|
||
|
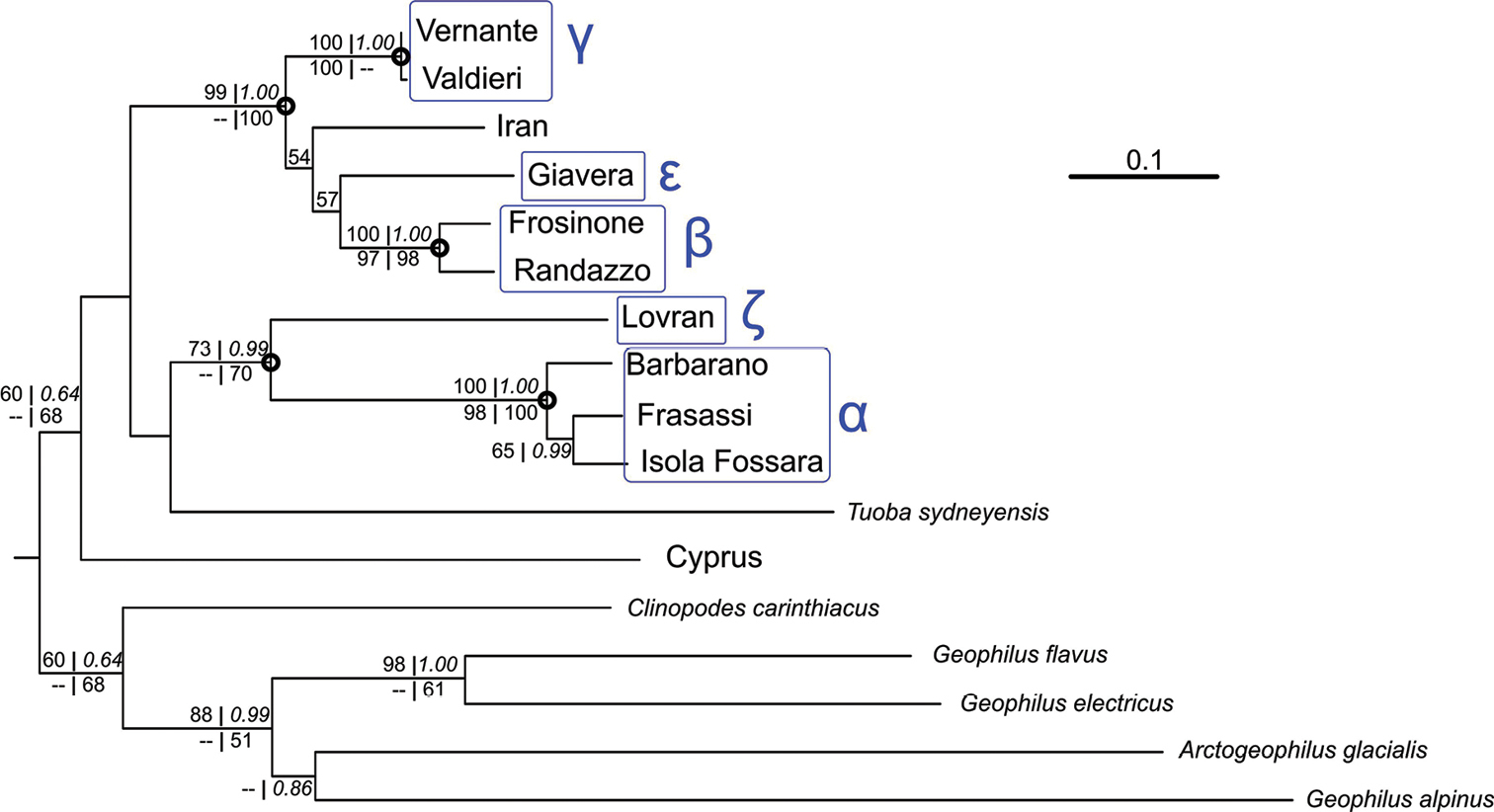
Maximum likelihood phylogeny. ML tree obtained from concatenated COI and 28S sequences, by the GTR+I+G model, and manually rooted. The following support values are indicated at the nodes (only for those present in the topology obtained from the concatenated sequences): ML bootstrap for the analysis of concatenated genes (upper left); Bayesian posterior probabilities (upper right, in italics); ML bootstrap for the analysis of COI sequences (lower left); ML bootstrap for the analysis of 28S sequences (lower right). Bootstrap values < 50% and posterior probabilities < 0.50 are not shown. Circles indicate ingroup nodes that are highly supported in the tree based on concatenated sequences. Terminal node groupings indicated by Greek letters refer to the species tentatively recognized (see text and Fig. 1). The specimen from Volpago (species δ) is absent because its 28S sequence was not obtained. |