|
||
|
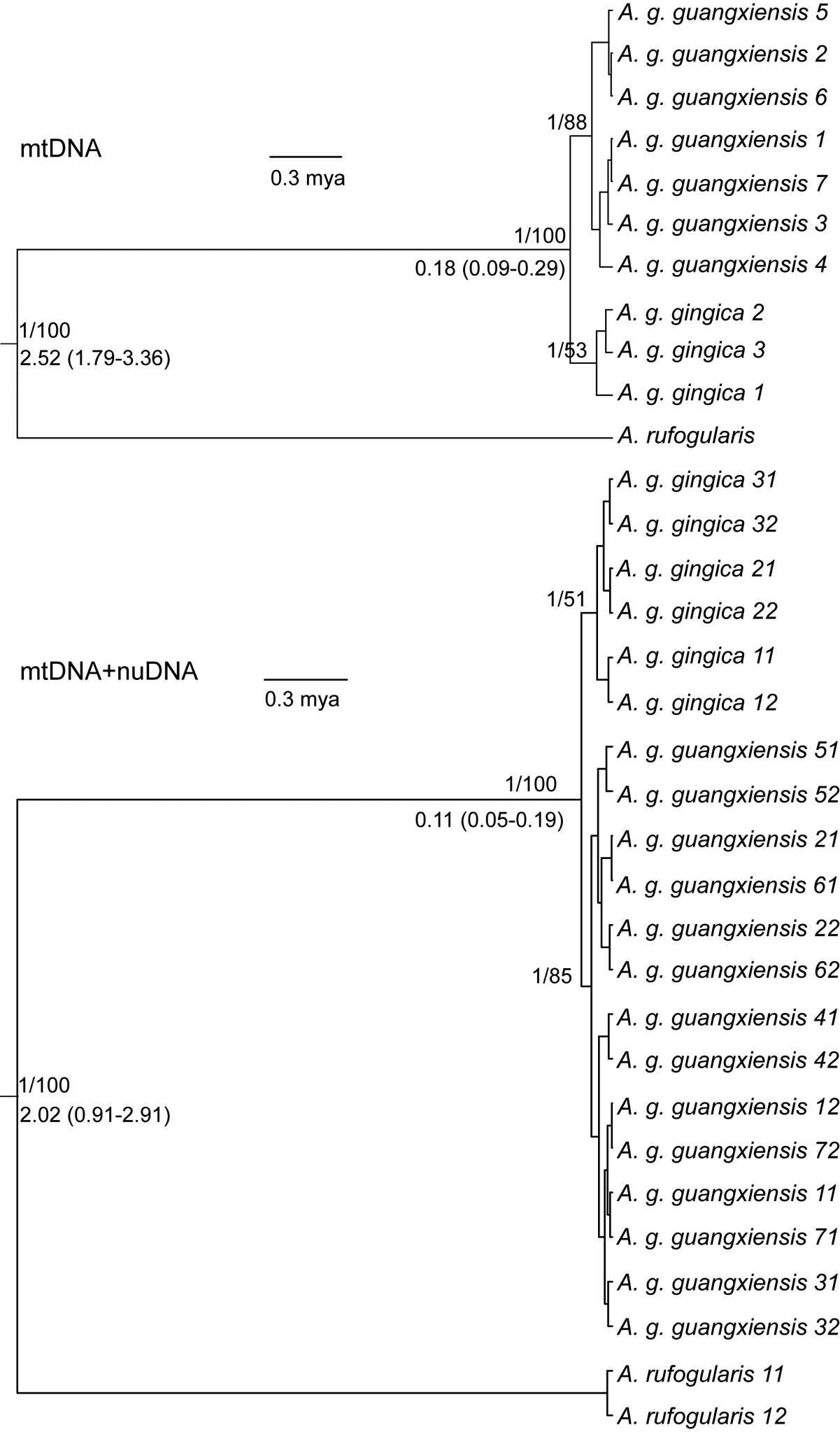
Phylogenetic consensus trees from the mtDNA matrix and complete data matrix. Node values above the branches represented the BI posterior probability and ML bootstrap support. Values below the branches represent the divergence times (median) and 95% highest posterior density (HPD) between lineage groups, note that the divergence times in the multi-locus tree were estimated by species tree analysis. The last number in tip labels in the multi-locus tree represent the two haplotypes phased from diploid nuclear sequences. |