(C) 2012 Takafumi Nakano. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
A new sexannulate species of the genus Orobdella Oka, 1895, Orobdella mononoke sp. n., is described on the basis of five specimens collected from Yakushima Island, Japan. Orobdella mononoke sp. n. differs from other sexannulate Orobdella species in its possessing the following combination of characters: dorsal surface bicolor in life, I–XIII, XXVII and caudal sucker grayish purple, XIV–XXVI amber, male gonopore at XI c11/c12, female gonopore at XIII b2, 8 + 1/2 between gonopores, tubular but bulbous at junction with crop gastroporal duct, epididymides in XV–XIX, and atrial cornua ovate. Phylogenetic analyses using nuclear 18S rDNA and histone H3, and mitochondrial COI, tRNACys, tRNAMet, 12S rDNA, tRNAVal and 16S rDNA markers show that Orobdella mononoke sp. n. is closely related to Orobdella esulcata Nakano, 2010 from Kyushu, Japan, and two species, Orobdella dolichopharynx Nakano, 2011 and Orobdella shimadae Nakano, 2011, from the Ryukyu Archipelago, Japan.
Hirudinida, Hirudinea, Orobdellidae, Orobdella, new species, molecular phylogeny, Japan
The genus Orobdella Oka, 1895 consists of nine terrestrial gastroporous leeches described from Japan (
The nine Orobdella species are split into three groups based on their mid-body somite annulation (
Leeches were collected from Yakushima Island, Japan (Fig. 1), under rocks along mountain or forest trails. Altitude and coordinates for localities were obtained using a Garmin eTrex GPS unit.
Botryoidal tissue was taken from every specimen for DNA extraction, and the rest of the bodies were fixed in 10% formalin and preserved in 70% ethanol. Two measurements were made: body length (BL) from the anterior margin of the oral sucker to the posterior margin of the caudal sucker, and maximum body width (BW). Examination, dissection, and drawings of the specimens were accomplished under a stereoscopic microscope with a drawing tube (Leica M125). Specimens used in this study have been deposited in the Zoological Collection of Kyoto University (KUZ).
The numbering convention is based on
Map showing the northern and the central parts of the Ryukyu Archipelago, Japan.
The extraction of genomic DNA followed
PCR and cycle sequencing (CS) primers used in this study.
| Gene | Primer name | Reaction | Primer sequence (5’ → 3’) | Source | |
|---|---|---|---|---|---|
| 18S | |||||
| 1 | A | PRC & CS | AACCTGGTTGATCCTGCCAGT |
|
|
| L | PRC & CS | CCAACTACGAGCTTTTTAACTG |
|
||
| 2 | C | PRC & CS | CGGTAATTCCAGCTCCAATAG |
|
|
| Y | PRC & CS | CAGACAAATCGCTCCACCAAC |
|
||
| 3 | O | PRC & CS | AAGGGCACCACCAGGAGTGGAG |
|
|
| B | PRC & CS | TGATCCTTCCGCAGGTTCACCT |
|
||
| Histone H3 | |||||
| H3aF | PRC & CS | ATGGCTCGTACCAAGCAGACVGC |
|
||
| H3bR | PRC & CS | ATATCCTTRGGCATRATRGTGAC |
|
||
| COI | |||||
| 1 | LCO1490 | PRC & CS | GGTCAACAAATCATAAAGATATTGG |
|
|
| HCO2198 | CS | TAAACTTCAGGGTGACCAAAAAATCA |
|
||
| 2 | LCO-in | CS | TCCAGAACGTATTCCATTATTTG |
| Nakano (2012) |
| HCO-outout | PCR & CS | TACACATCTGGATAGTCTGAAT | This study | ||
| tRNACys–16S | |||||
| 1 | 12SA-out | PCR & CS | TTGATGAACAACATTAAATTGC |
| Nakano (2012) |
| 12SB-in | CS | TAAGCTGCACTTTGACCTGA |
| Nakano (2012) | |
| 2 | 12SA-in | CS | AATTAAAACAAGGATTAGATACCC |
| Nakano (2012) |
| 12SB-out | PCR & CS | AACCCATAATGCAAAAGGTAC |
| Nakano (2012) | |
All samples were sequenced in both directions. Sequencing reactions were performed using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Each sequencing reaction mixture was incubated at 96°C for 2 min, followed by 40 cycles of 96°C (10 s), 50°C (5 s), and 60°C (42 s for 18S, 21 s for Histone H3, 45 s for COI, and 40 s for tRNACys-16S). The products were collected by ethanol precipitation and sequenced on an ABI 3130xl Genetic Analyzer (Applied Biosystems). The obtained sequences were edited using DNA BASER (Heracle Biosoft S.R.L.). In this study, the following DNA sequences were newly obtained and deposited in GenBank (Table 2): 1) 18S sequences from the holotype (KUZ Z224) of the new species, the holotype (KUZ Z156) of Orobdella koikei Nakano, 2012 and the topotype (KUZ Z181) of Orobdella octonaria Oka, 1895; 2) histone H3 sequences from ten Orobdella species, Erpobdella japonica Pawłowski, 1962 (Erpobdellidae), Gastrostomobdella monticola Moore, 1929 (Gastrostomobdellidae) and Mimobdella japonica Blanchard, 1897 (Salifidae); 3) COI and tRNACys–16S sequences from the holotype (KUZ Z224) and two of the paratypes (KUZ Z221, 223) of the new species. Among the new species, DNA sequences of the holotype (KUZ Z224) were analyzed in the present study. The other DNA sequences were taken from GenBank (Table 2).
Samples used for the phylogenetic analyses. The information on voucher, collection locality, and GenBank accession numbers is indicated. Acronym: UNIMAS, the Universiti Malaysia Sarawak. Sources: a
| Species | Voucher | 18S | Histone H3 | COI | tRNACys–16S |
|---|---|---|---|---|---|
| Orobdella esulcata | KUZ Z29 Holotype | AB663655b | AB698873 | AB679664a | AB679665a |
| Orobdella dolichopharynx | KUZ Z120 Holotype | AB663665b | AB698876 | AB679680a | AB679681a |
| Orobdella ijimai | KUZ Z110 Topotype | AB663659b | AB698877 | AB679672a | AB679673a |
| Orobdella kawakatsuorum | KUZ Z167 Topotype | AB663661b | AB698878 | AB679704a | AB679705a |
| Orobdella koikei | KUZ Z156 Holotype | AB698883 | AB698882 | AB679688a | AB679689a |
| Orobdella mononoke sp. n. | KUZ Z221 | AB698862 | AB698863 | ||
| Orobdella mononoke sp. n. | KUZ Z223 | AB698864 | AB698865 | ||
| Orobdella mononoke sp. n. | KUZ Z224 Holotype | AB698868 | AB698869 | AB698866 | AB698867 |
| Orobdella octonaria | KUZ Z181 Topotype | AB698870 | AB698871 | AB679708a | AB679709a |
| Orobdella shimadae | KUZ Z128 Holotype | AB663663b | AB698875 | AB679676a | AB679677a |
| Orobdella tsushimensis | KUZ Z134 Holotype | AB663653b | AB698872 | AB679662a | AB679663a |
| Orobdella whitmani | KUZ Z45 Topotype | AB663657b | AB698874 | AB679668a | AB679669a |
| Erpobdella japonica | KUZ Z178 | AB663648b | AB698879 | AB679654a | AB679655a |
| Gastrostomobdella monticola | UNIMAS/A3/BH01/10 | AB663649b | AB698880 | AB679656a | AB679657a |
| Mimobdella japonica | KUZ Z179 | AB663650b | AB698881 | AB679658a | AB679659a |
Histone H3 and COI sequences were aligned by eye since there were no indels. Nuclear 18S and mitochondrial tRNACys–16S sequences were aligned using MAFFT X-INS-i (
Phylogenetic trees were constructed using maximum likelihood (ML) and Bayesian inference (BI). ML phylogenies were calculated using TREEFINDER v October 2008 (
The nodes with bootstrap value (BS) higher than 70% were regarded as sufficiently resolved (
urn:lsid:zoobank.org:act:FA8333ED-8C17-41FD-AFC1-62A4F98D4AC1
urn:lsid:zoobank.org:act:8B4ED1DA-E1B9-49A8-8B58-014A0921695C
http://species-id.net/wiki/Orobdella_mononoke
Figs 2 –5In life, dorsal surface of somites I–XIII, XXVII and caudal sucker grayish purple and of somites XIV–XXVI amber, ventral surface grayish white. Somite VI quadrannulate on dorsal, b1 = b2 < a2 = a3, and triannulate on venter, a1 = a2 = a3. Somite VII quadrannulate, somites VIII–XXV sexannulate, somite XXVI quinquannulate. Pharynx reaching to XIV. Gastropore conspicucous at XIII b2 (slightly anterior to middle of annulus). Gastroporal duct, winding at junction with gastropore, tubular but slightly bulbous at junction with crop. Male gonopore at XI c11/c12, female gonopore at XIII b2, behind gastropore, gonopores separated by 8 + 1/2 annuli. Paired epididymides in XV–XIX (approximately four somites). Atrial cornua developed, ovate.
KUZ Z224, holotype, dissected, collected from under a rock along a mountain trail at Shiratani–unsuikyo, Yakushima, Kagoshima Pref. (Yakushima Island), Japan (30°22.78'N, 130°34.49'E; Alt. 648 m), by Takafumi Nakano on 29 October, 2011.
Four paratypes collected from under rocks along mountain trails in Yakushima, Kagoshima Pref. (Yakushima Island), Japan, by Takafumi Nakano. Two specimens from the type locality: KUZ Z221 (30°22.87'N, 130°34.68'E; Alt. 649 m), dissected, on 28 October, 2011, and KUZ Z225 (30°22.75'N, 130°34.49'E; Alt. 646 m), on 29 October, 2011. Two specimens from Kusugawa on 28 October, 2011: KUZ Z222 (30°23.76'N, 130°35.25'E; Alt. 363 m), and KUZ Z223 (30°23.75'N, 130°35.25'E; Alt. 363 m), dissected.
The specific name is from the Japanese animation movie title ‘Mononoke-hime (Princess Mononoke)’. The type locality of this new species is the origin of an epic forest in that movie. The specific name is a Japanese word, not a Latin or latinized word.
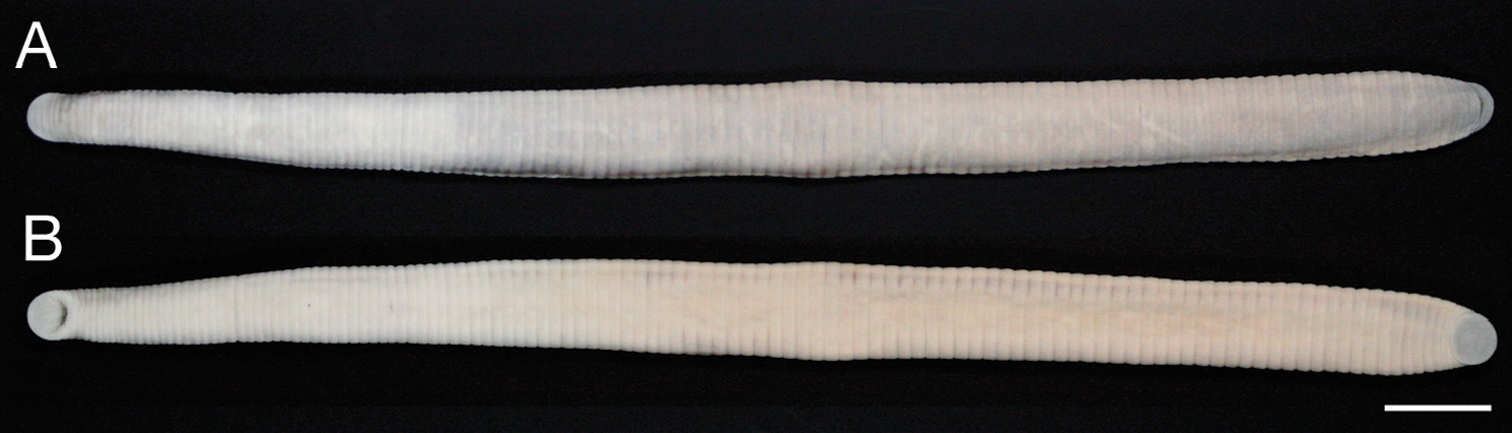
Body firm, muscular, elongated, gaining regularly in width in caudal direction, dorso-ventral depressed, sides nearly parallel from mid length to point just anterior to caudal sucker, BL 139.3 mm, BW 9.2 mm (Figs 2, 3). Caudal sucker ventral, oval, its diameter smaller than BW (Figs 3B, 4D). In life, dorsal surface of somites I–XIII, XXVII and caudal sucker grayish purple, and of somites XIV–XXVI amber (Fig. 2), ventral surface grayish white. Color faded in preservative, without any dark lines (Fig. 3).
Orobdella mononoke sp. n., holotype, KUZ Z224, taken of live animal, dorsal view.
Orobdella mononoke sp. n., holotype, KUZ Z224. A Dorsal and B ventral views. Scale bar, 1 cm.
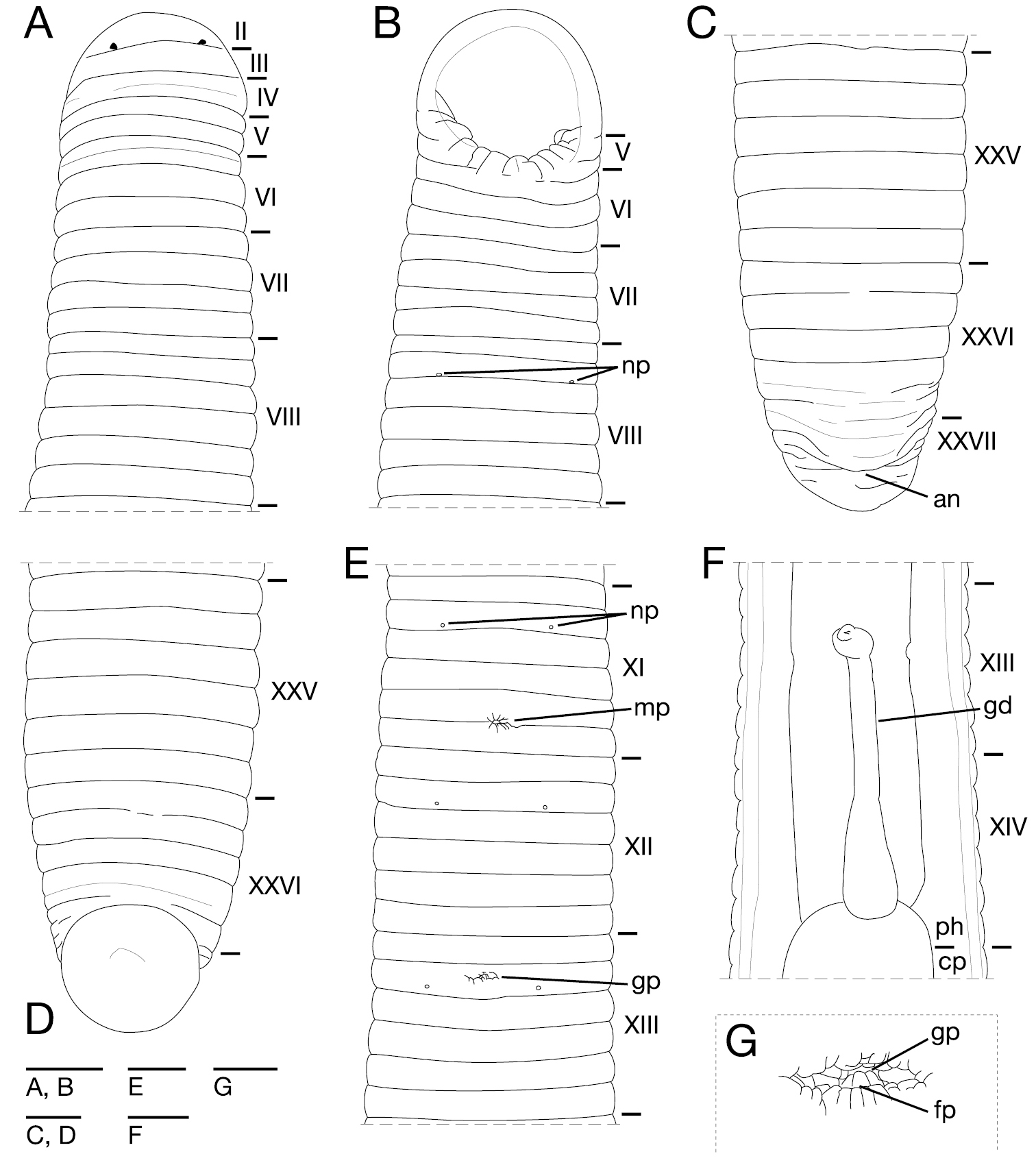
Orobdella mononoke sp. n., holotype, KUZ Z224. A Dorsal view of somites I–VIII B ventral view of somites I–VIII C dorsal view of somites XXV–XXVII and caudal sucker D ventral view of somites XXV–XXVII and caudal sucker E ventral view of somites XI–XIII F ventral view of gastroporal duct; and G ventral view of gastropore and female gonopore. Scale bars, 2 mm (A–F) and 0.5 mm (G). Abbreviations: an, anus; cp, crop; fp, female gonopore; gd, gastroporal duct; gp, gastropore; mp, male gonopore; np, nephridiopore; and ph, pharynx.
Somite I completely merged with prostomium (Fig. 4A). Somites II and III uniannulate (Fig. 4A). Somites IV and V biannulate, (a1+a2) = a3 (Fig. 4A), V a3 forming posterior margin of oral sucker (Fig. 4B). Somite VI quadrannulate on dorsal, b1 = b2 < a2 = a3, triannulate on venter, a1 = a2 = a3 (Fig. 4A–B). Somite VII quadrannulate, a1 = a2 = b5 = b6 (Fig. 4A–B). Somites VIII–XXV sexannulate. b1 = b2 = a2 = b5 = c11 = c12 (Fig. 4A–E). Somite XXVI quinquannulate, b1 = b2 = a2 < b5 = b6, b5 and b6 with slight furrows on dorsal (Fig. 4C–D), XXVI b5 being last complete annulus on venter (Fig. 4D). Somite XXVII comprising a few furrows; anus behind it with no post-anal annulus (Fig. 4C).
Anterior ganglionic mass in VI a2 and a3. Ganglion VII in a1 and a2. Ganglia VIII–XV, XXII and XXIII in a2 of each somite (Fig. 5A). Ganglia XVI–XXI and XXIV in b2 and a2 of each somite (Fig. 5A). Ganglion XXV in b2. Ganglion XXVI in XXV c12 and XXVI b1. Posterior ganglionic mass in XXVI a2 and b5.
Eyes three pairs, first pair dorsally on posterior margin of II (Fig. 4A), second pair dorsolaterally on middle of V (a1 + a2). Nephridiopores in 17 pairs, ventrally at posterior margin of a1 of each somite of VIII–XXIV (Fig. 4B, E). Papillae numerous, minute, hardly visible, one row on every annulus.
Pharynx agnathous, euthylaematous, reaching to XIV/XV (Fig. 4F). Crop tubular, acaecate, in XIV/XV to XXI b2/a2. Gastropore conspicuous, ventral, located slightly anterior to middle of XIII b2 (Fig. 4E, G). Gastroporal duct, winding at junction with gastropore, tubular but slightly bulbous at junction with crop, joining with crop in XIV c11 (Fig. 4F). Intestine tubular, acaecate, in XXI b2/a2 to XXIV b2/a2. Rectum, tubular, thin-walled.
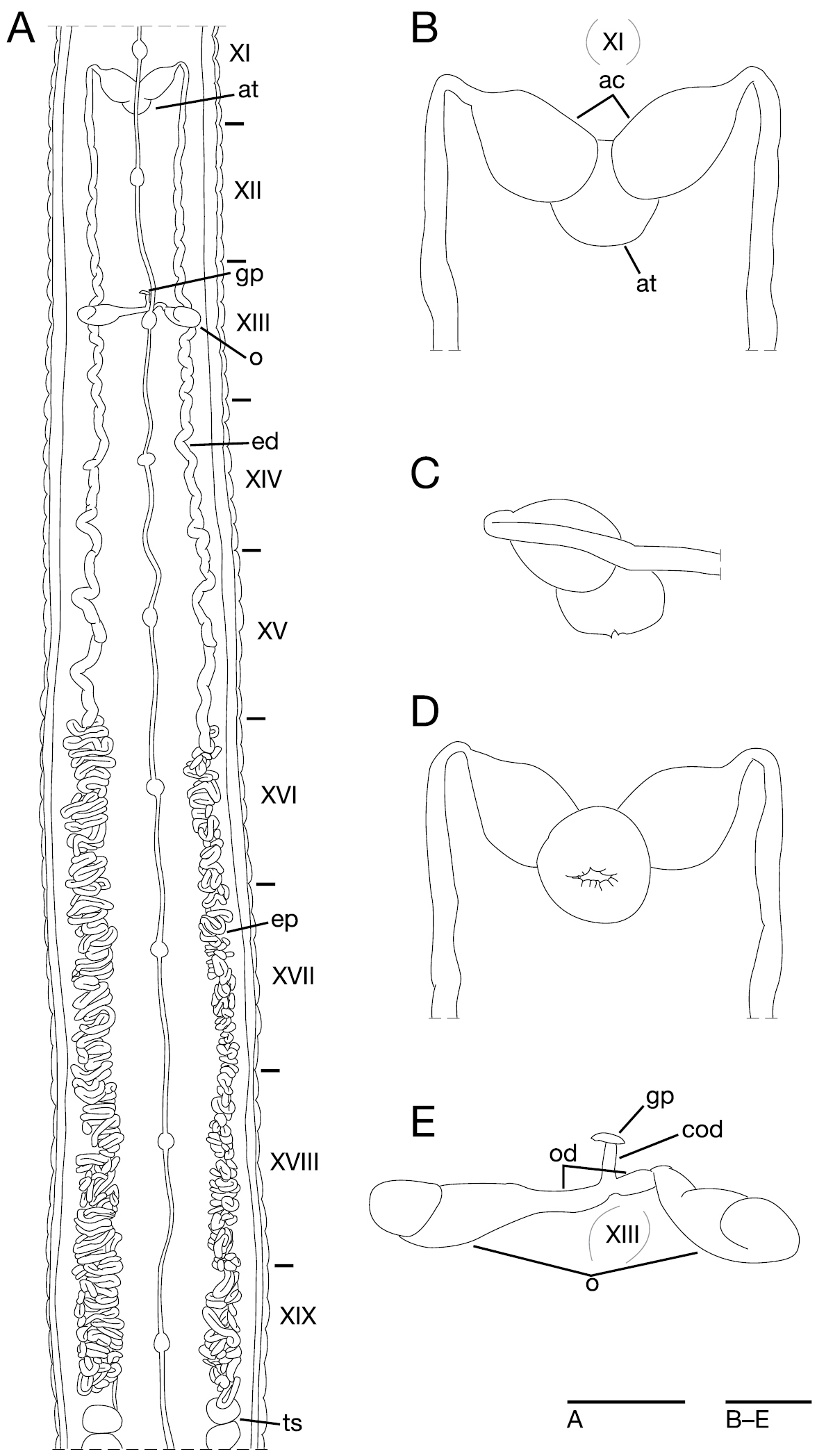
Male gonopore in the furrow of XI c11/c12 (Fig. 4E). Female gonopore located slightly anterior to middle of XIII b2, inconspicuous, located behind gastropore (Fig. 4E, G). Gonopores separated by 8 + 1/2 annuli (Fig. 4E). Testisacs multiple, one or two testisacs on each side in each annulus, in XIX c11 to XXV b5 (Fig. 5A). Paired epididymides in XVI b2 to XIX b5 (Fig. 5A). Ejaculatory bulbs absent. Ejaculatory ducts in XI b5 to XVI b2, loosely coiled, each winding from each junction with epididymis, narrowing at junction with atrial cornu, then turning sharply inward toward atrial cornu without pre-atrial loop (Fig. 5A–D). Pair of atrial cornua in XI b5 and c11, muscular, ovate (Fig. 5A–B, D). Atrium short, muscular, globular in XI c11 and c12 (Fig. 5B–D). Penis sheath and penis absent. Ovisacs one pair, thin-walled, globular, in XIII a2 and b5 (Fig. 5A, E). Oviducts thin-walled, right oviduct crossing ventrally beneath nerve cord, both oviducts converging into common oviduct in XIII b2 (Fig. 5A, E). Common oviduct thin-walled, short, directly ascending to female gonopore (Fig. 5E).
Orobdella mononoke sp. n., holotype, KUZ Z224. A Dorsal view of reproductive system including ventral nervous system B dorsal view of male atrium including position of ganglion XI C lateral view of male atrium D ventral view of male atrium; and E dorsal view of female reproductive system including position of ganglion XIII. Scale bars, 5 mm (A) and 1 mm (B–E). Abbreviations: ac, atrial cornu; at, atrium; cod, common oviduct; ed, ejaculatory duct; ep, epididymis; gp, gastropore; o, ovisac; od, oviduct; and ts, testisac.
In life, color generally same as holotype (Fig. 2). Somites III and IV uniannulate. Pharynx reaching to XIV b5/c11–XIV c11/c12. Crop reaching to XXI b2/a2–XXI a2. Gastroporal duct joining with crop in XIV b5; immature specimen (KUZ Z223), simple tubular. Intestine reaching to XXIV b1–XXIV b5. Testisacs in XIX b1 to XXIV c11. Epididymides in XV a2 to XVIII c11. Immature specimen (KUZ Z223), pair of atrial cornua in XI c11, fusiform. Left oviduct crossing ventrally beneath nerve cord.
Known from mountainous regions of Yakushima Island, Japan (Fig. 1).
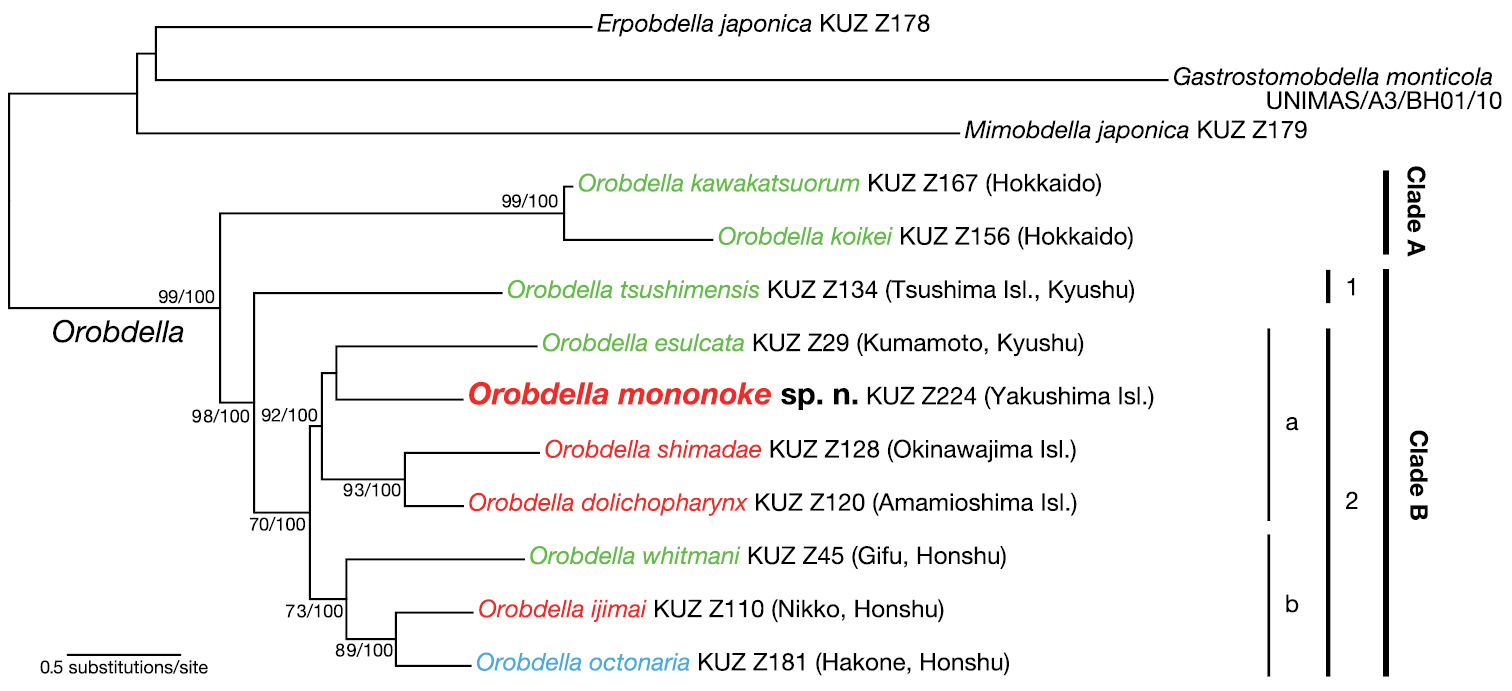
The ML tree with ln L = -14306.80 (Fig. 6) was nearly identical to the obtained BI tree (not shown). Monophyly of the genus Orobdella was confirmed (BS = 99 %, BPP = 100 %). The genus Orobdella then divided into two clades: clade A (BS = 99 %, BPP = 100 %) consisted of two species from Hokkaido, Japan, Orobdella kawakatsuorum Richardson, 1975 and Orobdella koikei; and clade B (BS = 98 %, BPP = 100 %) included all the other Orobdella species. Clade B comprised two subclades: subclade B1 was Orobdella tsushimensis Nakano, 2011 from Tsushima Island, Japan; and subclade B2 (BS = 70 %, BPP = 100 %) was further divided into two subclades. Subclade B2a (BS = 92 %, BPP = 100 %) included Orobdella mononoke sp. n., Orobdella esulcata Nakano, 2010 from Kyushu, and two Orobdella species from the Ryukyu Archipelago, Orobdella dolichopharynx and Orobdella shimadae. Subclade B2b (BS = 73 %, BPP = 100 %) consisted of three species from Honshu, Japan, Orobdella whitmani Oka, 1895, Orobdella ijimai and Orobdella octonaria.
The ML tree of 4167 bp of nuclear 18S rDNA and histone H3 and mitochondrial COI, tRNACys, tRNAMet, 12S rDNA, tRNAVal and 16S rDNA. A species name in green indicates a quadrannulate species; in red, sexannulate; and in blue, octannulate. The numbers associated with the nodes represent the bootstrap values for ML (BS)/ and Baysian posterior probabilities (BPPs). BS higher than 70% and/or BPP higher than 95% are indicated.
In subclade B2a, monophyly of Orobdella dolichopharynx and Orobdella shimadae was well supported (BS = 93 %, BPP = 100 %). However, the precise phylogenetic position of Orobdella mononoke sp. n. in the sublcade could not be determined. In the ML analysis, Orobdella mononoke sp. n. and Orobdella esulcata formed a monophyletic clade, but this clade was not supported well (BS = 30 %). In the BI analysis, Orobdella mononoke sp. n. and two Ryukyu Archipelago species formed a monophyletic clade, but this relationship was not also supported (BPP = 77 %).
Orobdella mononoke sp. n. differs from the three other sexannulate congeneric species, Orobdella ijimai, Orobdella dolichopharynx, and Orobdella shimadae, in the following characteristics (Table 3): 1) dorsal surface bicolor, I–XIII, XXVII and caudal sucker grayish purple, XIV–XXVI amber; 2) VI quadrannulate on dorsal; 3) VII quadrannulate; 4) VIII sexannulate; 5) gonopores separated by 8 + 1/2 annuli; 6) pharynx reaching to XIV; 7) gastroporal duct tubular but bulbous at junction with crop; 8) epididymides in XV–XIX (approximately four somites); and 9) atrial cornua ovate. Orobdella mononoke sp. n. is clearly distinguished from Orobdella esulcata, Orobdella kawakatsuorum, Orobdella koikei, Orobdella tsushimensis, Orobdella octonaria and Orobdella whitmani, in having mid-body somites that are sexannulate; they are quadrannulate in Orobdella esulcata, Orobdella kawakatsuorum, Orobdella koike, Orobdella tsushimensis and Orobdella whitmani, and octannulate in Orobdella octonaria.
Comparisons of morphological characters between Orobdella mononoke sp. n. and three sexannulate congeneric species.
| Character | Orobdella mononoke sp. n. | Orobdella dolichopharynx | Orobdella ijimai | Orobdella shimadae |
|---|---|---|---|---|
| Color of dorsal surface | bicolor, I–XIII, XXVII and caudal sucker grayish purple, XIV–XXVI amber | yellowish green | yellowish green | yellowish green |
| Annulation of VI | quadrannulate on dorsal | triannulate | triannulate | triannulate |
| Annulation of VII | quadrannulate | quadrannulate | quadrannulate | triannulate |
| Annulation of VIII | sexannulate | quinquannulate | sexannulate | quinquannulate |
| Number of annuli between gonopores | 8 + 1/2 | 8 | 1/2 + 7 + 1/2 | 9 |
| Pharynx | reaching to XIV | reaching to XVI | reaching to XIV | reaching to XVI |
| Gastroporal duct | tubular, but bulbous at junctions with crop | tubular, reaching to XVI | bulbous | tubular reaching to XV |
| Epididymides | in XV–XIX (about four somites) | absent | in XVI–XIX (about two and half somites) | absent |
| Atrial cornua | ovate | absent | ellipsoid | absent |
The trees obtained in this study are nearly identical to those obtained in other phylogenetic analyses of the genus Orobdella (
Orobdella mononoke sp. n. inhabits Yakushima Island, which is located in the northern part of the Ryukyu Archipelago (Fig. 1). In the Ryukyu Archipelago, two sexannulate Orobdella species have been described: 1) Orobdella dolichopharynx from Amamioshima Island; and 2) Orobdella shimadae from Okinawajima Island. These two species have the following characteristics in common: 1) long pharynx, reaching to somite XVI; 2) rudimentary gastroporal duct and absence of gastropore; 3) absence of epididymides; and 4) absence of male atrial cornua. Although Orobdella mononoke sp. n. is a sexannulate species, this species does not share such morphological characteristics. Orobdella mononoke sp. n. possesses 1) normal length pharynx for the genus Orobdella, 2) developed gastroporal duct and conspicuous gastropore, 3) epididymides in XV–XIX, 4) ovate atrial cornua. Molecular phylogenetic analyses in this study also could not show monophyly of the three species in the Ryukyu Archipelago, Orobdella mononoke sp. n., Orobdella dolichopharynx and Orobdella shimadae. These differences of morphological characteristics and molecular phylogenetic analyses suggest that Orobdella mononoke sp. n. is not closely related to Orobdella dolichoharynx and Orobdella shimadae. In vertebrates, the fauna of the Osumi Islands, in which Yaushima Island is included, is related to that of Kyushu (
The author is grateful to Professor Tsutomu Hikida (Kyoto University; KU) for his helpful comments and suggestions to improve this manuscript. I am also grateful to Koshiro Eto (KU) for his help in collecting specimens in Yakushima Island, to Dr Zainudin Ramlah (UNIMAS) for permitting me to use the Gastrostomobdella specimen for molecular analyses, to Dr Elizabeth Nakajima (KU) for checking the English of this text, to two anonymous reviewers and Dr Fredric R. Govedich (Southern Utah University) for their constructive comments on this manuscript, and to Hiroshi Noda (KU) and Eri Kawaguchi (KU) for their technical support. I also express my sincere thanks to Naoki Koike (KU), Dr Kanto Nishikawa (KU) and Taku Shimada (Ant Room) for providing specimens. This study was financially supported in part by a Grant-in Aid for Biodiversity and Evolutionary Research of Global COE (A06) from MEXT, Japan, to Kyoto University.