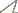(C) 2011 Hiromichi Sakai. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Mixed populations of Xiphinema americanum-group species were detected from a root zone soil sample of Japanese holly, Ilex crenata, during a survey for plant-parasitic nematodes of commercial ornamental plant nurseries in Chiba Prefecture, Japan. From the result of the morphological study, the species were identified as Xiphinema brevicolle and Xiphinema sp. This is the first record of Xiphinema brevicolle in Japan. Morphometrics of Xiphinema brevicolle generally agree with those of the type specimens and the topotype specimens. Xiphinema sp. morphometrically resembles Xiphinema paramonovi except for tail length. The mitochondrial COI region, the nuclear 18S rDNA and the nuclear large subunit rDNA D2/D3 region of the species were sequenced and compared in the molecular study. For the COI region, PCR primers were newly designed to obtain longer sequences, ca. 900 bp, than previously used. Sequence identities of COI, 18S and D2/D3 regions between these two populations were 84.0-84.1%, 99.9% and 98.1-98.2%, respectively. Phylogenetic analyses of maximum likelihood trees were carried out to compare genetic relationships among the group and some suggestions were made on the Xiphinema brevicolle-subgroup.
COI, Ilex crenata, Japanese holly, rDNA, Xiphinema americanum-group, Xiphinema brevicolle-subgroup
During a survey for plant-parasitic nematodes of commercial ornamental plant nurseries in Chiba Prefecture, Japan, we detected mixed populations of Xiphinema americanum-group species from a root zone soil sample of Japanese holly, Ilex crenata Thunb., one of the major garden tree species in Japan. This study was conducted using morphological characters of females and DNA sequences of the mitochondrial COI region and the nuclear ribosomal RNA (rDNA) regions to identify and characterize the species in the mixed populations.
Methods Collection of the nematode specimensThe soil sample containing the Xiphinema spp. (No. 001-001) was taken from the root zone of Japanese holly growing in a commercial plant nursery at Sosa City, Chiba, Japan. Nematodes were extracted with Cobb’s wet-sieving technique. Material collected on a 75 µm mesh sieve was placed on a Baermann funnel and nematodes were collected after one day at room temperature.
Morphological observationFemales of Xiphinema
spp. were removed from the nematode suspension using a dissecting
microscope and nematode pick. Female nematodes were transferred to a
small amount of water then killed by either heating at 60°C for 2 min or
by adding hot FP4:1 (
DNA extractions from single female specimens were carried out according to
DNA fragments of the mitochondrial COI region were amplified by PCR using the set of primers, CO1-F1 and CO1-R1 (Table 1), which were originally designed from sequence comparison of the COI region between Xiphinema americanum (
DNA fragments of the nuclear 18S rDNA region were amplified using the set of primers, 18S 39 and 18S 1573R (
DNA fragments of the nuclear large subunit rDNA D2/D3 region were amplified using the set of primers, D2Ab (
These PCR products were purified with QIAquick PCR
Purification Kit (Qiagen K.K., Japan), subjected to cycle sequencing
with BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies
Japan), purified with either DyeEx 2.0 Spin Kit (Qiagen) or Agencourt
CleanSEQ (Beckman Coulter Inc., USA), and sequenced on the ABI PRISM
3100 Genetic Analyzer (Life Technologies Japan Ltd., Japan). Primers
18S 599R, 18S 550, 18S 977R, 18S 965 (
Phylogenetic analyses including substitution model selections were performed using MEGA5 (
Primers designed and employed originally in this study for PCR amplification and sequencing of mitochondrial COI region and rDNA D2/D3 region.
| primer name | Sequence | Note |
|---|---|---|
| CO1-F1 | 5’-ATAATTTTTTTTATGGTAATACC-3’ | PCR, sequencing |
| CO1-R1 | 5’-ACTACATAATAAGTATCATG-3’ | PCR, sequencing |
| CO1-F2 | 5’-TATATTTTAATTTTACCTGG-3’ | sequencing |
| CO1-R2 | 5’-CCAGGTAAAATTAAAATATA-3’ | sequencing |
| D3A-R | 5’-AGACTCCTTGGTCCGTGTTTC-3’ | sequencing |
The DNA sequence analysis of the mitochondrial COI region for 9 specimens suggested two different Xiphinema americanum-group species were present. Morphometric data were used to identify them as Xiphinema brevicolle Lordello et Costa, 1961, and an undescribed Xiphinema americanum-group species.
Morphological observationshttp://species-id.net/wiki/Xiphinema_brevicolle
Figs 1A, 1C, 1E, 1G, 2, 4A, 5A, 6ASee Tables 2, 3.
Morphometrics of the specimens obtained here generally agree with those of the type specimens and topotype specimens (
The emended name of this species, Xiphinema brevicollum, was proposed by
See Tables 2, 3.
These specimens morphometrically resemble Xiphinema paramonovi Romanenko, 1981, except for the clearly different tail length (Table 2). The morphometrics of the specimens partly overlap those of Xiphinema brevicolle. General morphology and DNA information addressed below suggest that these specimens belong to some species related to Xiphinema brevicolle, though a specific species accommodating them was not found. We finally regarded these specimens as an unidentified Xiphinema americanum-group species. Further information is required to identify the specimens as a new species or determine if they represent intra-specific variation of species previously described. No male was detected.
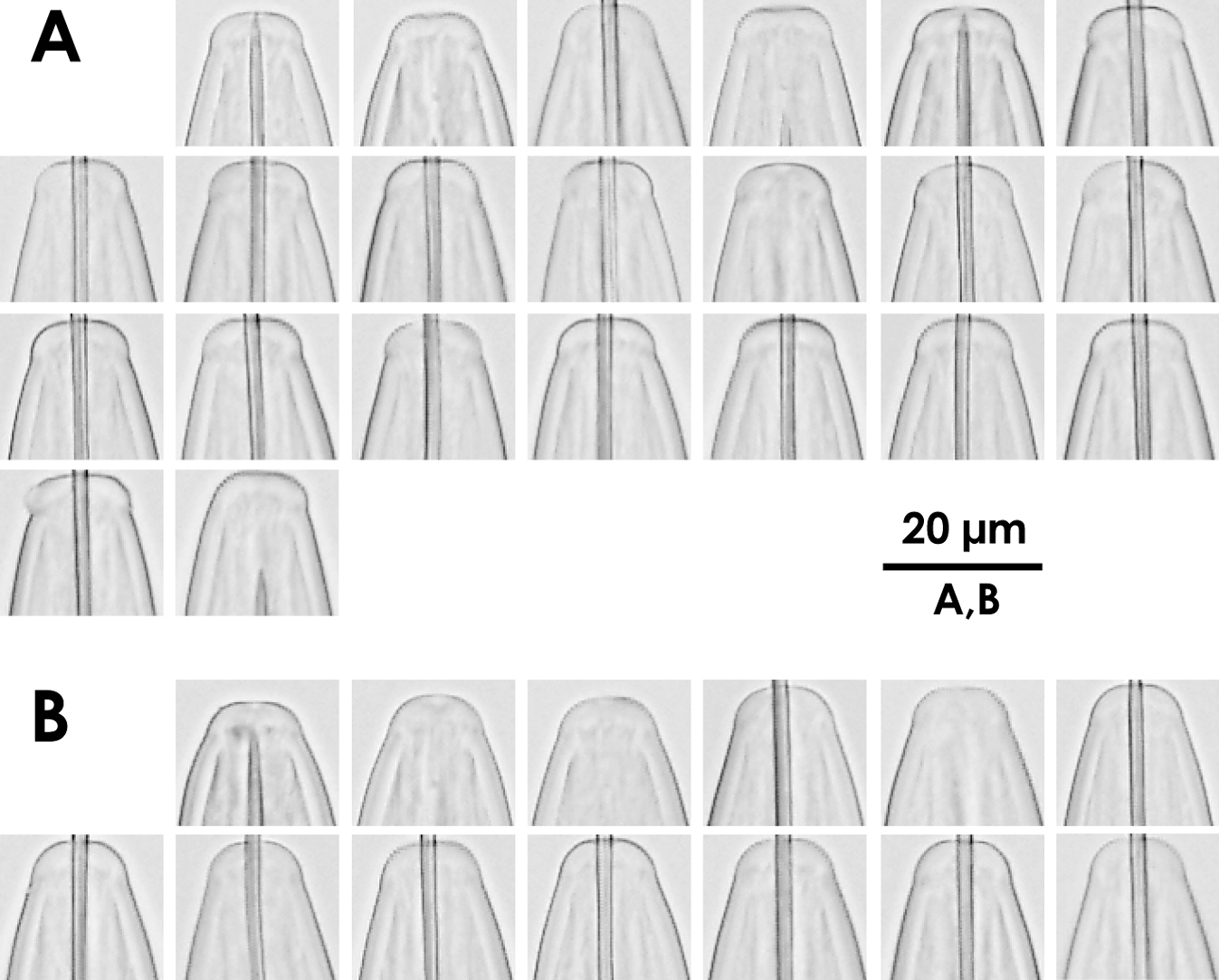
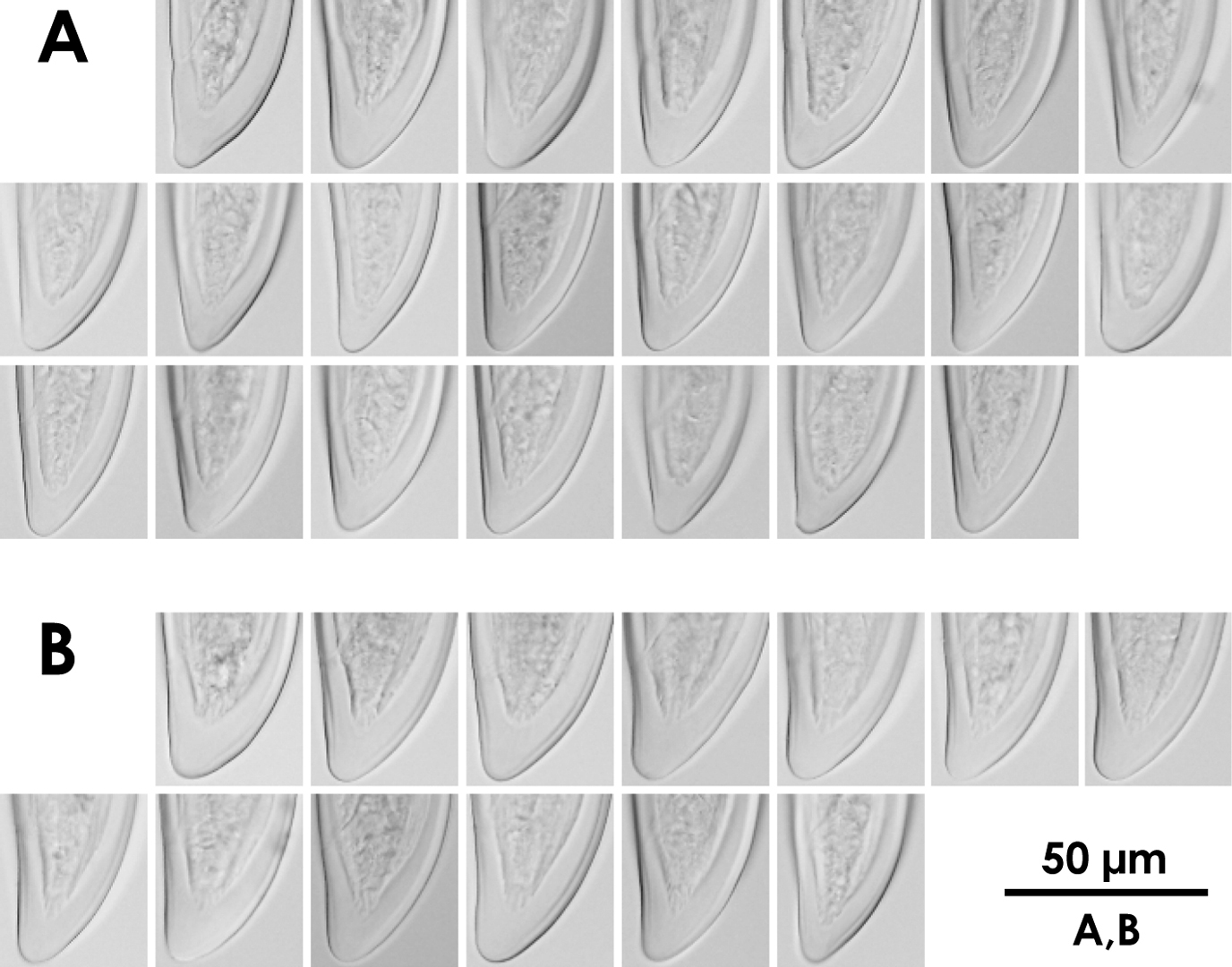
Morphometrics of Xiphinema brevicolle, Xiphinema sp., Xiphinema paramonovi and Xiphinema diffusum (female). Mean ± standard deviation (range) in μm, except for L in mm, and ratios.
| Xiphinema brevicolle | Xiphinema sp. (this study) |
Xiphinema paramonovi Paratypes(Romanenko 1981) |
Xiphinema diffusum Paratypes (Lamberti and Bleve-Zacheo 1979) |
||||
|---|---|---|---|---|---|---|---|
| Populationin this study | Topotypes | Types (Lordello and Costa 1961) | |||||
| ( |
( |
||||||
| n | 22 | 25 | 17 | - | 13 | 27 | 10 |
| L | 1.93 ± 0.11(1.71-2.10) | 1.92 ± 0.122(1.7-2.16) | 2.1 ± 0.1(1.8-2.2) | (1.82-2.20) | 2.30 ± 0.12(2.08-2.47) | 2.1(2.0-2.3) | 1.7(1.6-1.8) |
| a | 46.1 ± 2.6(40.5-50.5) | 46.1 ± 1.72(44-51) | 44.5 ± 2.3(40.7-50.1) | (36.0-42.2) | 48.8 ± 2.4(45.0-52.3) | 49.6(44-54.2) | 47(46-51) |
| b | 6.6 ± 0.6(5.5-8.2) | 5.92 ± 0.37(5.1-6.7) | 6.4 ± 0.6(5.6-7.7) | (7.0-10.5) | 7.0 ± 0.4(6.5-8.1) | 6.1(4.8-7.1) | 6.9(5.3-8.9) |
| c | 69.8 ± 4.4(60.5-79.1) | 76.9 ± 5.64(67.6-89.9) | 77.8 ± 0.6(60.3-94.0) | (62.5-93.0) | 79.6 ± 8.5(67.5-94.5) | 60.5(49.1-68.5) | 72(63-84) |
| c’ | 1.0 ± 0.1(0.9-1.2) | 0.96 ± 0.06(0.89-1.10) | 1.0 ± 0.06(0.9-1.1) | - | 0.9 ± 0.1(0.8-1.2) | 1.1(0.9-1.2) | 0.9(0.8-1.1 |
| V | 50.5 ± 1.2(48.1-53.6) | 53 ± 1.9(50-55) | 53 ± 0.9(51.0-54.0) | (50.0-54.0) | 52.4 ± 1.1(50.9-54.0) | 52.1(50.8-55.0) | 50(47-52) |
| Total stylet | 149.0 ± 4.4(144-161) | 159 ± 8.05(144-173) | - | (156.0-168.3) | 166.5 ± 2.6(161-171) | - | - |
| Odontostyle | 94.1 ± 3.3(88-102) | 101 ± 6.14(89-110) | 101.9 ± 7.2(84.7-108.2) | - | 107.3 ± 2.7(103-111) | 103.5(88.5-120.0) | 87(84-89) |
| Odontophore | 54.9 ± 2.1(51-59) | 59 ± 3.43(50-64) | 57.0 ± 2.9(48.8-60.0) | (61.2-62.7) | 59.2 ± 1.6(57-62) | 56.7(53.1-60.0) | 50(48-51) |
| Oral apertureto guide ring | 76.5 ± 3.8(67-83) | 86.0 ± 4.23(77-92) | 86.3 ± 5.6(72.3-92.3) | - | 85.5 ± 5.0(78-93) | 79.6(66.0-103.0) | 62(60-64) |
| Pharyngeal bulblength | 79.3 ± 4.0(73-86; n = 10) | - | - | (61-79) | 84.9 ± 2.7(82-90; n = 7) | 94.2(65.0-117) | - |
| Pharyngeal bulbwidth | 21.2 ± 1.2(18-23; n = 21) | - | - | (12-19) | 22.9 ± 1.7(20-26) | 21.1(18.0-24.0) | - |
| Tail | 27.8 ± 2.0(25-32) | 26.0 ± 1.71(23-28) | 26.8 ± 2.0(24.1-31.2) | (24.5-29.0) | 29.2 ± 2.8(24-34) | 36.1(33.0-47.0) | 24(21-28) |
| Hyaline portionof tail (J) | 10.4 ± 1.5(8-13) | - | 8.0 ±0.9(5.9-9.4) | - | 11.4 ± 1.5(9-14) | 9(7-11) | 12(10-14) |
| Body diam.at lip region | 12.5 ± 0.4(12-13) | 11.5(11-13) | 11.5 ± 0.5(10.6-12.3) | - | 12.4 ± 0.4(11-13) | 14.6(13.5-15.0) | 11(10-12) |
| Body diam.at guide ring | 30.4 ± 0.8(29-32) | - | 29.8 ± 1.5(27.1-31.8) | - | 32.6 ± 0.8(31-34) | 31.7(30.0-36.0) | 26(26-27) |
| Body diam.at base of pharynx | 37.9 ± 1.2(35-40) | - | 39.4 ± 2.9(35.3-45.3) | - | 41.3 ± 2.2(38-45) | 40.1(36.0-42.0) | 33(31-35) |
| Body diam.at vulva | 42.0 ± 2.0(38-46) | - | 46.6 ± 3.4(39.4-50.0) | (49.0-59.7) | 47.2 ± 2.5(44-52) | 43.4(39.0-47.2) | 36(33-38) |
| Body diam.at anus | 27.0 ± 1.6(24-30) | 26(22-29) | 26.6 ± 1.7(21.8-29.4) | (30.6-36.7) | 29.7 ± 1.1(28-32) | 32.4(27.0-41.0) | 25(23-28) |
| Body diam.at beginning of J | 16.2 ± 2.2(12-20) | - | 13.7 ± 1.1(11.2-14.7) | - | 18.4 ± 2.2(15-22) | 7(6-8) | 17(15-20) |
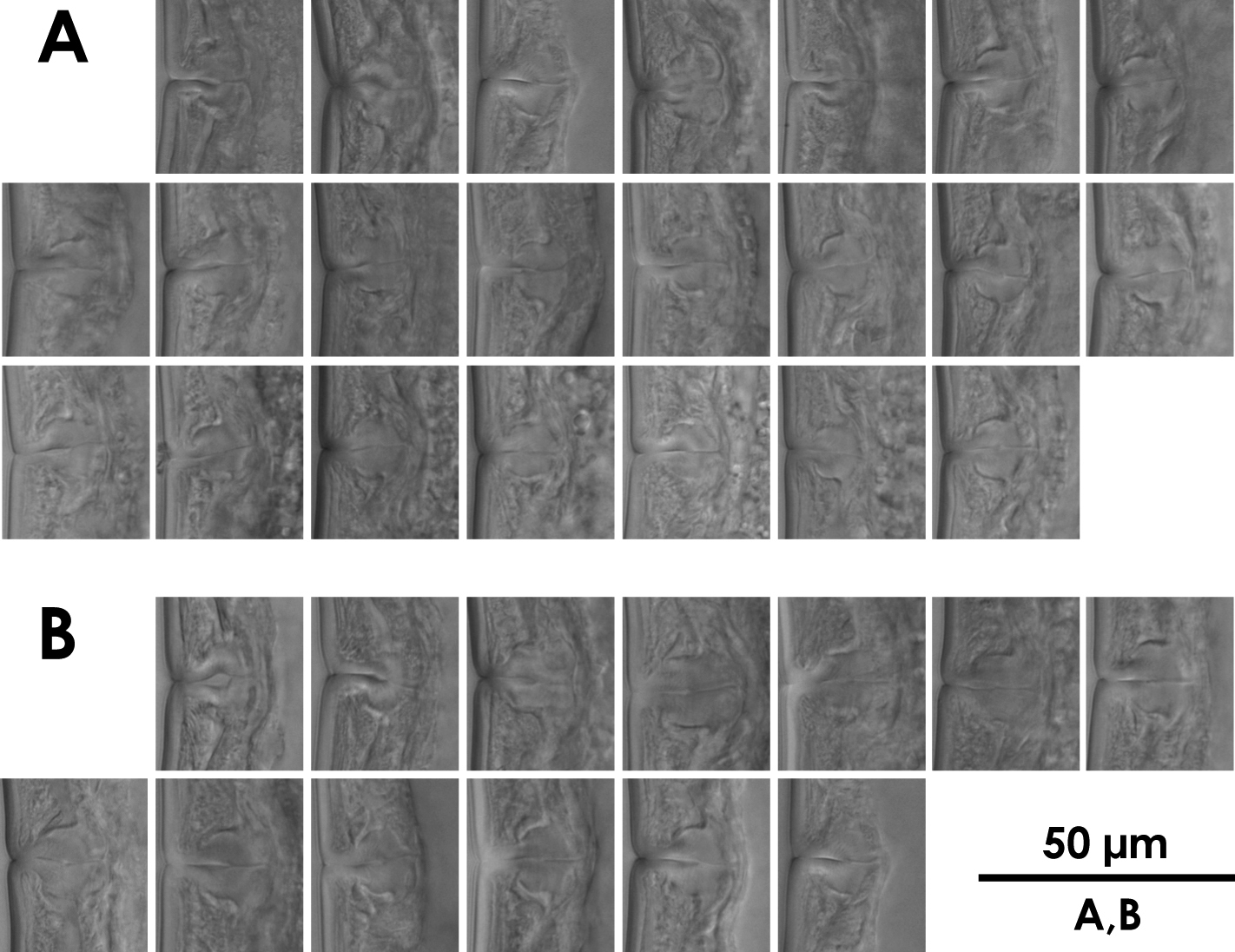
Uterine and vaginal region lengths of Xiphinema brevicolle and Xiphinema sp. measured in this study (female). Mean ± standard deviation (range; number of specimens) in μm, except where indicated.
| Xiphinema brevicolle | Xiphinema sp. | |
|---|---|---|
| Anterior uterus length | 44.3 ± 4.8(34-54; n = 14) | 35.1 ± 2.9(32-40; n = 9) |
| Posterior uterus length | 41.1 ± 2.9(37-46; n = 9) | 32.7 ± 2.7(29-37; n = 9) |
| Pars proximalis vaginae | 5.9 ± 0.7(5-7; n = 22) | 7.6 ± 1.5(5-10; n = 13) |
| Pars distalis vaginae | 10.5 ± 0.7(9-12; n = 22) | 11.3 ± 1.0(9-13; n = 13) |
| Vagina length | 16.4 ± 1.0(14-18; n = 22) | 18.9 ± 1.5(17-22; n = 13) |
| Vagina length /body diam. at vulva (%) | 39.2 ± 3.2(31.3-44.6; n = 22) | 40.1 ± 2.9(34.8-44.2; n = 13) |
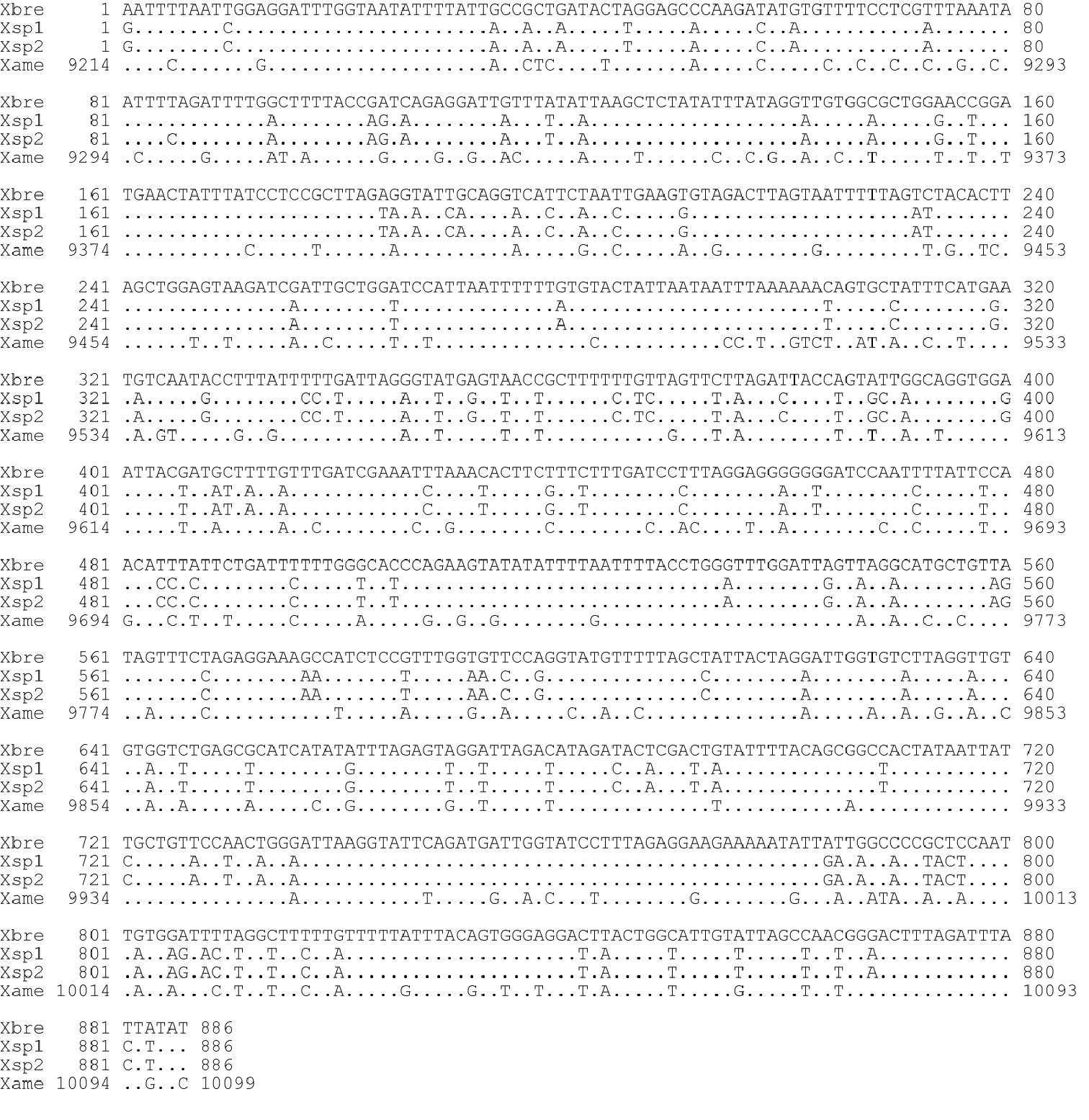
DNA sequences of 886 bp except for primer regions were obtained for the mitochondrial COI region. The five Xiphinema brevicolle specimens observed had identical sequences, whereas a single nucleotide in the sequence differed among the four specimens of Xiphinema
sp. though this variation resulted in no difference between the
translated amino acid sequences within the specimens. Sequence identity
between these two species was 84.0-84.1%, whereas the sequences of Xiphinema brevicolle and Xiphinema sp. were 80.7 and 80.4-80.5% identical to that of Xiphinema americanum (
ML trees inferred for the 18S rDNA and D2/D3 regions placed our specimens in similar clades, which include Xiphinema brevicolle and its junior synonyms by
This study reports the occurrence of Xiphinema brevicolle in Japan for the first time. The only member species of the Xiphinema americanum-group recorded to date in Japan was Xiphinema incognitum
Lamberti & Bleve-Zacheo, 1979, described as a new species for
nematode specimens detected from soil of bonsai trees exported from
Japan to England (
The specimens of this study were obtained from mixed populations of the Xiphinema americanum-group.
Information on juveniles was not included because the coexistence of
the two closely related nematodes requires special care to separate
juvenile specimens of the different species. Situations like this will
make matters worse if one is to identify the species, since it is
difficult enough to identify even a single population of the group in
many cases. Detection of mixed populations of Xiphinema americanum Cobb, 1913 and Xiphinema rivesi Dalmasso, 1969 were reported (
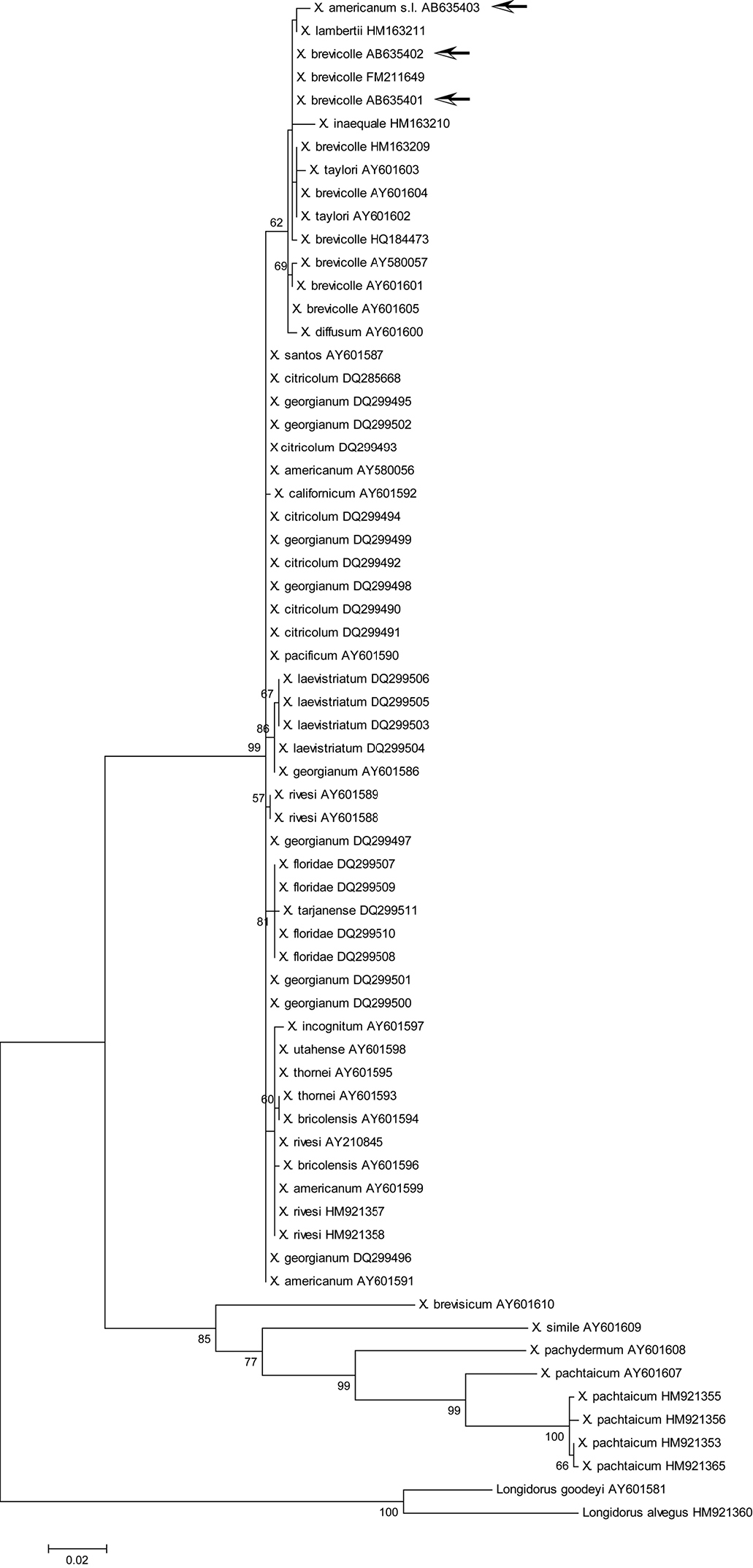
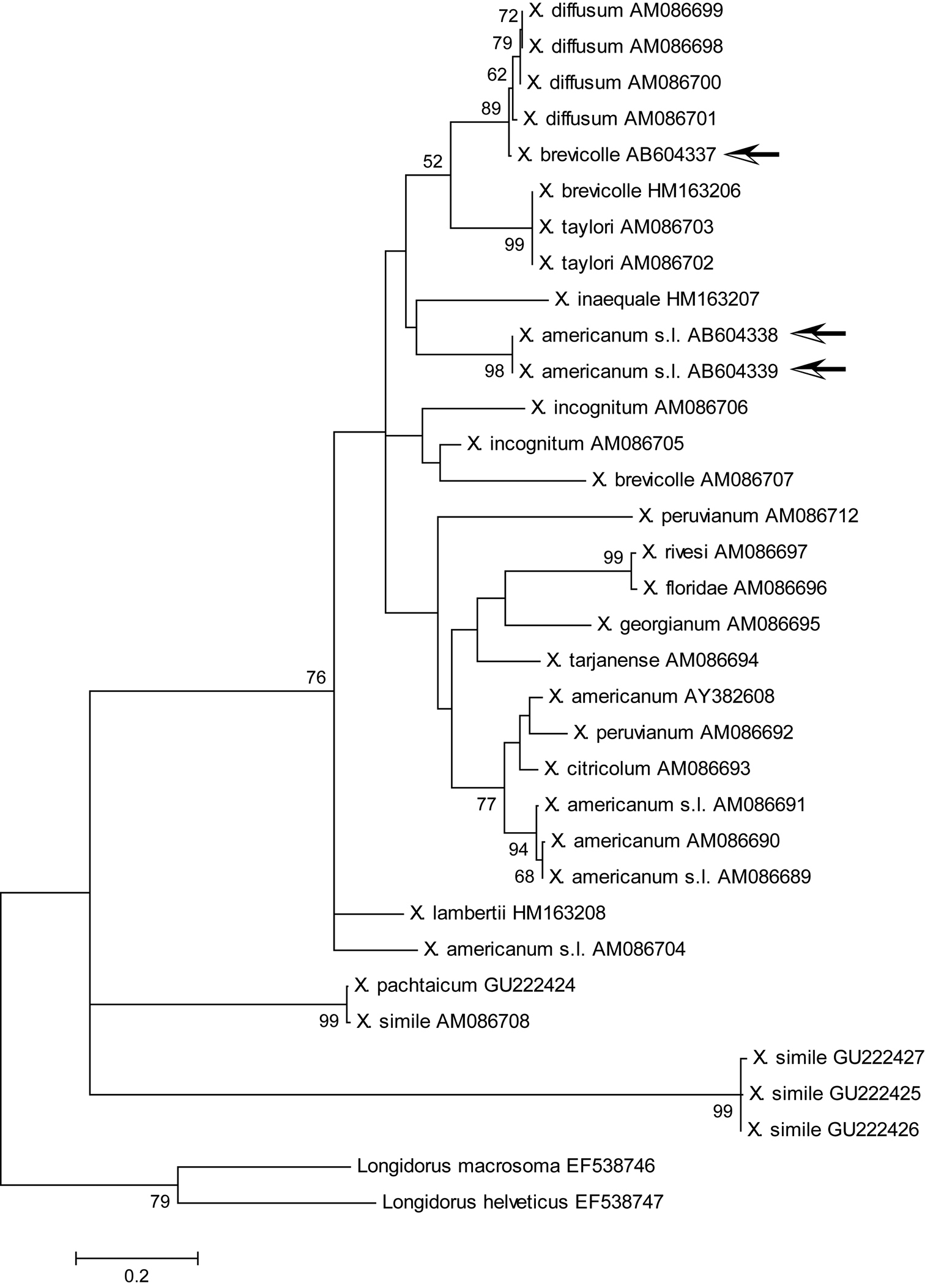
ML phylogenetic analyses of 18S rDNA and D2/D3 regions moderately supported the clade including Xiphinema brevicolle, Xiphinema diffusum, Xiphinema phinema inaequale, Xiphinema incognitum, Xiphinema lamberti, and Xiphinema taylori with our materials (Figs 8, 9), whereas such a clade was not so highly supported in the ML tree of COI (Fig. 10). Member species of Xiphinema americanum-group harbor endosymbionts (
In this study, specimens from mixed populations of the Xiphinema americanum-group, present in the root zone of Japanese holly in Japan, were identified as Xiphinema brevicolle and Xiphinema sp. This record of Xiphinema brevicolle is the first for Japan. PCR primers to amplify longer sequences of the mitochondrial COI regions were originally designed and used to efficiently differentiate the specimens. Phylogenetic analyses using 18S rDNA, D2/D3, and COI regions supported a close relationship among our specimens and species related to Xiphinema brevicolle or the Xiphinema brevicolle-subgroup.
The first author thanks Tom Prior for useful suggestions and information and Dr. Nobuhiro Minaka for helpful comments on phylogenetic analyses. The authors thank Dr. Jerome T. Gaspard for improving the manuscript. This study was supported by Research and development projects for application in promoting new policy of Agriculture Forestry and Fisheries (21043) from the Ministry of Agriculture, Forestry and Fisheries of Japan.
Female morphology of Xiphinema brevicolle (A, C, E, G) and Xiphinema sp. (B, D, F, H) A–B Anterior region C–D Tail E–F Reproductive system G–H Entire body.
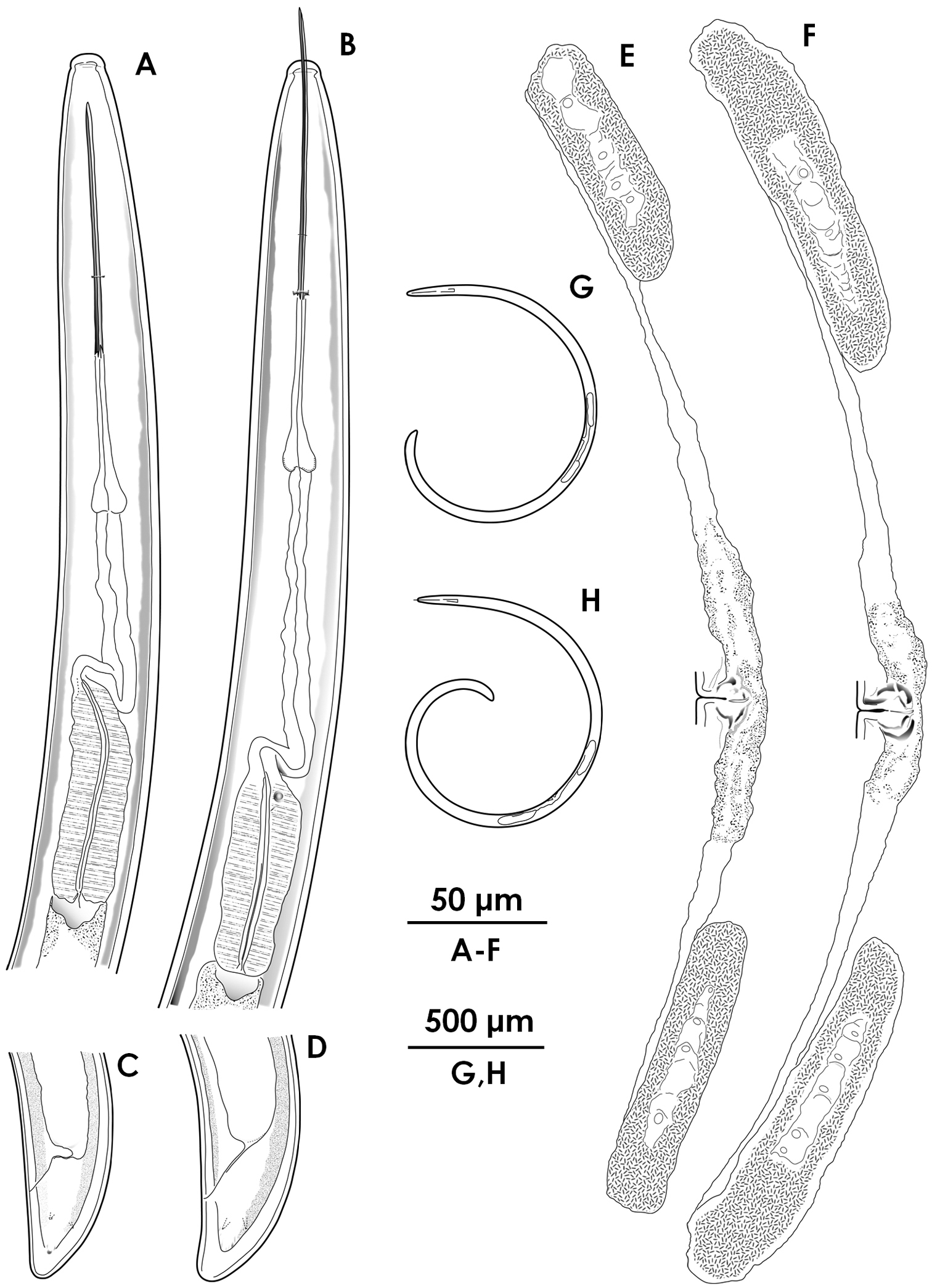
Xiphinema brevicolle (female) A–C Anterior region D–G Tail H–K Vulval region L–P Entire body.
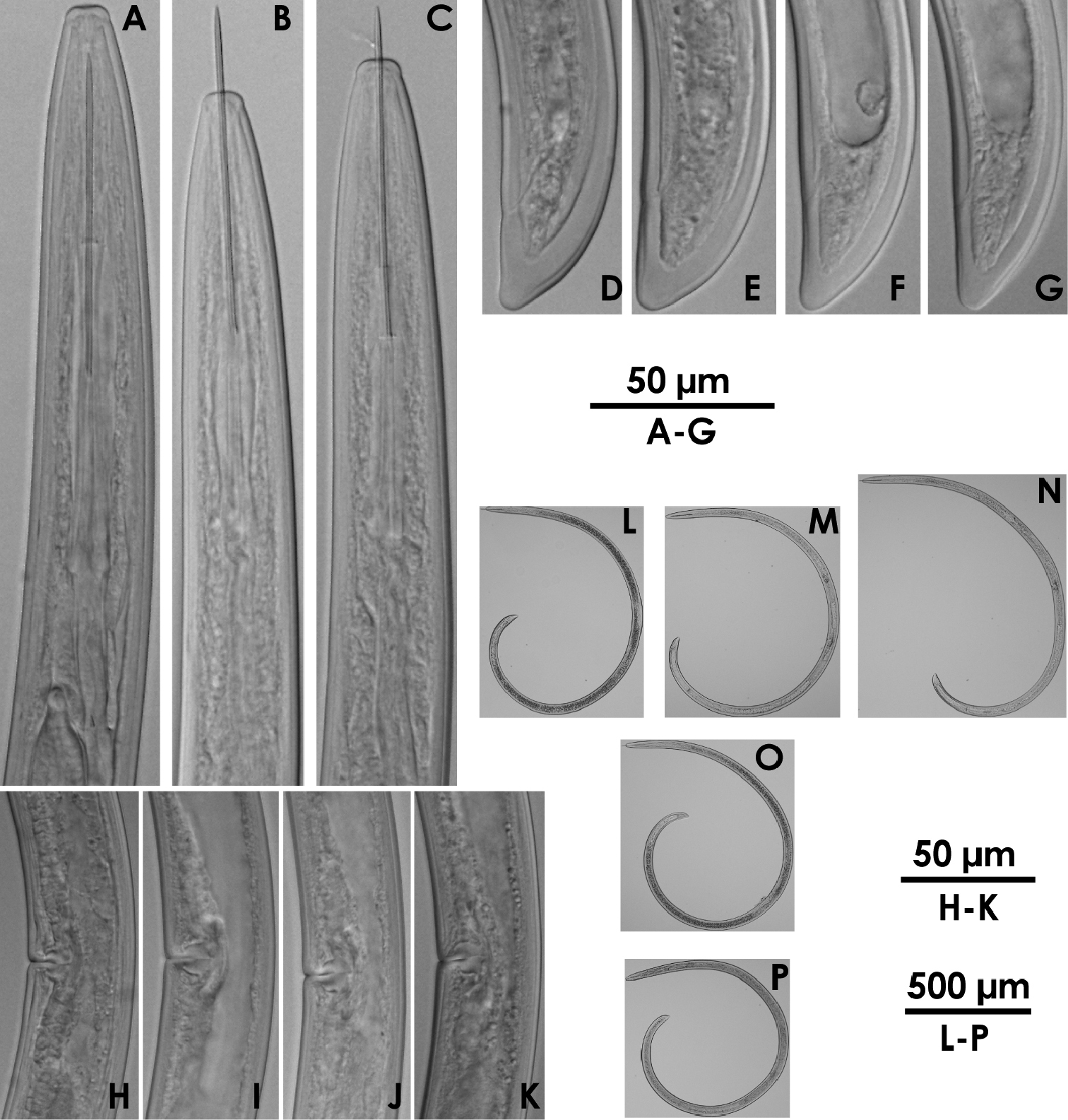
Xiphinema sp.(female) A–C Anterior region D–F Tail G–I Vulval region J–M Entire body.
Intra-population variation of lip region in females A Xiphinema brevicolle B Xiphinema sp.
Intra-population variation of tail in females A Xiphinema brevicolle B Xiphinema sp.
Intra-population variation of vagina in females A Xiphinema brevicolle B Xiphinema sp.
Sequence comparison of mitochondrial COI region. Xbre: Xiphinema brevicolle (this study: GenBank accession AB604337); Xsp1, Xsp2: Xiphinema sp. (this study: AB604338 and AB604339 respectively); Xame: X. americanum (He et al. 2005: usAY382608).
Maximum likelihood tree for 18S rDNA region. Bootstrap values higher than 50 are shown. Arrows indicate specimens examined in this study: Xiphinema brevicolle GenBank accession AB604340; Xiphinema sp. AB604341. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
Maximum likelihood tree for D2/D3 region. Bootstrap values higher than 50 are shown. Arrows indicate specimens examined in this study: Xiphinema brevicolle GenBank accession AB635401 and AB635402; Xiphinema sp. AB635403. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
Maximum likelihood tree for COI region. Bootstrap values higher than 50 are shown. Arrows indicate specimens examined in this study. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.