 | 0000000110 | 1100100000 | 1110200000 | 00000-1010 | 0100010101 | 00000011 |
| Pyramisternum | 0001110000 | 0101001000 | 10102--210 | 0001000110 | 1000000000 | 1100000000 | 0000000010 | 0--------- | -------- |
| Quilta | 2000010000 | 0101001000 | 101101--00 | 0011101110 | 2000000010 | 0110000000 | 10010-1000 | 0111001001 | 020100-0 |
| Qinlingacris | 0000010110 | 0001000000 | 001000--11 | 0000000010 | 03---00000 | 1020001000 | 0000000010 | 002000-100 | 01000100 |
| Ranacris | 0000000000 | 0001100000 | 200000--11 | 0000011010 | 03---00000 | 1010000000 | 0000000--- | ---------- | -------- |
| Shirakiacris | 0000010000 | 010100000- | 01001-0-10 | 1000011010 | 0000000000 | 100001-111 | 00000-1000 | 0111001000 | 10010100 |
| Sinacris | 0000010000 | 0101001000 | 10102--110 | 0001001110 | 1000000000 | 1100000000 | 0000010010 | 0010001101 | 01001100 |
| Sinopodisma | 0000-10000 | 0001001000 | 101000--10 | 0000001010 | 0100000000 | 10100001-- | 0000100010 | 0000-1-100 | 00000-00 |
| Sinstauchira | 0002-10000 | 0101001000 | 10102--010 | 1000001110 | 1000100000 | 11-0000000 | 0000000010 | 0100000001 | 00001000 |
| Spathosternum | 0000010000 | 0101000001 | 01002--010 | 1000000110 | 0000100100 | 1010001000 | 0000000000 | 0110011000 | 01101121 |
| Squaroplatacris | 0000010000 | 0101000000 | 01101-0-10 | 1000001020 | 0000000000 | 101001-111 | 0000010000 | 0011000100 | 00001000 |
| Stenocatantops | 0000210000 | 0101000000 | 00001-0-10 | 1000011010 | 0000000000 | 1000000110 | 1000000000 | 0100000100 | 10000000 |
| Stolzia | 0000210000 | -101101000 | 001000--11 | 0001001110 | 0000001000 | 1110000000 | 0000000010 | 0----0--01 | 00000-1- |
| Tauchira | 0001010000 | 0101001000 | 10102--110 | 0001001010 | 1000100000 | 1110000000 | 0000000010 | 0100000101 | 01001100 |
| Toacris | 0002010000 | 0101001000 | 10101-0-10 | 1001001010 | 1000100000 | 1110000000 | 0000000010 | 0101000101 | 00000100 |
| Tonkinacris | 0000210000 | 1101001000 | -01000--10 | 0000001010 | 0000000000 | 10100----0 | 0000100010 | 0100010100 | 00000100 |
| Traulia | 0000001000 | -101000000 | 101000--1- | 0000011010 | 0-00000000 | 10-00000-- | 00000000-0 | 01--000001 | 00000100 |
| Tristria | 2000010000 | 0101000000 | 11103---12 | 0000000120 | 0000000000 | 1020000100 | 1001000000 | 0101001100 | 00000000 |
| Traulitonkinacris | 0000011000 | 1101001000 | 101000--10 | 0000011010 | 1000000001 | 001021-031 | 0000000010 | 0--------- | -------- |
| Tylotropidius | 0000010000 | 0101000000 | 01101-2-10 | 1000011020 | 0000000000 | 1010200000 | 0000000000 | 0011000100 | 10001001 |
| Xenacanthippus | 2212010001 | 0101001000 | 20101-0-12 | 1000000110 | 0000000010 | 0010000000 | 2010000010 | 0011210200 | 120100-2 |
| Xenocatantops | 0000210100 | 0100000000 | 00101-0-10 | 0000011010 | 0000000000 | 1010000010 | 0000000000 | 0110000000 | 100000-0 |
| Yupodisma | 0000010110 | 0001001000 | 101000--11 | 0000001010 | 0100000001 | 0021000000 | 0000100010 | 1001001100 | 00000001 |
| Yunnanacris | 0000210000 | 0001001000 | 100000--11 | 0000001010 | 0100000000 | 1010000010 | 0000100010 | 0000111100 | 01000101 |
| Zubovskia | 0000110000 | 0001001000 | 20-000--11 | 0000000010 | 03---1--00 | 10-0000010 | 0000-00010 | 1000111100 | 01000100 |